Abstract
The title triangulo-triruthenium(0) compound, [Ru3(C30H32P2)(CO)10], contains a triangle of singly bonded Ru atoms. The phosphane-bridged Ru—Ru distance [2.9531 (2) Å] is significantly longer than the non-bridged Ru—Ru distances [2.8842 (2) and 2.8876 (2) Å] . The bis(diphenylphosphanyl)hexane ligand bridges the Ru—Ru bond. Each phosphane-substituted Ru atom bears one equatorial and two axial terminal carbonyl ligands, whereas the unsubstituted Ru atom bears two equatorial and two axial terminal carbonyl ligands. The dihedral angles between the benzene rings attached to each P atom are 72.75 (7) and 82.02 (7)°. The molecular structure is stabilized by an intramolecular C—H⋯O hydrogen bond involving a methylene group of the phosphane ligand and an axial carbonyl O atom, which generates an S(6) ring motif. In the crystal, molecules are linked via C—H⋯O hydrogen bonds into layers parallel to (100).
Related literature
For general background to triangulo-triruthenium clusters with structures of the general type Ru3(CO)10
L (where L is a group 15 bidentate ligand), see: Bruce et al. (1982 ▶); Coleman et al. (1984 ▶); Teoh et al. (1990 ▶); Diz et al. (2001 ▶); Shawkataly et al. (2006 ▶, 2011 ▶); Churchill et al. (1977 ▶). For the preparation of the title compound, see: Bruce et al. (1983 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶). For the stability of the temperature controller used in the data collection, see: Cosier & Glazer (1986 ▶).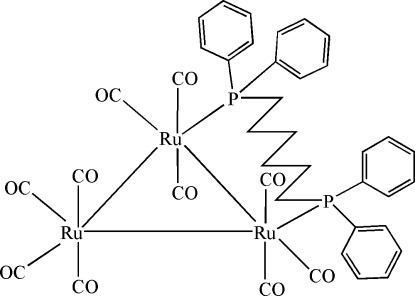
Experimental
Crystal data
[Ru3(C30H32P2)(CO)10]
M r = 1037.81
Monoclinic,

a = 13.4836 (6) Å
b = 21.270 (1) Å
c = 16.1025 (6) Å
β = 122.295 (3)°
V = 3903.7 (3) Å3
Z = 4
Mo Kα radiation
μ = 1.29 mm−1
T = 100 K
0.51 × 0.26 × 0.11 mm
Data collection
Bruker APEXII DUO CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.562, T max = 0.869
53262 measured reflections
14125 independent reflections
12773 reflections with I > 2σ(I)
R int = 0.023
Refinement
R[F 2 > 2σ(F 2)] = 0.019
wR(F 2) = 0.046
S = 1.04
14125 reflections
496 parameters
H-atom parameters constrained
Δρmax = 0.54 e Å−3
Δρmin = −0.58 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812014614/sj5228sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812014614/sj5228Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C9—H9A⋯O3i | 0.93 | 2.48 | 3.3421 (17) | 154 |
| C14—H14A⋯O6 | 0.97 | 2.58 | 3.2007 (19) | 122 |
| C20—H20A⋯O6i | 0.93 | 2.59 | 3.495 (2) | 164 |
| C21—H21A⋯O9ii | 0.93 | 2.51 | 3.329 (2) | 147 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
ObS would like to thank Universiti Sains Malaysia (USM) for the Research University Grant No. 1001/PJJAUH/811188. SSS thanks USM for the award of a USM fellowship and for a scholarship under the Postgraduate Research Grant Scheme (PRGS) No. 1001/PJJAUH/834064. HKF thanks USM for the Research University Grant No. 1001/PFIZIK/811160.
supplementary crystallographic information
Comment
Synthesis and structural reports on substitutued triangulo-triruthenium clusters with group 15 ligands are of interest because of the observed structural variations and their potential catalytic activity. There are several reports of substituted derivatives, of the type Ru3(CO)10(L)[where L= bidentate phosphine ligand] (Coleman et al., 1984; Bruce et al., 1982; Teoh et al., 1990; Diz et al., 2001; Shawkataly et al., 2006, 2011).
The title triangulo-triruthenium(0) compound, [Ru3(CO)10(Ph2P(CH2)6PPh2)], contains triangle of singly bonded Ru atoms. These type of structures are derived from that of [Ru3(CO)12] (Churchill et al., 1977) by replacement of an equatorial carbonyl group on each of two Ru atoms by the Ph2P groups of the diphosphane ligands. The phosphane bridged Ru-Ru distance [Ru2—Ru3 = 2.9531 (2) Å ] is significantly longer than the non-bridged Ru-Ru distances [Ru1 —Ru2 = 2.8842 (2) Å and Ru1 — Ru3 = 2.8876 (2) Å]. The bis(diphenylphosphanyl) hexane ligand bridges the Ru2– Ru3 bond. These Ru-Ru distances 2.8842 (2), 2.8876 (2) and 2.9531 (2) Å agree well with those observed in Ru3(CO)10Ph2P(CH2)2PPh2 (2.847 (1), 2.855 (1) and 2.856 (1) Å) (Bruce et al., 1982) andRu3(CO)10(F-dppe) [where F-dppe = bis(perfluoro-diphenylphosphanyl)ethane] (2.842 (4), 2.849 (4) and 2.868 (4) Å) (Diz et al., 2001) whereby the longest Ru-Ru bond is bridged by the bidentate phosphane ligand. The Ru1 atom carries two equatorial and two axial terminal carbonyl ligands whereas the Ru2 and Ru3 atoms each carries one equatorial and two axial terminal carbonyl ligands. The dihedral angles between the two benzene rings (C1–C6/C7–C12 and C19–C24/C25–C30) are 72.75 (7) and 82.02 (7)°, respectively. The molecular structure is stabilized by an intramolecular C14–H14A···O6 (Table 1) hydrogen bond, which generates an S(6) ring motif (Fig. 1, Bernstein et al., 1995).
In the crystal structure, Fig. 2, molecules are linked via intermolecular C9–H9A···O3, C20–H20A···O6 and C21–H21A···O9 hydrogen bonds (Table 1) into two-dimensional planes parallel to (100).
Experimental
All manipulations were performed under a dry, oxygen-free nitrogen atmosphere using standard Schlenk techniques. Tetrahydrofuran was dried over sodium and distilled from sodium benzophenone ketyl under nitrogen. The Ru3(CO)12 (Aldrich) and Ph2P(CH2)6PPh2 (Strem Chemicals) were used as received. Ru3(CO)10(Ph2P(CH2)6PPh2) was prepared by a reported procedure (Bruce et al., 1983). The title compound was obtained by reacting equimolar quantities of Ru3(CO)12 with Ph2P(CH2)6PPh2 in 25 ml THF. Crystals suitable for X-ray diffraction were grown by slow solvent/solvent diffusion of CH3OH into CH2Cl2.
Refinement
All H atoms were positioned geometrically and refined using a riding model with C–H = 0.93 or 0.97 Å and Uiso(H) = 1.2 Ueq(C).
Figures
Fig. 1.
The molecular structure of the title compound showing 40% probability displacement ellipsoids for non-H atoms. Intramolecular hydrogen bond is shown as dash line.
Fig. 2.
The crystal structure of the title compound, viewed along the c axis. H atoms not involved in hydrogen bonds (dashed lines) have been omitted for clarity.
Crystal data
| [Ru3(C30H32P2)(CO)10] | F(000) = 2056 |
| Mr = 1037.81 | Dx = 1.766 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 9965 reflections |
| a = 13.4836 (6) Å | θ = 3.1–32.6° |
| b = 21.270 (1) Å | µ = 1.29 mm−1 |
| c = 16.1025 (6) Å | T = 100 K |
| β = 122.295 (3)° | Block, brown |
| V = 3903.7 (3) Å3 | 0.51 × 0.26 × 0.11 mm |
| Z = 4 |
Data collection
| Bruker APEXII DUO CCD area-detector diffractometer | 14125 independent reflections |
| Radiation source: fine-focus sealed tube | 12773 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.023 |
| φ and ω scans | θmax = 32.7°, θmin = 1.9° |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | h = −20→20 |
| Tmin = 0.562, Tmax = 0.869 | k = −15→32 |
| 53262 measured reflections | l = −24→24 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.019 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.046 | H-atom parameters constrained |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0182P)2 + 1.6109P] where P = (Fo2 + 2Fc2)/3 |
| 14125 reflections | (Δ/σ)max = 0.004 |
| 496 parameters | Δρmax = 0.54 e Å−3 |
| 0 restraints | Δρmin = −0.58 e Å−3 |
Special details
| Experimental. The crystal was placed in the cold stream of an Oxford Cryosystems Cobra open-flow nitrogen cryostat (Cosier & Glazer, 1986) operating at 100.0 (1) K. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Ru1 | 0.234282 (8) | 0.486493 (4) | 0.439613 (7) | 0.01568 (2) | |
| Ru2 | 0.007592 (7) | 0.536230 (4) | 0.293260 (7) | 0.01454 (2) | |
| Ru3 | 0.221591 (7) | 0.613182 (4) | 0.370118 (6) | 0.01222 (2) | |
| P1 | −0.17243 (2) | 0.578706 (14) | 0.16853 (2) | 0.01642 (5) | |
| P2 | 0.21229 (2) | 0.713604 (13) | 0.30678 (2) | 0.01351 (5) | |
| O1 | 0.16758 (11) | 0.35305 (5) | 0.46383 (10) | 0.0385 (3) | |
| O2 | 0.49388 (9) | 0.47616 (6) | 0.59809 (8) | 0.0305 (2) | |
| O3 | 0.28204 (10) | 0.43668 (5) | 0.28450 (8) | 0.0287 (2) | |
| O4 | 0.19513 (9) | 0.54598 (5) | 0.59416 (7) | 0.0278 (2) | |
| O5 | −0.08834 (9) | 0.40860 (5) | 0.29939 (8) | 0.0266 (2) | |
| O6 | 0.05342 (9) | 0.48931 (5) | 0.13661 (7) | 0.02588 (19) | |
| O7 | −0.05743 (9) | 0.58536 (5) | 0.43750 (7) | 0.0266 (2) | |
| O8 | 0.47322 (8) | 0.62400 (5) | 0.54079 (7) | 0.02603 (19) | |
| O9 | 0.27197 (8) | 0.56475 (4) | 0.21625 (7) | 0.02217 (17) | |
| O10 | 0.13472 (8) | 0.67573 (4) | 0.49164 (7) | 0.02337 (18) | |
| C1 | −0.33183 (12) | 0.47903 (7) | 0.12075 (11) | 0.0274 (3) | |
| H1A | −0.2955 | 0.4605 | 0.0915 | 0.033* | |
| C2 | −0.42138 (13) | 0.44728 (7) | 0.12156 (12) | 0.0338 (3) | |
| H2A | −0.4448 | 0.4078 | 0.0926 | 0.041* | |
| C3 | −0.47589 (12) | 0.47432 (8) | 0.16546 (12) | 0.0327 (3) | |
| H3A | −0.5356 | 0.4529 | 0.1661 | 0.039* | |
| C4 | −0.44125 (12) | 0.53312 (8) | 0.20821 (11) | 0.0293 (3) | |
| H4A | −0.4780 | 0.5514 | 0.2373 | 0.035* | |
| C5 | −0.35137 (11) | 0.56491 (7) | 0.20765 (10) | 0.0237 (2) | |
| H5A | −0.3279 | 0.6043 | 0.2371 | 0.028* | |
| C6 | −0.29594 (10) | 0.53843 (6) | 0.16344 (9) | 0.0198 (2) | |
| C7 | −0.32163 (10) | 0.68222 (6) | 0.08869 (9) | 0.0218 (2) | |
| H7A | −0.3698 | 0.6547 | 0.0379 | 0.026* | |
| C8 | −0.35592 (11) | 0.74400 (6) | 0.08411 (10) | 0.0229 (2) | |
| H8A | −0.4253 | 0.7583 | 0.0293 | 0.027* | |
| C9 | −0.28679 (11) | 0.78458 (6) | 0.16140 (10) | 0.0224 (2) | |
| H9A | −0.3105 | 0.8260 | 0.1590 | 0.027* | |
| C10 | −0.18240 (11) | 0.76352 (6) | 0.24221 (9) | 0.0215 (2) | |
| H10A | −0.1366 | 0.7907 | 0.2942 | 0.026* | |
| C11 | −0.14562 (10) | 0.70156 (6) | 0.24601 (9) | 0.0177 (2) | |
| H11A | −0.0748 | 0.6879 | 0.3000 | 0.021* | |
| C12 | −0.21474 (9) | 0.66040 (5) | 0.16923 (8) | 0.01632 (19) | |
| C13 | −0.20318 (11) | 0.56961 (6) | 0.04288 (9) | 0.0227 (2) | |
| H13A | −0.2003 | 0.5252 | 0.0304 | 0.027* | |
| H13B | −0.2823 | 0.5842 | −0.0034 | 0.027* | |
| C14 | −0.11829 (13) | 0.60517 (7) | 0.02351 (11) | 0.0276 (3) | |
| H14A | −0.0394 | 0.5994 | 0.0801 | 0.033* | |
| H14B | −0.1212 | 0.5856 | −0.0322 | 0.033* | |
| C15 | −0.13912 (11) | 0.67564 (7) | 0.00301 (9) | 0.0232 (2) | |
| H15A | −0.1500 | 0.6949 | 0.0522 | 0.028* | |
| H15B | −0.2105 | 0.6820 | −0.0607 | 0.028* | |
| C16 | −0.03673 (11) | 0.70793 (7) | 0.00415 (9) | 0.0231 (2) | |
| H16A | −0.0293 | 0.6900 | −0.0476 | 0.028* | |
| H16B | −0.0552 | 0.7522 | −0.0106 | 0.028* | |
| C17 | 0.08159 (10) | 0.70214 (6) | 0.10166 (9) | 0.0194 (2) | |
| H17A | 0.1005 | 0.6580 | 0.1169 | 0.023* | |
| H17B | 0.1421 | 0.7209 | 0.0944 | 0.023* | |
| C18 | 0.08221 (10) | 0.73398 (6) | 0.18724 (8) | 0.0172 (2) | |
| H18A | 0.0124 | 0.7215 | 0.1864 | 0.021* | |
| H18B | 0.0795 | 0.7792 | 0.1785 | 0.021* | |
| C19 | 0.12991 (12) | 0.81696 (6) | 0.36458 (10) | 0.0215 (2) | |
| H19A | 0.0584 | 0.8121 | 0.3055 | 0.026* | |
| C20 | 0.14418 (14) | 0.86478 (6) | 0.42961 (11) | 0.0289 (3) | |
| H20A | 0.0820 | 0.8917 | 0.4135 | 0.035* | |
| C21 | 0.24985 (16) | 0.87246 (7) | 0.51764 (11) | 0.0324 (3) | |
| H21A | 0.2591 | 0.9048 | 0.5601 | 0.039* | |
| C22 | 0.34173 (15) | 0.83192 (7) | 0.54228 (11) | 0.0326 (3) | |
| H22A | 0.4129 | 0.8368 | 0.6017 | 0.039* | |
| C23 | 0.32780 (12) | 0.78409 (7) | 0.47855 (10) | 0.0263 (3) | |
| H23A | 0.3895 | 0.7565 | 0.4962 | 0.032* | |
| C24 | 0.22258 (10) | 0.77667 (5) | 0.38820 (9) | 0.0170 (2) | |
| C25 | 0.32775 (12) | 0.79391 (6) | 0.24798 (10) | 0.0244 (2) | |
| H25A | 0.2639 | 0.8203 | 0.2284 | 0.029* | |
| C26 | 0.41702 (13) | 0.81267 (7) | 0.23448 (11) | 0.0294 (3) | |
| H26A | 0.4117 | 0.8510 | 0.2045 | 0.035* | |
| C27 | 0.51388 (13) | 0.77433 (8) | 0.26570 (11) | 0.0299 (3) | |
| H27A | 0.5740 | 0.7872 | 0.2575 | 0.036* | |
| C28 | 0.52104 (11) | 0.71679 (7) | 0.30923 (10) | 0.0267 (3) | |
| H28A | 0.5864 | 0.6913 | 0.3307 | 0.032* | |
| C29 | 0.43030 (10) | 0.69707 (6) | 0.32084 (9) | 0.0203 (2) | |
| H29A | 0.4347 | 0.6581 | 0.3487 | 0.024* | |
| C30 | 0.33307 (10) | 0.73567 (6) | 0.29084 (9) | 0.0172 (2) | |
| C31 | 0.18777 (12) | 0.40334 (6) | 0.45353 (11) | 0.0252 (3) | |
| C32 | 0.39740 (11) | 0.48128 (6) | 0.53746 (10) | 0.0212 (2) | |
| C33 | 0.26107 (11) | 0.45749 (6) | 0.33856 (10) | 0.0217 (2) | |
| C34 | 0.20400 (11) | 0.52583 (6) | 0.53304 (10) | 0.0208 (2) | |
| C35 | −0.05373 (11) | 0.45692 (6) | 0.29581 (9) | 0.0201 (2) | |
| C36 | 0.04399 (10) | 0.50930 (6) | 0.19821 (10) | 0.0203 (2) | |
| C37 | −0.02635 (10) | 0.56878 (6) | 0.38771 (9) | 0.0189 (2) | |
| C38 | 0.37863 (10) | 0.61975 (5) | 0.47485 (9) | 0.0177 (2) | |
| C39 | 0.24923 (10) | 0.57992 (5) | 0.27257 (9) | 0.0168 (2) | |
| C40 | 0.16108 (10) | 0.64892 (5) | 0.44471 (9) | 0.0170 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ru1 | 0.01584 (4) | 0.01339 (4) | 0.02068 (4) | 0.00335 (3) | 0.01167 (3) | 0.00438 (3) |
| Ru2 | 0.01199 (4) | 0.01355 (4) | 0.01822 (4) | −0.00056 (3) | 0.00816 (3) | −0.00232 (3) |
| Ru3 | 0.01112 (4) | 0.01189 (4) | 0.01314 (4) | 0.00063 (3) | 0.00614 (3) | 0.00121 (3) |
| P1 | 0.01275 (11) | 0.01649 (12) | 0.01796 (13) | −0.00106 (9) | 0.00683 (10) | −0.00405 (10) |
| P2 | 0.01276 (11) | 0.01292 (11) | 0.01512 (12) | 0.00080 (9) | 0.00762 (10) | 0.00159 (9) |
| O1 | 0.0567 (7) | 0.0181 (4) | 0.0655 (8) | 0.0020 (5) | 0.0492 (7) | 0.0052 (5) |
| O2 | 0.0213 (4) | 0.0415 (6) | 0.0276 (5) | 0.0099 (4) | 0.0124 (4) | 0.0101 (4) |
| O3 | 0.0399 (5) | 0.0225 (4) | 0.0327 (5) | 0.0099 (4) | 0.0254 (5) | 0.0058 (4) |
| O4 | 0.0331 (5) | 0.0297 (5) | 0.0264 (5) | 0.0078 (4) | 0.0197 (4) | 0.0051 (4) |
| O5 | 0.0330 (5) | 0.0193 (4) | 0.0354 (5) | −0.0065 (4) | 0.0235 (4) | −0.0062 (4) |
| O6 | 0.0247 (4) | 0.0281 (5) | 0.0280 (5) | −0.0006 (4) | 0.0163 (4) | −0.0075 (4) |
| O7 | 0.0257 (4) | 0.0307 (5) | 0.0302 (5) | −0.0051 (4) | 0.0195 (4) | −0.0077 (4) |
| O8 | 0.0165 (4) | 0.0280 (5) | 0.0237 (4) | −0.0011 (3) | 0.0041 (3) | 0.0029 (4) |
| O9 | 0.0284 (4) | 0.0205 (4) | 0.0219 (4) | 0.0029 (3) | 0.0162 (4) | 0.0022 (3) |
| O10 | 0.0276 (4) | 0.0204 (4) | 0.0293 (5) | −0.0031 (3) | 0.0200 (4) | −0.0045 (4) |
| C1 | 0.0203 (5) | 0.0245 (6) | 0.0328 (7) | −0.0057 (5) | 0.0110 (5) | −0.0068 (5) |
| C2 | 0.0249 (6) | 0.0293 (7) | 0.0364 (8) | −0.0113 (5) | 0.0091 (6) | −0.0031 (6) |
| C3 | 0.0169 (5) | 0.0404 (8) | 0.0320 (7) | −0.0055 (5) | 0.0071 (5) | 0.0094 (6) |
| C4 | 0.0207 (6) | 0.0371 (7) | 0.0312 (7) | 0.0030 (5) | 0.0146 (5) | 0.0097 (6) |
| C5 | 0.0203 (5) | 0.0249 (6) | 0.0263 (6) | 0.0009 (4) | 0.0127 (5) | 0.0021 (5) |
| C6 | 0.0129 (4) | 0.0204 (5) | 0.0225 (5) | −0.0009 (4) | 0.0071 (4) | −0.0007 (4) |
| C7 | 0.0151 (5) | 0.0237 (5) | 0.0205 (5) | 0.0001 (4) | 0.0054 (4) | −0.0020 (5) |
| C8 | 0.0159 (5) | 0.0252 (6) | 0.0238 (6) | 0.0045 (4) | 0.0082 (4) | 0.0051 (5) |
| C9 | 0.0219 (5) | 0.0189 (5) | 0.0283 (6) | 0.0035 (4) | 0.0147 (5) | 0.0033 (5) |
| C10 | 0.0228 (5) | 0.0183 (5) | 0.0223 (5) | −0.0006 (4) | 0.0112 (5) | −0.0032 (4) |
| C11 | 0.0157 (5) | 0.0187 (5) | 0.0167 (5) | 0.0003 (4) | 0.0073 (4) | −0.0011 (4) |
| C12 | 0.0138 (4) | 0.0169 (5) | 0.0174 (5) | 0.0004 (4) | 0.0077 (4) | −0.0015 (4) |
| C13 | 0.0215 (5) | 0.0253 (6) | 0.0201 (5) | −0.0037 (4) | 0.0102 (5) | −0.0075 (5) |
| C14 | 0.0314 (7) | 0.0268 (6) | 0.0312 (7) | −0.0001 (5) | 0.0212 (6) | −0.0033 (5) |
| C15 | 0.0183 (5) | 0.0305 (6) | 0.0167 (5) | 0.0021 (5) | 0.0067 (4) | 0.0030 (5) |
| C16 | 0.0210 (5) | 0.0313 (6) | 0.0147 (5) | 0.0041 (5) | 0.0080 (4) | 0.0049 (5) |
| C17 | 0.0177 (5) | 0.0247 (5) | 0.0158 (5) | 0.0024 (4) | 0.0089 (4) | 0.0019 (4) |
| C18 | 0.0160 (5) | 0.0188 (5) | 0.0162 (5) | 0.0028 (4) | 0.0082 (4) | 0.0026 (4) |
| C19 | 0.0272 (6) | 0.0168 (5) | 0.0267 (6) | 0.0036 (4) | 0.0186 (5) | 0.0034 (4) |
| C20 | 0.0468 (8) | 0.0183 (5) | 0.0377 (7) | 0.0062 (5) | 0.0333 (7) | 0.0035 (5) |
| C21 | 0.0619 (10) | 0.0191 (6) | 0.0292 (7) | −0.0046 (6) | 0.0331 (7) | −0.0038 (5) |
| C22 | 0.0443 (8) | 0.0274 (7) | 0.0223 (6) | −0.0061 (6) | 0.0152 (6) | −0.0059 (5) |
| C23 | 0.0269 (6) | 0.0243 (6) | 0.0217 (6) | 0.0002 (5) | 0.0090 (5) | −0.0031 (5) |
| C24 | 0.0208 (5) | 0.0140 (4) | 0.0189 (5) | −0.0004 (4) | 0.0123 (4) | 0.0007 (4) |
| C25 | 0.0263 (6) | 0.0203 (5) | 0.0320 (7) | −0.0015 (4) | 0.0192 (5) | 0.0029 (5) |
| C26 | 0.0352 (7) | 0.0259 (6) | 0.0363 (7) | −0.0090 (5) | 0.0252 (6) | 0.0001 (6) |
| C27 | 0.0265 (6) | 0.0389 (8) | 0.0334 (7) | −0.0117 (6) | 0.0221 (6) | −0.0068 (6) |
| C28 | 0.0179 (5) | 0.0369 (7) | 0.0292 (6) | −0.0018 (5) | 0.0151 (5) | −0.0036 (6) |
| C29 | 0.0164 (5) | 0.0248 (5) | 0.0213 (5) | −0.0001 (4) | 0.0111 (4) | 0.0004 (4) |
| C30 | 0.0170 (5) | 0.0178 (5) | 0.0188 (5) | −0.0024 (4) | 0.0110 (4) | −0.0008 (4) |
| C31 | 0.0302 (6) | 0.0196 (5) | 0.0371 (7) | 0.0043 (5) | 0.0255 (6) | 0.0035 (5) |
| C32 | 0.0226 (5) | 0.0222 (5) | 0.0243 (6) | 0.0064 (4) | 0.0162 (5) | 0.0071 (5) |
| C33 | 0.0240 (5) | 0.0163 (5) | 0.0269 (6) | 0.0053 (4) | 0.0150 (5) | 0.0062 (4) |
| C34 | 0.0193 (5) | 0.0190 (5) | 0.0256 (6) | 0.0045 (4) | 0.0129 (5) | 0.0066 (4) |
| C35 | 0.0198 (5) | 0.0186 (5) | 0.0246 (6) | −0.0009 (4) | 0.0138 (5) | −0.0048 (4) |
| C36 | 0.0159 (5) | 0.0198 (5) | 0.0251 (6) | −0.0004 (4) | 0.0109 (4) | −0.0025 (4) |
| C37 | 0.0147 (4) | 0.0186 (5) | 0.0221 (5) | −0.0024 (4) | 0.0089 (4) | −0.0025 (4) |
| C38 | 0.0174 (5) | 0.0168 (5) | 0.0191 (5) | 0.0006 (4) | 0.0099 (4) | 0.0023 (4) |
| C39 | 0.0173 (5) | 0.0140 (4) | 0.0177 (5) | 0.0013 (4) | 0.0085 (4) | 0.0030 (4) |
| C40 | 0.0159 (4) | 0.0155 (5) | 0.0196 (5) | −0.0012 (4) | 0.0095 (4) | 0.0018 (4) |
Geometric parameters (Å, º)
| Ru1—C32 | 1.9050 (13) | C8—H8A | 0.9300 |
| Ru1—C31 | 1.9293 (14) | C9—C10 | 1.3849 (18) |
| Ru1—C34 | 1.9459 (13) | C9—H9A | 0.9300 |
| Ru1—C33 | 1.9461 (14) | C10—C11 | 1.3980 (17) |
| Ru1—Ru2 | 2.8842 (2) | C10—H10A | 0.9300 |
| Ru1—Ru3 | 2.8876 (2) | C11—C12 | 1.3899 (16) |
| Ru2—C35 | 1.8887 (13) | C11—H11A | 0.9300 |
| Ru2—C36 | 1.9271 (13) | C13—C14 | 1.5366 (19) |
| Ru2—C37 | 1.9354 (13) | C13—H13A | 0.9700 |
| Ru2—P1 | 2.3522 (3) | C13—H13B | 0.9700 |
| Ru2—Ru3 | 2.9531 (2) | C14—C15 | 1.528 (2) |
| Ru3—C38 | 1.8792 (12) | C14—H14A | 0.9700 |
| Ru3—C39 | 1.9319 (12) | C14—H14B | 0.9700 |
| Ru3—C40 | 1.9322 (12) | C15—C16 | 1.5333 (19) |
| Ru3—P2 | 2.3413 (3) | C15—H15A | 0.9700 |
| P1—C12 | 1.8308 (12) | C15—H15B | 0.9700 |
| P1—C6 | 1.8353 (12) | C16—C17 | 1.5326 (17) |
| P1—C13 | 1.8458 (13) | C16—H16A | 0.9700 |
| P2—C24 | 1.8285 (12) | C16—H16B | 0.9700 |
| P2—C18 | 1.8334 (11) | C17—C18 | 1.5315 (17) |
| P2—C30 | 1.8399 (12) | C17—H17A | 0.9700 |
| O1—C31 | 1.1376 (17) | C17—H17B | 0.9700 |
| O2—C32 | 1.1402 (16) | C18—H18A | 0.9700 |
| O3—C33 | 1.1387 (16) | C18—H18B | 0.9700 |
| O4—C34 | 1.1368 (16) | C19—C24 | 1.3902 (17) |
| O5—C35 | 1.1429 (15) | C19—C20 | 1.3974 (19) |
| O6—C36 | 1.1472 (16) | C19—H19A | 0.9300 |
| O7—C37 | 1.1409 (16) | C20—C21 | 1.381 (2) |
| O8—C38 | 1.1468 (15) | C20—H20A | 0.9300 |
| O9—C39 | 1.1480 (15) | C21—C22 | 1.382 (2) |
| O10—C40 | 1.1445 (15) | C21—H21A | 0.9300 |
| C1—C2 | 1.390 (2) | C22—C23 | 1.385 (2) |
| C1—C6 | 1.3954 (18) | C22—H22A | 0.9300 |
| C1—H1A | 0.9300 | C23—C24 | 1.3959 (17) |
| C2—C3 | 1.388 (2) | C23—H23A | 0.9300 |
| C2—H2A | 0.9300 | C25—C26 | 1.3910 (19) |
| C3—C4 | 1.383 (2) | C25—C30 | 1.4012 (17) |
| C3—H3A | 0.9300 | C25—H25A | 0.9300 |
| C4—C5 | 1.3920 (19) | C26—C27 | 1.386 (2) |
| C4—H4A | 0.9300 | C26—H26A | 0.9300 |
| C5—C6 | 1.3969 (18) | C27—C28 | 1.388 (2) |
| C5—H5A | 0.9300 | C27—H27A | 0.9300 |
| C7—C8 | 1.3818 (18) | C28—C29 | 1.3973 (17) |
| C7—C12 | 1.4061 (16) | C28—H28A | 0.9300 |
| C7—H7A | 0.9300 | C29—C30 | 1.3961 (17) |
| C8—C9 | 1.3869 (19) | C29—H29A | 0.9300 |
| C32—Ru1—C31 | 98.85 (6) | C12—C11—H11A | 120.0 |
| C32—Ru1—C34 | 90.69 (5) | C10—C11—H11A | 120.0 |
| C31—Ru1—C34 | 95.10 (6) | C11—C12—C7 | 118.84 (11) |
| C32—Ru1—C33 | 91.44 (5) | C11—C12—P1 | 122.69 (9) |
| C31—Ru1—C33 | 91.35 (6) | C7—C12—P1 | 118.47 (9) |
| C34—Ru1—C33 | 172.82 (5) | C14—C13—P1 | 114.55 (9) |
| C32—Ru1—Ru2 | 161.50 (4) | C14—C13—H13A | 108.6 |
| C31—Ru1—Ru2 | 99.37 (4) | P1—C13—H13A | 108.6 |
| C34—Ru1—Ru2 | 84.58 (4) | C14—C13—H13B | 108.6 |
| C33—Ru1—Ru2 | 91.27 (4) | P1—C13—H13B | 108.6 |
| C32—Ru1—Ru3 | 100.25 (4) | H13A—C13—H13B | 107.6 |
| C31—Ru1—Ru3 | 160.89 (4) | C15—C14—C13 | 117.11 (11) |
| C34—Ru1—Ru3 | 84.54 (4) | C15—C14—H14A | 108.0 |
| C33—Ru1—Ru3 | 88.33 (4) | C13—C14—H14A | 108.0 |
| Ru2—Ru1—Ru3 | 61.547 (3) | C15—C14—H14B | 108.0 |
| C35—Ru2—C36 | 92.98 (5) | C13—C14—H14B | 108.0 |
| C35—Ru2—C37 | 90.71 (5) | H14A—C14—H14B | 107.3 |
| C36—Ru2—C37 | 176.27 (5) | C14—C15—C16 | 112.10 (11) |
| C35—Ru2—P1 | 95.60 (4) | C14—C15—H15A | 109.2 |
| C36—Ru2—P1 | 91.18 (4) | C16—C15—H15A | 109.2 |
| C37—Ru2—P1 | 87.95 (4) | C14—C15—H15B | 109.2 |
| C35—Ru2—Ru1 | 86.55 (4) | C16—C15—H15B | 109.2 |
| C36—Ru2—Ru1 | 86.07 (4) | H15A—C15—H15B | 107.9 |
| C37—Ru2—Ru1 | 94.67 (4) | C17—C16—C15 | 114.47 (10) |
| P1—Ru2—Ru1 | 176.602 (9) | C17—C16—H16A | 108.6 |
| C35—Ru2—Ru3 | 145.79 (4) | C15—C16—H16A | 108.6 |
| C36—Ru2—Ru3 | 83.75 (4) | C17—C16—H16B | 108.6 |
| C37—Ru2—Ru3 | 93.48 (3) | C15—C16—H16B | 108.6 |
| P1—Ru2—Ru3 | 118.456 (9) | H16A—C16—H16B | 107.6 |
| Ru1—Ru2—Ru3 | 59.283 (4) | C18—C17—C16 | 112.87 (10) |
| C38—Ru3—C39 | 98.20 (5) | C18—C17—H17A | 109.0 |
| C38—Ru3—C40 | 93.46 (5) | C16—C17—H17A | 109.0 |
| C39—Ru3—C40 | 168.09 (5) | C18—C17—H17B | 109.0 |
| C38—Ru3—P2 | 95.48 (4) | C16—C17—H17B | 109.0 |
| C39—Ru3—P2 | 88.20 (3) | H17A—C17—H17B | 107.8 |
| C40—Ru3—P2 | 88.26 (3) | C17—C18—P2 | 112.48 (8) |
| C38—Ru3—Ru1 | 85.21 (4) | C17—C18—H18A | 109.1 |
| C39—Ru3—Ru1 | 88.63 (3) | P2—C18—H18A | 109.1 |
| C40—Ru3—Ru1 | 94.79 (3) | C17—C18—H18B | 109.1 |
| P2—Ru3—Ru1 | 176.823 (8) | P2—C18—H18B | 109.1 |
| C38—Ru3—Ru2 | 142.89 (4) | H18A—C18—H18B | 107.8 |
| C39—Ru3—Ru2 | 91.49 (3) | C24—C19—C20 | 119.99 (13) |
| C40—Ru3—Ru2 | 80.56 (3) | C24—C19—H19A | 120.0 |
| P2—Ru3—Ru2 | 120.673 (8) | C20—C19—H19A | 120.0 |
| Ru1—Ru3—Ru2 | 59.170 (4) | C21—C20—C19 | 120.57 (13) |
| C12—P1—C6 | 99.50 (5) | C21—C20—H20A | 119.7 |
| C12—P1—C13 | 102.59 (6) | C19—C20—H20A | 119.7 |
| C6—P1—C13 | 103.60 (6) | C20—C21—C22 | 119.71 (13) |
| C12—P1—Ru2 | 122.83 (4) | C20—C21—H21A | 120.1 |
| C6—P1—Ru2 | 110.90 (4) | C22—C21—H21A | 120.1 |
| C13—P1—Ru2 | 114.87 (4) | C21—C22—C23 | 119.98 (14) |
| C24—P2—C18 | 104.06 (5) | C21—C22—H22A | 120.0 |
| C24—P2—C30 | 100.32 (5) | C23—C22—H22A | 120.0 |
| C18—P2—C30 | 102.40 (5) | C22—C23—C24 | 121.02 (13) |
| C24—P2—Ru3 | 113.02 (4) | C22—C23—H23A | 119.5 |
| C18—P2—Ru3 | 118.14 (4) | C24—C23—H23A | 119.5 |
| C30—P2—Ru3 | 116.62 (4) | C19—C24—C23 | 118.68 (12) |
| C2—C1—C6 | 120.65 (14) | C19—C24—P2 | 122.90 (9) |
| C2—C1—H1A | 119.7 | C23—C24—P2 | 118.41 (9) |
| C6—C1—H1A | 119.7 | C26—C25—C30 | 120.65 (13) |
| C3—C2—C1 | 120.18 (14) | C26—C25—H25A | 119.7 |
| C3—C2—H2A | 119.9 | C30—C25—H25A | 119.7 |
| C1—C2—H2A | 119.9 | C27—C26—C25 | 120.05 (13) |
| C4—C3—C2 | 119.89 (13) | C27—C26—H26A | 120.0 |
| C4—C3—H3A | 120.1 | C25—C26—H26A | 120.0 |
| C2—C3—H3A | 120.1 | C26—C27—C28 | 119.99 (12) |
| C3—C4—C5 | 119.99 (14) | C26—C27—H27A | 120.0 |
| C3—C4—H4A | 120.0 | C28—C27—H27A | 120.0 |
| C5—C4—H4A | 120.0 | C27—C28—C29 | 120.18 (13) |
| C4—C5—C6 | 120.80 (13) | C27—C28—H28A | 119.9 |
| C4—C5—H5A | 119.6 | C29—C28—H28A | 119.9 |
| C6—C5—H5A | 119.6 | C30—C29—C28 | 120.31 (12) |
| C1—C6—C5 | 118.49 (12) | C30—C29—H29A | 119.8 |
| C1—C6—P1 | 120.81 (10) | C28—C29—H29A | 119.8 |
| C5—C6—P1 | 120.58 (10) | C29—C30—C25 | 118.81 (11) |
| C8—C7—C12 | 120.79 (11) | C29—C30—P2 | 122.63 (9) |
| C8—C7—H7A | 119.6 | C25—C30—P2 | 118.56 (9) |
| C12—C7—H7A | 119.6 | O1—C31—Ru1 | 175.53 (12) |
| C7—C8—C9 | 119.91 (11) | O2—C32—Ru1 | 176.94 (12) |
| C7—C8—H8A | 120.0 | O3—C33—Ru1 | 174.44 (11) |
| C9—C8—H8A | 120.0 | O4—C34—Ru1 | 173.73 (11) |
| C10—C9—C8 | 120.02 (12) | O5—C35—Ru2 | 178.36 (13) |
| C10—C9—H9A | 120.0 | O6—C36—Ru2 | 171.84 (11) |
| C8—C9—H9A | 120.0 | O7—C37—Ru2 | 173.03 (10) |
| C9—C10—C11 | 120.31 (12) | O8—C38—Ru3 | 177.79 (11) |
| C9—C10—H10A | 119.8 | O9—C39—Ru3 | 173.87 (10) |
| C11—C10—H10A | 119.8 | O10—C40—Ru3 | 171.83 (10) |
| C12—C11—C10 | 120.08 (11) | ||
| C32—Ru1—Ru2—C35 | −170.75 (13) | C12—P1—C6—C1 | 152.39 (11) |
| C31—Ru1—Ru2—C35 | −0.70 (6) | C13—P1—C6—C1 | 46.84 (12) |
| C34—Ru1—Ru2—C35 | −94.99 (5) | Ru2—P1—C6—C1 | −76.90 (12) |
| C33—Ru1—Ru2—C35 | 90.88 (5) | C12—P1—C6—C5 | −31.60 (11) |
| Ru3—Ru1—Ru2—C35 | 178.29 (4) | C13—P1—C6—C5 | −137.14 (11) |
| C32—Ru1—Ru2—C36 | 96.01 (13) | Ru2—P1—C6—C5 | 99.11 (10) |
| C31—Ru1—Ru2—C36 | −93.93 (6) | C12—C7—C8—C9 | −2.6 (2) |
| C34—Ru1—Ru2—C36 | 171.78 (5) | C7—C8—C9—C10 | 1.2 (2) |
| C33—Ru1—Ru2—C36 | −2.35 (5) | C8—C9—C10—C11 | 0.6 (2) |
| Ru3—Ru1—Ru2—C36 | 85.06 (4) | C9—C10—C11—C12 | −1.06 (19) |
| C32—Ru1—Ru2—C37 | −80.33 (13) | C10—C11—C12—C7 | −0.26 (18) |
| C31—Ru1—Ru2—C37 | 89.73 (6) | C10—C11—C12—P1 | 179.77 (9) |
| C34—Ru1—Ru2—C37 | −4.56 (5) | C8—C7—C12—C11 | 2.09 (19) |
| C33—Ru1—Ru2—C37 | −178.69 (5) | C8—C7—C12—P1 | −177.93 (10) |
| Ru3—Ru1—Ru2—C37 | −91.28 (4) | C6—P1—C12—C11 | 119.28 (11) |
| C32—Ru1—Ru2—P1 | 59.97 (19) | C13—P1—C12—C11 | −134.37 (10) |
| C31—Ru1—Ru2—P1 | −129.98 (15) | Ru2—P1—C12—C11 | −3.29 (12) |
| C34—Ru1—Ru2—P1 | 135.73 (15) | C6—P1—C12—C7 | −60.70 (11) |
| C33—Ru1—Ru2—P1 | −38.40 (15) | C13—P1—C12—C7 | 45.66 (11) |
| Ru3—Ru1—Ru2—P1 | 49.01 (14) | Ru2—P1—C12—C7 | 176.73 (8) |
| C32—Ru1—Ru2—Ru3 | 10.95 (12) | C12—P1—C13—C14 | 71.06 (11) |
| C31—Ru1—Ru2—Ru3 | −178.99 (4) | C6—P1—C13—C14 | 174.24 (10) |
| C34—Ru1—Ru2—Ru3 | 86.72 (4) | Ru2—P1—C13—C14 | −64.66 (11) |
| C33—Ru1—Ru2—Ru3 | −87.41 (4) | P1—C13—C14—C15 | −79.49 (14) |
| C32—Ru1—Ru3—C38 | −7.39 (5) | C13—C14—C15—C16 | 169.24 (11) |
| C31—Ru1—Ru3—C38 | 172.15 (13) | C14—C15—C16—C17 | −59.62 (15) |
| C34—Ru1—Ru3—C38 | 82.32 (5) | C15—C16—C17—C18 | −63.05 (15) |
| C33—Ru1—Ru3—C38 | −98.55 (5) | C16—C17—C18—P2 | 168.47 (9) |
| Ru2—Ru1—Ru3—C38 | 169.10 (4) | C24—P2—C18—C17 | 159.32 (9) |
| C32—Ru1—Ru3—C39 | 90.96 (5) | C30—P2—C18—C17 | 55.19 (10) |
| C31—Ru1—Ru3—C39 | −89.51 (13) | Ru3—P2—C18—C17 | −74.45 (9) |
| C34—Ru1—Ru3—C39 | −179.33 (5) | C24—C19—C20—C21 | −0.1 (2) |
| C33—Ru1—Ru3—C39 | −0.20 (5) | C19—C20—C21—C22 | −1.0 (2) |
| Ru2—Ru1—Ru3—C39 | −92.56 (3) | C20—C21—C22—C23 | 0.4 (2) |
| C32—Ru1—Ru3—C40 | −100.47 (5) | C21—C22—C23—C24 | 1.3 (2) |
| C31—Ru1—Ru3—C40 | 79.06 (13) | C20—C19—C24—C23 | 1.69 (18) |
| C34—Ru1—Ru3—C40 | −10.76 (5) | C20—C19—C24—P2 | −179.12 (10) |
| C33—Ru1—Ru3—C40 | 168.37 (5) | C22—C23—C24—C19 | −2.3 (2) |
| Ru2—Ru1—Ru3—C40 | 76.02 (3) | C22—C23—C24—P2 | 178.48 (12) |
| C32—Ru1—Ru3—P2 | 95.40 (15) | C18—P2—C24—C19 | 16.52 (12) |
| C31—Ru1—Ru3—P2 | −85.07 (19) | C30—P2—C24—C19 | 122.23 (11) |
| C34—Ru1—Ru3—P2 | −174.89 (15) | Ru3—P2—C24—C19 | −112.87 (10) |
| C33—Ru1—Ru3—P2 | 4.24 (15) | C18—P2—C24—C23 | −164.28 (10) |
| Ru2—Ru1—Ru3—P2 | −88.11 (14) | C30—P2—C24—C23 | −58.58 (11) |
| C32—Ru1—Ru3—Ru2 | −176.49 (4) | Ru3—P2—C24—C23 | 66.33 (11) |
| C31—Ru1—Ru3—Ru2 | 3.05 (13) | C30—C25—C26—C27 | −1.6 (2) |
| C34—Ru1—Ru3—Ru2 | −86.78 (4) | C25—C26—C27—C28 | 0.9 (2) |
| C33—Ru1—Ru3—Ru2 | 92.35 (4) | C26—C27—C28—C29 | 0.6 (2) |
| C35—Ru2—Ru3—C38 | −21.23 (9) | C27—C28—C29—C30 | −1.5 (2) |
| C36—Ru2—Ru3—C38 | −107.34 (7) | C28—C29—C30—C25 | 0.78 (19) |
| C37—Ru2—Ru3—C38 | 75.17 (7) | C28—C29—C30—P2 | −178.39 (10) |
| P1—Ru2—Ru3—C38 | 164.72 (6) | C26—C25—C30—C29 | 0.7 (2) |
| Ru1—Ru2—Ru3—C38 | −18.20 (6) | C26—C25—C30—P2 | 179.95 (11) |
| C35—Ru2—Ru3—C39 | 84.48 (8) | C24—P2—C30—C29 | 118.01 (11) |
| C36—Ru2—Ru3—C39 | −1.62 (5) | C18—P2—C30—C29 | −134.96 (10) |
| C37—Ru2—Ru3—C39 | −179.12 (5) | Ru3—P2—C30—C29 | −4.39 (12) |
| P1—Ru2—Ru3—C39 | −89.57 (3) | C24—P2—C30—C25 | −61.16 (11) |
| Ru1—Ru2—Ru3—C39 | 87.51 (3) | C18—P2—C30—C25 | 45.87 (11) |
| C35—Ru2—Ru3—C40 | −104.44 (8) | Ru3—P2—C30—C25 | 176.43 (9) |
| C36—Ru2—Ru3—C40 | 169.45 (5) | C32—Ru1—C31—O1 | −27.5 (18) |
| C37—Ru2—Ru3—C40 | −8.04 (5) | C34—Ru1—C31—O1 | −119.0 (18) |
| P1—Ru2—Ru3—C40 | 81.51 (4) | C33—Ru1—C31—O1 | 64.1 (18) |
| Ru1—Ru2—Ru3—C40 | −101.41 (3) | Ru2—Ru1—C31—O1 | 155.6 (18) |
| C35—Ru2—Ru3—P2 | 173.27 (7) | Ru3—Ru1—C31—O1 | 152.9 (17) |
| C36—Ru2—Ru3—P2 | 87.17 (4) | C31—Ru1—C32—O2 | −25 (2) |
| C37—Ru2—Ru3—P2 | −90.32 (4) | C34—Ru1—C32—O2 | 70 (2) |
| P1—Ru2—Ru3—P2 | −0.775 (13) | C33—Ru1—C32—O2 | −117 (2) |
| Ru1—Ru2—Ru3—P2 | 176.308 (10) | Ru2—Ru1—C32—O2 | 145 (2) |
| C35—Ru2—Ru3—Ru1 | −3.03 (7) | Ru3—Ru1—C32—O2 | 155 (2) |
| C36—Ru2—Ru3—Ru1 | −89.14 (4) | C32—Ru1—C33—O3 | 48.8 (13) |
| C37—Ru2—Ru3—Ru1 | 93.37 (4) | C31—Ru1—C33—O3 | −50.1 (13) |
| P1—Ru2—Ru3—Ru1 | −177.083 (10) | C34—Ru1—C33—O3 | 155.9 (11) |
| C35—Ru2—P1—C12 | 141.36 (6) | Ru2—Ru1—C33—O3 | −149.5 (13) |
| C36—Ru2—P1—C12 | −125.53 (6) | Ru3—Ru1—C33—O3 | 149.0 (13) |
| C37—Ru2—P1—C12 | 50.85 (6) | C32—Ru1—C34—O4 | −27.4 (11) |
| Ru1—Ru2—P1—C12 | −89.57 (15) | C31—Ru1—C34—O4 | 71.6 (11) |
| Ru3—Ru2—P1—C12 | −42.00 (5) | C33—Ru1—C34—O4 | −134.6 (10) |
| C35—Ru2—P1—C6 | 24.19 (6) | Ru2—Ru1—C34—O4 | 170.5 (11) |
| C36—Ru2—P1—C6 | 117.30 (6) | Ru3—Ru1—C34—O4 | −127.6 (11) |
| C37—Ru2—P1—C6 | −66.32 (6) | C36—Ru2—C35—O5 | 100 (4) |
| Ru1—Ru2—P1—C6 | 153.26 (14) | C37—Ru2—C35—O5 | −81 (4) |
| Ru3—Ru2—P1—C6 | −159.17 (4) | P1—Ru2—C35—O5 | −169 (4) |
| C35—Ru2—P1—C13 | −92.83 (6) | Ru1—Ru2—C35—O5 | 14 (4) |
| C36—Ru2—P1—C13 | 0.28 (6) | Ru3—Ru2—C35—O5 | 17 (4) |
| C37—Ru2—P1—C13 | 176.66 (6) | C35—Ru2—C36—O6 | 37.9 (8) |
| Ru1—Ru2—P1—C13 | 36.24 (16) | C37—Ru2—C36—O6 | −134.1 (8) |
| Ru3—Ru2—P1—C13 | 83.81 (5) | P1—Ru2—C36—O6 | −57.7 (8) |
| C38—Ru3—P2—C24 | −67.34 (6) | Ru1—Ru2—C36—O6 | 124.3 (8) |
| C39—Ru3—P2—C24 | −165.41 (5) | Ru3—Ru2—C36—O6 | −176.2 (8) |
| C40—Ru3—P2—C24 | 25.97 (5) | C35—Ru2—C37—O7 | −48.1 (10) |
| Ru1—Ru3—P2—C24 | −169.85 (14) | C36—Ru2—C37—O7 | 123.9 (10) |
| Ru2—Ru3—P2—C24 | 103.93 (4) | P1—Ru2—C37—O7 | 47.4 (10) |
| C38—Ru3—P2—C18 | 170.88 (6) | Ru1—Ru2—C37—O7 | −134.7 (9) |
| C39—Ru3—P2—C18 | 72.82 (6) | Ru3—Ru2—C37—O7 | 165.8 (9) |
| C40—Ru3—P2—C18 | −95.80 (6) | C39—Ru3—C38—O8 | −165 (3) |
| Ru1—Ru3—P2—C18 | 68.38 (15) | C40—Ru3—C38—O8 | 17 (3) |
| Ru2—Ru3—P2—C18 | −17.85 (5) | P2—Ru3—C38—O8 | 106 (3) |
| C38—Ru3—P2—C30 | 48.17 (6) | Ru1—Ru3—C38—O8 | −78 (3) |
| C39—Ru3—P2—C30 | −49.90 (5) | Ru2—Ru3—C38—O8 | −62 (3) |
| C40—Ru3—P2—C30 | 141.48 (5) | C38—Ru3—C39—O9 | −55.2 (10) |
| Ru1—Ru3—P2—C30 | −54.34 (16) | C40—Ru3—C39—O9 | 112.9 (9) |
| Ru2—Ru3—P2—C30 | −140.56 (4) | P2—Ru3—C39—O9 | 40.1 (10) |
| C6—C1—C2—C3 | −0.3 (2) | Ru1—Ru3—C39—O9 | −140.2 (10) |
| C1—C2—C3—C4 | 0.2 (2) | Ru2—Ru3—C39—O9 | 160.7 (10) |
| C2—C3—C4—C5 | −0.4 (2) | C38—Ru3—C40—O10 | 50.2 (7) |
| C3—C4—C5—C6 | 0.6 (2) | C39—Ru3—C40—O10 | −118.0 (7) |
| C2—C1—C6—C5 | 0.5 (2) | P2—Ru3—C40—O10 | −45.2 (7) |
| C2—C1—C6—P1 | 176.64 (11) | Ru1—Ru3—C40—O10 | 135.7 (7) |
| C4—C5—C6—C1 | −0.7 (2) | Ru2—Ru3—C40—O10 | −166.7 (7) |
| C4—C5—C6—P1 | −176.80 (10) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C9—H9A···O3i | 0.93 | 2.48 | 3.3421 (17) | 154 |
| C14—H14A···O6 | 0.97 | 2.58 | 3.2007 (19) | 122 |
| C20—H20A···O6i | 0.93 | 2.59 | 3.495 (2) | 164 |
| C21—H21A···O9ii | 0.93 | 2.51 | 3.329 (2) | 147 |
Symmetry codes: (i) −x, y+1/2, −z+1/2; (ii) x, −y+3/2, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SJ5228).
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruce, M. I., Hambley, T. W., Nicholson, B. K. & Snow, M. R. (1982). J. Organomet. Chem. 235, 83–91.
- Bruce, M. I., Matisons, J. G. & Nicholson, B. K. (1983). J. Organomet. Chem. 247, 321–343.
- Bruker (2009). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Churchill, M. R., Hollander, F. J. & Hutchinson, J. P. (1977). Inorg. Chem. 16, 2655–2659.
- Coleman, A. W., Jones, D. F., Dixneuf, P. H., Brisson, C., Bonnet, J. J. & Lavigne, G. (1984). Inorg. Chem. 23, 952–956.
- Cosier, J. & Glazer, A. M. (1986). J. Appl. Cryst. 19, 105–107.
- Diz, E. L., Neels, A., Stoeckli-Evans, H. & Suss-Fink, G. (2001). Polyhedron, 20, 2771–2780.
- Shawkataly, O. bin, Chong, M.-L., Fun, H.-K., Didierjean, C. & Aubry, A. (2006). Acta Cryst. E62, m168–m169.
- Shawkataly, O. bin, Khan, I. A., Hafiz Malik, H. A., Yeap, C. S. & Fun, H.-K. (2011). Acta Cryst E67, m197–m198. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Teoh, S.-G., Fun, H.-K. & Shawkataly, O. bin (1990). Z. Kristallogr. 190, 287–293.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812014614/sj5228sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812014614/sj5228Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




