Abstract
BCL6 is the product of a proto-oncogene implicated in the pathogenesis of human B-cell lymphomas1,2. By binding specific DNA sequences, BCL6 controls the transcription of a variety of genes involved in B-cell development, differentiation and activation. BCL6 is overexpressed in the majority of patients with aggressive diffuse large B-cell lymphoma (DLBCL), the most common lymphoma in adulthood, and transgenic mice constitutively expressing BCL6 in B cells develop DLBCLs similar to the human disease3,4. In many DLBCL patients, BCL6 overexpression is achieved through translocation ( 40%) or hypermutation of its promoter ( 15%). However, many other DLBCLs overexpress BCL6 through an unknown mechanism. Here we show that BCL6 is targeted for ubiquitylation and proteasomal degradation by a SKP1–CUL1–F-box protein (SCF) ubiquitin ligase complex that contains the orphan F-box protein FBXO11 (refs 5, 6). The gene encoding FBXO11 was found to be deleted or mutated in multiple DLBCL cell lines, and this inactivation of FBXO11 correlated with increased levels and stability of BCL6. Similarly, FBXO11 was either deleted or mutated in primary DLBCLs. Notably, tumour-derived FBXO11 mutants displayed an impaired ability to induce BCL6 degradation. Reconstitution of FBXO11 expression in FBXO11-deleted DLBCL cells promoted BCL6 ubiquitylation and degradation, inhibited cell proliferation, and induced cell death. FBXO11-deleted DLBCL cells generated tumours in immunodeficient mice, and the tumorigenicity was suppressed by FBXO11 reconstitution. We reveal a molecular mechanism controlling BCL6 stability and propose that mutations and deletions in FBXO11 contribute to lymphomagenesis through BCL6 stabilization. The deletions/mutations found in DLBCLs are largely monoallelic, indicating that FBXO11 is a haplo-insufficient tumour suppressor gene.
Because the degradation of BCL6 has been reported to be phosphorylation-dependent7,8, we examined the hypothesis that BCL6 is degraded by an SCF ubiquitin ligase complex, as most SCF ligases target phosphorylated substrates5. The expression of a dominant-negative CUL1 mutant (CUL1(1–242) or CUL1(1–385)) results in the accumulation of SCF substrates9–11. Western blotting of extracts from Ramos cells (a Burkitt’s lymphoma cell line) infected with a retrovirus expressing CUL1(1–385) revealed that BCL6 level was increased compared to control cells, indicating that BCL6 is an SCF substrate (Supplementary Fig. 1a). Similarly, silencing RBX1, another SCF subunit, induced the accumulation of BCL6 (Supplementary Fig. 1b). Therefore, we investigated the ability of BCL6 to be recruited to the SCF via a panel of F-box proteins. Screening of a library of F-box proteins (ten shown) revealed that BCL6 specifically bound FBXO11 (Fig. 1a and Supplementary Fig. 2a), and this binding was confirmed with endogenous proteins in both Ramos cells (Fig. 1b) and primary B cells from mice (Supplementary Fig. 2b). Moreover, BCL6 was found to interact with endogenous SKP1 and neddylated CUL1, the form of CUL1 that preferentially binds SCF substrates12 (Supplementary Fig. 2a). We also found that BCL6 and FBXO11 co-localized in the nucleus where they showed overlapping, punctate staining throughout the nucleoplasm (Supplementary Fig. 3). Moreover, expression of FBXO11 resulted in a marked reduction of BCL6 levels (Supplementary Fig. 4). This reduction in protein level was due to enhanced proteolysis, as shown by the decrease in BCL6 half-life (Supplementary Fig. 4a, b) and the rescue of BCL6 levels by either CUL1(1–242) (compare lanes 2 and 3 in whole-cell extract of Fig. 1a) or the addition of MG132, a proteasome inhibitor (Supplementary Fig. 3). In contrast, none of the other 20 F-box proteins tested promoted a reduction in BCL6 levels or stability (Supplementary Fig. 4 and data not shown).
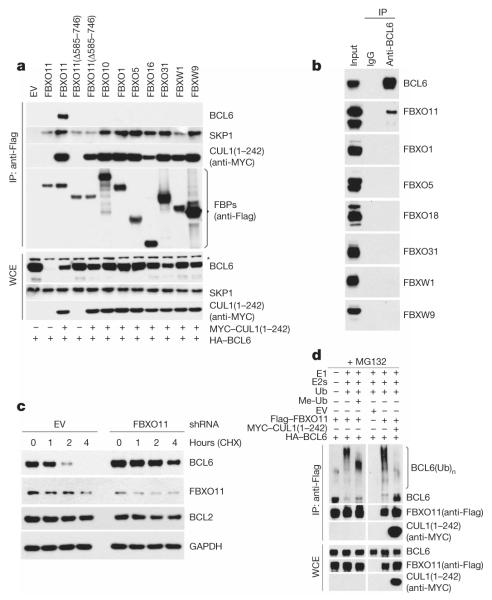
Figure 1. FBXO11 controls the ubiquitylation and degradation of BCL6.
a, HEK-293T cells were transfected with HA-tagged BCL6 and the indicated Flag-tagged F-box proteins (FBPs) or an empty vector (EV). Where indicated, MYC-tagged CUL1(1–242) was also transfected. Twenty-four hours after transfection, cells were harvested and lysed. Whole-cell extracts (WCE) were subjected to immunoprecipitation (IP) with anti-Flag resin and immunoblotting as indicated. The asterisk denotes a nonspecific band present in the anti-BCL6 blot. b, Ramos cells were treated with MG132 during the last 5 h before lysis. Lysates were immunoprecipitated with either an antibody against BCL6 or a nonspecific IgG and analysed by immunoblotting as indicated. c, SU-DHL6 cells were infected with viruses expressing two different FBXO11 shRNAs (in combination) or a control shRNA, selected, and treated with cycloheximide for the indicated times. Protein extracts were immunoblotted for the indicated proteins. d, HEK-293T cells were transfected with HA-tagged BCL6, Flag-tagged FBXO11, MYC-tagged CUL1(1–242), and/or an empty vector (EV) as indicated. After immunopurification with anti-Flag resin, in vitro ubiquitylation of BCL6 was performed in the presence or absence of E1, E2s and ubiquitin (Ub). Where indicated, methylated ubiquitin (Me-Ub) was added. Samples were analysed by immunoblotting with an anti-BCL6 antibody. The bracket on the right marks a ladder of bands >86 kDa corresponding to ubiquitylated BCL6. Immunoblots of whole-cell extracts are shown at the bottom.
To test further whether FBXO11 regulates the degradation of BCL6, we used two short hairpin RNA (shRNA) constructs to reduce FBXO11 expression in Ramos and SU-DHL6 cells. Depletion of FBXO11 by both shRNAs induced an increase in the steady-state levels and stability of BCL6 (Fig. 1c and Supplementary Fig. 5a, b). Similar effects were observed when the cellular expression of FBXO11 was silenced using three additional shRNAs or four different short interfering RNAs (siRNAs; Supplementary Fig. 5c–e).
We also observed that treatment of cells expressing FBXO11 with MG132 induced the appearance of high molecular mass species of BCL6 (Supplementary Fig. 6). These high molecular mass species are strongly induced by the expression of FBXO11 and are probably ubiquitylated forms of BCL6 as they became more evident in the presence of overexpressed ubiquitin. Moreover, we reconstituted the ubiquitylation of BCL6 in vitro. Immunopurified FBXO11 promoted the in vitro ubiquitylation of BCL6 only when the E1 and E2 enzymes were present in the reaction, and the ubiquitylation was inhibited by the presence of CUL1(1–242) (Fig. 1d). Methylated ubiquitin inhibited the formation of the highest molecular mass forms of BCL6, demonstrating that the high molecular mass forms of BCL6 are indeed polyubiquitylated. These results support the hypothesis that FBXO11 directly controls the ubiquitin-mediated degradation of BCL6.
Notably, FBXO11 silencing induced BCL6 stabilization in the absence of ligand-mediated receptor activation (Fig. 1c and Supplementary Fig. 5), which has been reported to accelerate BCL6 degradation by stimulating its ERK-dependent phosphorylation7. Accordingly, the binding of FBXO11 to BCL6 was unaffected in Ramos cells incubated with anti-IgM antibodies (a treatment that mimics B-cell antigen-receptor signalling and activates ERK2 (ref. 7)) both in the presence or absence of PD98059, a MEK inhibitor (Supplementary Fig. 7a). Similarly, when immunopurified BCL6 was phosphorylated in vitro by ERK2 or treated with λ-phosphatase, its binding to FBXO11 was unaffected (Supplementary Fig. 7b, c). Finally, a BCL6 mutant lacking the residues phosphorylated by ERK1/2 still bound FBXO11 efficiently (Supplementary Fig. 7d). Together, these results indicate that the FBXO11-dependent degradation of BCL6 is ERK-independent.
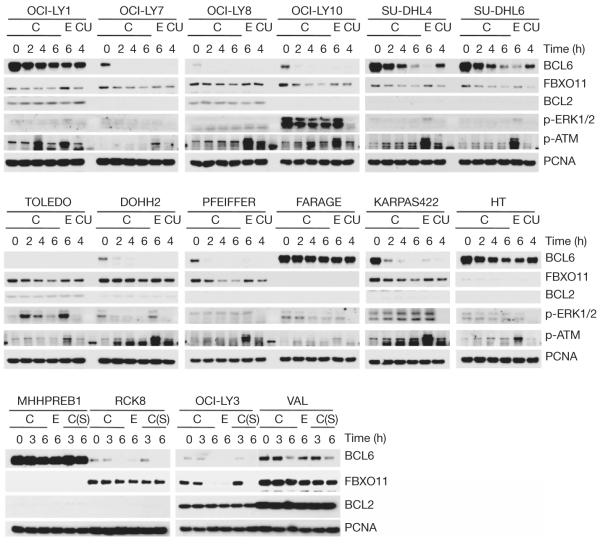
Together, the results in Fig. 1 and Supplementary Figs 1–6 demonstrate that FBXO11 mediates the ubiquitylation and degradation of BCL6. As the levels of BCL6 are elevated in B-cell malignancies and increased BCL6 expression has a crucial role in the pathogenesis of DLBCL, we determined whether the loss of FBXO11 might account for the elevated levels of BCL6. We examined the expression of FBXO11 and BCL6 in 22 B-cell lymphoma cell lines, including 16 DLBCL cell lines. We found that FBXO11 was absent or expressed at low levels in three DLBCL cell lines (FARAGE, MHHPREB1 and HT), and the expression was inversely correlated with BCL6 expression (Supplementary Fig. 8a). Using quantitative real-time polymerase chain reaction (qRT–PCR) on genomic DNA, we found that the FBXO11 gene was homozygously deleted in FARAGE and MHHPREB1 cells and hemizygously deleted in HT cells (Supplementary Fig. 8b, c). We then measured the stability of BCL6 in the 16 DLBCL cell lines by treating cells with either cycloheximide, which blocks protein synthesis, or etoposide, which enhances BCL6 degradation8. In some samples, UO126, a MEK inhibitor, was also added. First, we observed that treatment of cells with UO126 did not stabilize BCL6 (compare the 4-h time point with cycloheximide in the presence or absence of UO126 in Fig. 2), further supporting the conclusion that the FBXO11-mediated degradation of BCL6 is ERK-independent. Importantly, we found that in FARAGE, MHHPREB1 and HT cells, BCL6 was not only expressed at high levels, but it was also more stable than in other cell lines (Fig. 2). Notably, the OCI-LY1 cell line also showed increased BCL6 stability, and sequencing of the FBXO11 locus in OCI-LY1 cells revealed a point mutation in one of the CASH domains (Supplementary Fig. 9), the presumptive substrate recognition domains of FBXO11 (ref. 13).
Figure 2. DLBCL cell lines with FBXO11 deletions or an FBXO11 mutation display increased levels and stability of BCL6.
Sixteen DLBCL cell lines were incubated with either cycloheximide (C), etoposide (E), or cycloheximide and UO126 (CU) for the indicated hours before immunoblotting for the indicated proteins. Where indicated, the intra S-phase (S) checkpoint was activated by incubating cells with thymidine for 20 h prior to treatment with cycloheximide.
We then used high-density, single nucleotide polymorphism (HD-SNP) genotyping in a cohort of 27 DLBCL cell lines and confirmed the presence of FBXO11 deletions (including a focal deletion) in 14.8% of the samples (Supplementary Fig. 10). Public databases also indicate that the FBXO11 locus is deleted in B-cell malignancies. For example, the GSK cancer cell line genomic profiling database shows that deletions in the FBXO11 locus are present in six of 77 lymphoid cell lines, whereas only three of 255 non-lymphoid cell lines display FBXO11 deletions (Supplementary Fig. 11). (Of these six lines, three are DLBCLs and one is a Burkitt’s lymphoma.) Additionally, gene copy analyses in two different studies show FBXO11 deletions in 6 of 69 primary DLBCLs (Supplementary Fig. 12) (refs 14, 15).
To investigate FBXO11 gene inactivation in tumour samples further, we screened for mutations of FBXO11 in 100 primary DLBCL samples (Supplementary Table 1). After filtering for known polymorphisms and synonymous mutations, potential inactivating mutations were further analysed and confirmed. This strategy identified a total of six sequence changes in four tumours, of which two were in the same codon (Supplementary Fig. 9). None of these four samples displayed either BCL6 translocation or mutation of its promoter (Supplementary Table 2), and the analysis of paired normal DNA demonstrated the somatic origin of these events. Five of the six identified mutations are in the CASH domains, and one of these five changes creates a frame shift, resulting in chain termination and elimination of all three CASH domains. The fact that the point mutations are in highly conserved residues (Supplementary Fig. 9c) suggests that these FBXO11 mutants may be non-functional. Our analyses show that FBXO11 is deleted (in most cases hemizygously) in 14.8% of DLBCL cell lines (n = 27) and mutated in 14.3% of DLBCL cell lines (n = 7) and 4% of primary DLBCLs (n = 100). Moreover, data available in public databases report FBXO11 deletions in 8.7% of human DLBCLs (n = 69).
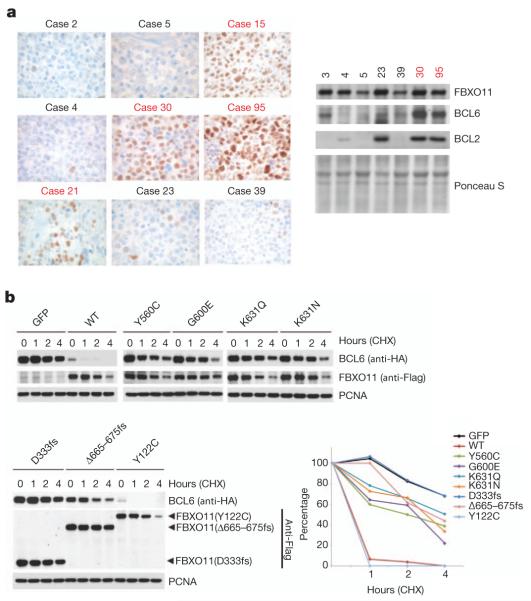
Notably, the DLBCL tumours with mutations in FBXO11 expressed much higher BCL6 levels than control samples, as detected by immunohistochemistry (Fig. 3a). High BCL6 protein levels were also confirmed using immunoblotting when sufficient frozen tissue was available (Fig. 3a).
Figure 3. Human DLBCLs with FBXO11 mutations display increased levels of BCL6, and FBXO11 tumour-derived mutants have impaired abilities to induce BCL6 degradation.
a, Immunohistochemical stains for BCL6 from eight DLBCLs, including four tumours with mutations in FBXO11, are shown (brown) (×100). The right panel shows lysates from human DLBCLs that were analysed by immunoblotting for the indicated proteins. The red colour denotes DLBCL cases with FBXO11 mutations. b, HEK-293T cells were transfected with BCL6 in combination with GFP, Flag-tagged wild type (WT) FBXO11, or the indicated Flag-tagged FBXO11 tumour-derived mutants. Twenty-four hours after transfection, cells were treated with cycloheximide (CHX) and samples were collected at the indicated time points for immunoblotting. The graph shows the quantification of BCL6 over the time course. The intensity of the bands from the experiment shown on the left was measured, and the ratio between the relative levels of BCL6 and PCNA in each time 0 was set as 100%.
To investigate whether the FBXO11 mutations in DLBCLs and OCI-LY1 cells interfere with FBXO11 activity, we generated complementary DNA constructs containing these mutations. Wild-type FBXO11 or FBXO11 mutants were expressed in HEK-293T cells to evaluate their ability to decrease BCL6 stability. Compared to wild-type FBXO11, tumour-derived FBXO11 mutants displayed an impaired ability to promote BCL6 degradation (Fig. 3b and Supplementary Fig. 13). Notably, FBXO11(Y122C), which has wild-type CASH domains, was the only mutant with activity similar to wild-type FBXO11. However, in the original sample this mutation was coupled with a deletion in FBXO11 that creates a frame shift and a chain termination (Δ666–675fs), which impaired the ability of FBXO11 to promote BCL6 degradation (Fig. 3b and Supplementary Fig. 13). We also found that mutations in FBXO11 modify its subcellular localization, preventing co-localization with BCL6 (Supplementary Fig. 14a, b). Moreover, FBXO11 mutants bind BCL6 less efficiently than wild-type FBXO11 (Supplementary Fig. 14c). These results indicate that the high levels of BCL6 observed in DLBCLs with FBXO11 mutations are due to a decreased ability of FBXO11 mutants to induce the proteolysis of BCL6.
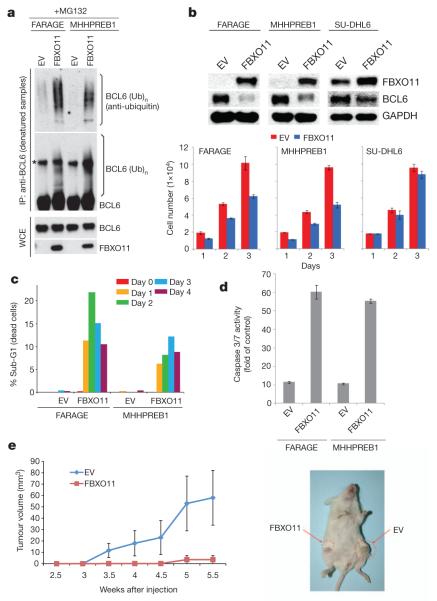
To study the role of FBXO11 in tumorigenesis further, we used doxycycline-inducible retroviral expression vectors to express FBXO11 in two cell lines (FARAGE and MHHPREB1) with biallelic FBXO11 deletion. BCL6 was poorly ubiquitylated in FARAGE and MHHPREB1 cells before FBXO11 reconstitution (Fig. 4a). Moreover, in parallel with a reduction in BCL6 levels, FBXO11 reconstitution inhibited cell proliferation in the FBXO11-null cell lines, but not a control line with an intact FBXO11 locus (Fig. 4b). FBXO11 expression also induced apoptosis, as determined by a significant increase in the percentage of the sub-G1 population and activation of caspases 3 and 7 (Fig. 4c, d). Finally, FARAGE cells transduced with FBXO11 or an empty virus were injected subcutaneously into NOD/SCID mice. Induction of FBXO11 expression with doxycycline inhibited tumour growth, as revealed by measuring tumour mass (Fig. 4e).
Figure 4. Expression of FBXO11 in FBXO11-null cells promotes BCL6 ubiquitylation and degradation, inhibits cell proliferation and induces apoptosis.
a, Denatured extracts from doxycycline- and MG132-treated FARAGE and MHHPREB1 cells stably transduced with a doxycycline-inducible FBXO11 construct or an empty virus (EV) were immunoprecipitated with an anti-BCL6 antibody and immunoblotted as indicated. The asterisk denotes a nonspecific band. Immunoblots of whole-cell extracts are also shown at the bottom. b, Extracts from doxycycline-treated FARAGE, MHHPREB1 and SU-DHL6 cells stably transduced with a doxycycline-inducible FBXO11 construct or an empty virus were immunoblotted as indicated. The graphs at the bottom show the number of cells measured in three different experiments (±s.d.) at different days after addition of doxycycline. c, Cells treated as in b for the indicated days were analysed by FACS. The graph represents the percentage of dying cells in the population (sub-G1 DNA content). d, Caspase 3 and Caspase 7 activity as measured in four different experiments (±s.d.) by the cleavage of a luminescent substrate in cells treated as in b for 2 days. The y axis indicates the caspase 3 and caspase 7 activity over cell number. The value given for the caspase activity in non-infected cells was set as 1. e, 1 × 107 FARAGE cells, stably transduced with a doxycycline-inducible FBXO11 construct (left flank) or an empty virus (right flank), were re-suspended in matrigel and injected subcutaneously into NOD/SCID mice. Two days after injection, doxycycline was administered in drinking water. Tumour growth was measured using a caliper at the indicated times after injection. n = 4 for each group; P < 0.05. Error bars indicate standard error of the mean (s.e.m.). The image on the right shows a representative mouse injected with the indicated cells.
Our results show that SCFFBXO11 is the ubiquitin ligase for BCL6 and that deregulation of this interaction may represent an oncogenic event. FBXO11 loss or mutation may contribute to DLBCL pathogenesis by allowing the accumulation of BCL6, an oncoprotein that has a critical role in lymphomagenesis. Because the deletions and mutations observed in our DLBCL samples were typically mono-allelic, we propose that FBXO11 is the product of a haplo-insufficient tumour suppressor gene.
METHODS SUMMARY
Biochemical methods
Extract preparation, immunoprecipitation and immunoblotting have previously been described16. Whole-cell lysates were generated by lysing cells in SDS lysis buffer (50 mM Tris pH 6.8, 2% SDS, 10% glycerol, 1 mM Na3VO4, 1 mM NaF and protease inhibitor mix (Roche)). In vitro ubiquitylation assays were preformed as described16,17. Briefly, haemagglutinin (HA)-tagged BCL6 was transfected into HEK-293T cells together with either Flag-tagged FBXO11 or an empty vector. Where indicated, MYC-tagged CUL1(1–242) was also transfected. Twenty-four hours after transfection, cells were incubated with MG132 for three hours before harvesting. Anti-Flag M2 agarose beads were used to immunoprecipitate the SCFFBXO11 complex. The beads were washed four times in lysis buffer and twice in ubiquitylation reaction buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 0.5 mM dithiothreitol). Beads were then used for in vitro ubiquitylation assays in a volume of 30 μl, containing 0.1 μM E1 (Boston Biochem), 0.25 μM Ubch3, 0.25 μM Ubch5c, 1 μM ubiquitin aldehyde, 2.5 μg μl−1 ubiquitin, and 1× magnesium/ATP cocktail. Samples were incubated for 2 h at 30 °C and analysed by immunoblotting. In vivo ubiquitylation assays were preformed as described18.
Mutation analysis
The complete coding sequences and exon/intron junctions of FBXO11 were analysed by PCR amplification and sequencing. The reference sequences for all annotated exons and flanking introns of FBXO11 were obtained from the UCSC Human Genome database using the mRNA accession NM_025133.4. PCR primers (see Supplementary Table 3), located ≥50 bp upstream or downstream of the target exon boundaries, were designed in the Primer 3 program (http://frodo.wi.mit.edu/primer3/). Sequences were compared to the corresponding germline sequences using the Mutation Surveyor Version 2.41 software package (Softgenetics; http://www.softgenetics.com). Synonymous mutations, previously reported polymorphisms (Human dbSNP Database at NCBI, build 130, and Ensembl Database) and changes present in the matched normal DNA were excluded.
Supplementary Material
Acknowledgements
We thank W. Carroll, S. Shaham and L. Staudt for sharing unpublished results; A. Melnick, Y. Sun and Y. Xiong for reagents; L. Cerchietti, Y. Cheng, E. Dehan, V. Donato, N. V. Dorello, S. N. Yang and Z. Yao for advice and/or contributions to this work. M.P. is grateful to T. M. Thor and K. E. Davidson, and L.C. to Zuzana S. for continuous support. This work was supported by grants from the National Institutes of Health to M.P. (R01-GM57587, R37-CA76584 and R21-CA161108) and M.S. (PO1-CA092625), a grant from Susan G. Komen for the Cure to S.D., a Lymphoma Research Foundation Fellowship to J.K.P. and grants from AIRC and ERC (ERC-2009-StG-Proposal No242965-LUNELY) to R.C. M.P. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions S.D., L.C. and J.K.P. planned and performed most experiments and helped to write the manuscript. M.P. coordinated the study, oversaw the results and wrote the manuscript. C.M., P.F.d.C. and R.C. provided the DLBCL tumour samples and some DLBCL cell lines, and performed some experiments. M.R. generated several constructs. B.C. and M.S. provided the HD-SNP data in Supplementary Fig. 10. All authors discussed the results and commented on the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
The authors declare no competing financial interests.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Ci W, Polo JM, Melnick A. B-cell lymphoma 6 and the molecular pathogenesis of diffuse large B-cell lymphoma. Curr. Opin. Hematol. 2008;15:381–390. doi: 10.1097/MOH.0b013e328302c7df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staudt LM, Dave S. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv. Immunol. 2005;87:163–208. doi: 10.1016/S0065-2776(05)87005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron BW, et al. The human BCL6 transgene promotes the development of lymphomas in the mouse. Proc. Natl Acad. Sci. USA. 2004;101:14198–14203. doi: 10.1073/pnas.0406138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cattoretti G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 5.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nature Rev. Mol. Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 6.Skaar JR, D’Angiolella V, Pagan JK, Pagano M. SnapShot: F Box Proteins II. Cell. 2009;137:1358. doi: 10.1016/j.cell.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 7.Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12:1953–1961. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phan RT, Saito M, Kitagawa Y, Means AR, Dalla-Favera R. Genotoxic stress regulates expression of the proto-oncogene Bcl6 in germinal center B cells. Nature Immunol. 2007;8:1132–1139. doi: 10.1038/ni1508. [DOI] [PubMed] [Google Scholar]
- 9.Benmaamar R, Pagano M. Involvement of the SCF complex in the control of Cdh1 degradation in S-phase. Cell Cycle. 2005;4:1230–1232. doi: 10.4161/cc.4.9.2048. [DOI] [PubMed] [Google Scholar]
- 10.Piva R, et al. In vivo interference with Skp1 function leads to genetic instability and neoplastic transformation. Mol. Cell. Biol. 2002;22:8375–8387. doi: 10.1128/MCB.22.23.8375-8387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- 12.Skaar JR, Pagano M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr. Opin. Cell Biol. 2009;21:816–824. doi: 10.1016/j.ceb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin J, et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato M, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 16.D’Angiolella V, et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan S, et al. mTOR generates an auto-amplification loop by triggering the βTrCP- and CK1α-dependent degradation of DEPTOR. Mol. Cell. 2011;44:317–324. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell. 2003;115:71–82. doi: 10.1016/s0092-8674(03)00755-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.