Abstract
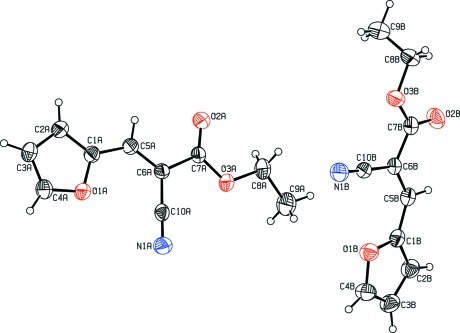
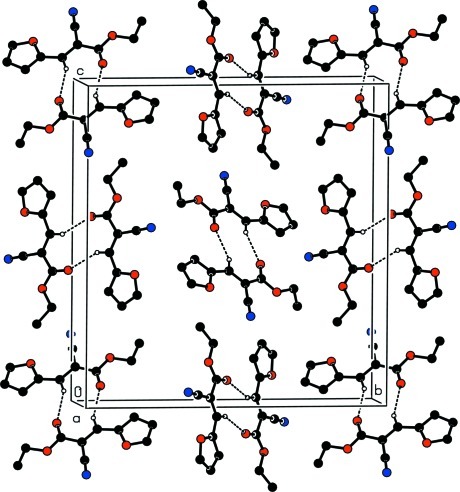
There are two independent molecules in the asymmetric unit of the title compound, C10H9NO3, in both of which, all non-H atoms except for the methyl C atom lie nearly in the same plane [maximum deviations = 0.094 (3) and 0.043 (2) Å]. In the crystal, each independent molecules is linked by pairs of C—H⋯O interactions, generating inversion dimers with R 2 2(10) ring motifs.
Related literature
For the synthesis of related compounds, see: Yadav et al. (2004 ▶). For related structures, see: Wang & Jian (2008 ▶); Zhang et al. (2009 ▶); Ye et al. (2009 ▶); Yuvaraj et al. (2011 ▶).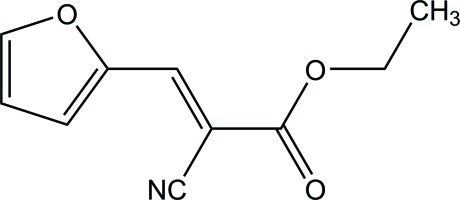
Experimental
Crystal data
C10H9NO3
M r = 191.18
Monoclinic,
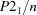
a = 4.6611 (2) Å
b = 19.8907 (9) Å
c = 20.9081 (9) Å
β = 91.988 (4)°
V = 1937.28 (15) Å3
Z = 8
Mo Kα radiation
μ = 0.10 mm−1
T = 293 K
0.30 × 0.20 × 0.10 mm
Data collection
Oxford Diffraction Xcalibur Sapphire3 diffractometer
Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2010 ▶) T min = 0.933, T max = 0.990
12870 measured reflections
4568 independent reflections
2407 reflections with I > 2σ(I)
R int = 0.034
Refinement
R[F 2 > 2σ(F 2)] = 0.066
wR(F 2) = 0.209
S = 1.04
4568 reflections
253 parameters
H-atom parameters constrained
Δρmax = 0.26 e Å−3
Δρmin = −0.24 e Å−3
Data collection: CrysAlis PRO (Oxford Diffraction, 2010 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis RED (Oxford Diffraction, 2010 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: PLATON.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812016510/is5119sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812016510/is5119Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812016510/is5119Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C5A—H5A⋯O2Ai | 0.93 | 2.40 | 3.242 (3) | 151 |
| C5B—H5B⋯O2Bii | 0.93 | 2.46 | 3.320 (3) | 153 |
Symmetry codes: (i) 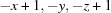 ; (ii)
; (ii) 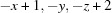 .
.
Acknowledgments
YTJ is thankful for the support provided by the second stage of the BK-21 program. The authors thank the Director, USIC University of Jammu, Jammu Tawi, India, for the X-ray data collection.
supplementary crystallographic information
Comment
Knoevenagel condensation is an important carbon-carbon bond forming reaction in organic synthesis (Yadav et al., 2004). In continuation of our work on nitrogen and oxygen based heterocycles, we herein report the crystal structure of the title compound.
The title compound crystallizes in monoclinic system with two molecules in the asymmetric unit. Bond lengths and bond angles are comparable with the similar crystal structures solved earlier (Zhang et al., 2009; Wang & Jian, 2008; Ye et al., 2009; Yuvaraj et al., 2011). All the non-hydrogen atoms, except the methyl group, lie nearly in the same plane with a maximum out-of-plane deviation of 0.094 (3) and 0.043 (2) Å (r.m.s deviation = 0.04 and 0.024 Å), respectively, for molecules A and B. Difference in the torsion angles C7A—O3A—C8A—C9A [-167.4 (3)°] and C7B—O3B—C8B—C9B [125.3 (4)°] has been observed, indicating the flexibility of the methyl group. The crystal packing is stabilized by C—H···O intermolecular interactions generating the centrosymmetric dimer of R22(10) ring motif.
Experimental
A solution of furan-2-aldehyde (1 mol), ethyl cyanoacetate (1.2 mol) and piperidine (0.1 ml) in ethanol (20 ml) was stirred at room temperature for 8 h. After removal of the volatiles in vacuo, orange solid was obtained in quantitative yield. A sample for analysis was obtained by recrystallization from EtOAc as pale yellow needles: 1H NMR (300 MHz, CDCl3) δ p.p.m.: 1.42 (t, 3H, CH3), 4.40 (q, 2H, CH2), 6.61 (m, 1H, CH), 6.80 (m, 1H, CH), 7.28 (m, 1H, CH), 7.98 (s, 1H, HC=C).
Refinement
All H-atoms were refined using a riding model [C—H = 0.93 Å and Uiso(H) = 1.2Ueq(C) for aromatic, C—H = 0.97 Å and Uiso = 1.2Ueq (C) for CH2, and C—H = 0.96 Å and Uiso = 1.5Ueq(C) for CH3].
Figures
Fig. 1.
The molecular structure of title compound, showing 30% probability displacement ellipsoids.
Fig. 2.
A molecular packing view of the title compound, showing intermolecular interactions. For clarity, hydrogen atoms not involved in the hydrogen bonding have been omitted.
Crystal data
| C10H9NO3 | F(000) = 800 |
| Mr = 191.18 | Dx = 1.311 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 4295 reflections |
| a = 4.6611 (2) Å | θ = 3.6–29.1° |
| b = 19.8907 (9) Å | µ = 0.10 mm−1 |
| c = 20.9081 (9) Å | T = 293 K |
| β = 91.988 (4)° | Needle, pale yellow |
| V = 1937.28 (15) Å3 | 0.30 × 0.20 × 0.10 mm |
| Z = 8 |
Data collection
| Oxford Diffraction Xcalibur Sapphire3 diffractometer | 4568 independent reflections |
| Radiation source: fine-focus sealed tube | 2407 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.034 |
| Detector resolution: 16.1049 pixels mm-1 | θmax = 29.2°, θmin = 3.6° |
| ω scans | h = −6→6 |
| Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2010) | k = −26→24 |
| Tmin = 0.933, Tmax = 0.990 | l = −27→27 |
| 12870 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.066 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.209 | H-atom parameters constrained |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0908P)2 + 0.185P] where P = (Fo2 + 2Fc2)/3 |
| 4568 reflections | (Δ/σ)max < 0.001 |
| 253 parameters | Δρmax = 0.26 e Å−3 |
| 0 restraints | Δρmin = −0.24 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1A | 1.1671 (4) | 0.14995 (9) | 0.41737 (9) | 0.0642 (5) | |
| O2A | 0.5561 (4) | 0.03506 (10) | 0.58448 (9) | 0.0724 (6) | |
| O3A | 0.8013 (4) | 0.10818 (9) | 0.64695 (8) | 0.0617 (5) | |
| N1A | 1.2730 (5) | 0.19852 (12) | 0.56530 (12) | 0.0695 (7) | |
| C1A | 0.9635 (5) | 0.10093 (13) | 0.41935 (12) | 0.0524 (6) | |
| C2A | 0.9058 (6) | 0.07717 (15) | 0.35931 (13) | 0.0658 (8) | |
| H2A | 0.7747 | 0.0437 | 0.3478 | 0.079* | |
| C3A | 1.0809 (7) | 0.11270 (15) | 0.31803 (14) | 0.0709 (8) | |
| H3A | 1.0900 | 0.1074 | 0.2740 | 0.085* | |
| C4A | 1.2328 (7) | 0.15591 (16) | 0.35497 (14) | 0.0747 (9) | |
| H4A | 1.3663 | 0.1861 | 0.3397 | 0.090* | |
| C5A | 0.8457 (5) | 0.08207 (13) | 0.47874 (12) | 0.0520 (6) | |
| H5A | 0.7104 | 0.0477 | 0.4761 | 0.062* | |
| C6A | 0.9001 (5) | 0.10636 (12) | 0.53795 (11) | 0.0468 (6) | |
| C7A | 0.7339 (6) | 0.07857 (13) | 0.59153 (12) | 0.0516 (6) | |
| C8A | 0.6382 (7) | 0.08504 (17) | 0.70119 (14) | 0.0781 (9) | |
| H8A1 | 0.4352 | 0.0835 | 0.6894 | 0.094* | |
| H8A2 | 0.6995 | 0.0402 | 0.7137 | 0.094* | |
| C9A | 0.6886 (10) | 0.13165 (19) | 0.75412 (17) | 0.1063 (13) | |
| H9A1 | 0.5820 | 0.1173 | 0.7902 | 0.159* | |
| H9A2 | 0.6274 | 0.1759 | 0.7413 | 0.159* | |
| H9A3 | 0.8896 | 0.1324 | 0.7658 | 0.159* | |
| C10A | 1.1070 (6) | 0.15760 (13) | 0.55250 (12) | 0.0511 (6) | |
| O1B | 1.1874 (4) | 0.12570 (9) | 0.88858 (9) | 0.0650 (5) | |
| O2B | 0.5452 (4) | −0.07433 (10) | 0.94337 (10) | 0.0763 (6) | |
| O3B | 0.7859 (4) | −0.11775 (9) | 0.86263 (10) | 0.0737 (6) | |
| N1B | 1.2343 (6) | −0.00911 (12) | 0.79884 (12) | 0.0752 (8) | |
| C1B | 0.9833 (5) | 0.11024 (13) | 0.93153 (12) | 0.0526 (6) | |
| C2B | 0.9368 (6) | 0.16376 (14) | 0.96945 (14) | 0.0654 (8) | |
| H2B | 0.8081 | 0.1659 | 1.0024 | 0.078* | |
| C3B | 1.1182 (7) | 0.21538 (15) | 0.95001 (15) | 0.0717 (8) | |
| H3B | 1.1340 | 0.2584 | 0.9672 | 0.086* | |
| C4B | 1.2645 (7) | 0.19017 (15) | 0.90153 (17) | 0.0732 (8) | |
| H4B | 1.4018 | 0.2139 | 0.8795 | 0.088* | |
| C5B | 0.8538 (5) | 0.04531 (13) | 0.93108 (12) | 0.0545 (7) | |
| H5B | 0.7222 | 0.0384 | 0.9629 | 0.065* | |
| C6B | 0.8908 (5) | −0.00742 (13) | 0.89236 (11) | 0.0492 (6) | |
| C7B | 0.7208 (6) | −0.06891 (14) | 0.90287 (13) | 0.0571 (7) | |
| C8B | 0.6333 (9) | −0.18142 (18) | 0.86891 (18) | 0.0999 (12) | |
| H8B1 | 0.7679 | −0.2160 | 0.8829 | 0.120* | |
| H8B2 | 0.4903 | −0.1769 | 0.9013 | 0.120* | |
| C9B | 0.4999 (11) | −0.2007 (2) | 0.8116 (2) | 0.1386 (19) | |
| H9B1 | 0.4030 | −0.2428 | 0.8173 | 0.208* | |
| H9B2 | 0.6412 | −0.2057 | 0.7796 | 0.208* | |
| H9B3 | 0.3631 | −0.1671 | 0.7981 | 0.208* | |
| C10B | 1.0847 (6) | −0.00751 (13) | 0.84084 (12) | 0.0541 (7) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1A | 0.0780 (12) | 0.0690 (13) | 0.0461 (11) | −0.0191 (10) | 0.0102 (9) | −0.0059 (9) |
| O2A | 0.0919 (14) | 0.0708 (13) | 0.0550 (12) | −0.0275 (11) | 0.0091 (10) | 0.0000 (10) |
| O3A | 0.0759 (12) | 0.0698 (12) | 0.0399 (10) | −0.0140 (10) | 0.0093 (8) | −0.0011 (8) |
| N1A | 0.0813 (16) | 0.0642 (16) | 0.0636 (16) | −0.0126 (14) | 0.0113 (13) | −0.0078 (12) |
| C1A | 0.0597 (15) | 0.0511 (15) | 0.0467 (15) | 0.0011 (12) | 0.0061 (11) | −0.0023 (11) |
| C2A | 0.0812 (19) | 0.0655 (18) | 0.0510 (16) | −0.0147 (15) | 0.0053 (14) | −0.0076 (13) |
| C3A | 0.091 (2) | 0.079 (2) | 0.0434 (16) | −0.0073 (17) | 0.0117 (15) | −0.0046 (14) |
| C4A | 0.095 (2) | 0.081 (2) | 0.0499 (17) | −0.0172 (17) | 0.0225 (16) | −0.0014 (15) |
| C5A | 0.0601 (15) | 0.0480 (15) | 0.0480 (15) | −0.0011 (12) | 0.0049 (11) | −0.0014 (11) |
| C6A | 0.0537 (14) | 0.0419 (14) | 0.0450 (14) | 0.0002 (11) | 0.0042 (11) | −0.0007 (10) |
| C7A | 0.0632 (16) | 0.0495 (15) | 0.0421 (14) | −0.0021 (13) | 0.0044 (11) | 0.0029 (11) |
| C8A | 0.098 (2) | 0.091 (2) | 0.0464 (17) | −0.0161 (18) | 0.0209 (15) | 0.0028 (15) |
| C9A | 0.150 (4) | 0.111 (3) | 0.060 (2) | −0.021 (3) | 0.036 (2) | −0.0106 (19) |
| C10A | 0.0634 (15) | 0.0515 (16) | 0.0390 (13) | 0.0055 (13) | 0.0098 (11) | 0.0003 (11) |
| O1B | 0.0693 (12) | 0.0633 (13) | 0.0633 (13) | −0.0045 (10) | 0.0132 (9) | −0.0107 (9) |
| O2B | 0.0828 (14) | 0.0810 (14) | 0.0665 (14) | −0.0095 (11) | 0.0257 (11) | 0.0105 (11) |
| O3B | 0.1015 (15) | 0.0592 (12) | 0.0617 (13) | −0.0246 (11) | 0.0199 (11) | −0.0061 (10) |
| N1B | 0.0915 (18) | 0.0755 (18) | 0.0601 (16) | −0.0159 (14) | 0.0255 (14) | −0.0079 (13) |
| C1B | 0.0568 (15) | 0.0572 (17) | 0.0438 (14) | 0.0064 (13) | 0.0024 (11) | −0.0014 (11) |
| C2B | 0.0736 (18) | 0.0630 (19) | 0.0596 (18) | 0.0083 (15) | 0.0041 (14) | −0.0094 (14) |
| C3B | 0.085 (2) | 0.0586 (18) | 0.071 (2) | 0.0061 (16) | −0.0049 (16) | −0.0118 (15) |
| C4B | 0.0759 (19) | 0.0613 (19) | 0.083 (2) | −0.0098 (16) | 0.0064 (16) | −0.0059 (16) |
| C5B | 0.0578 (15) | 0.0630 (18) | 0.0429 (15) | 0.0028 (13) | 0.0027 (11) | 0.0000 (12) |
| C6B | 0.0553 (14) | 0.0531 (15) | 0.0392 (13) | −0.0010 (12) | 0.0028 (10) | 0.0030 (11) |
| C7B | 0.0659 (17) | 0.0623 (17) | 0.0433 (15) | −0.0005 (14) | 0.0016 (12) | 0.0049 (13) |
| C8B | 0.151 (3) | 0.075 (2) | 0.074 (2) | −0.051 (2) | 0.011 (2) | 0.0059 (18) |
| C9B | 0.220 (5) | 0.098 (3) | 0.096 (3) | −0.083 (3) | −0.014 (3) | 0.009 (2) |
| C10B | 0.0702 (17) | 0.0485 (15) | 0.0438 (15) | −0.0059 (12) | 0.0039 (12) | −0.0017 (11) |
Geometric parameters (Å, º)
| O1A—C4A | 1.356 (3) | O1B—C4B | 1.357 (3) |
| O1A—C1A | 1.362 (3) | O1B—C1B | 1.366 (3) |
| O2A—C7A | 1.204 (3) | O2B—C7B | 1.203 (3) |
| O3A—C7A | 1.328 (3) | O3B—C7B | 1.327 (3) |
| O3A—C8A | 1.461 (3) | O3B—C8B | 1.461 (4) |
| N1A—C10A | 1.148 (3) | N1B—C10B | 1.140 (3) |
| C1A—C2A | 1.360 (3) | C1B—C2B | 1.349 (4) |
| C1A—C5A | 1.425 (3) | C1B—C5B | 1.425 (4) |
| C2A—C3A | 1.400 (4) | C2B—C3B | 1.399 (4) |
| C2A—H2A | 0.9300 | C2B—H2B | 0.9300 |
| C3A—C4A | 1.341 (4) | C3B—C4B | 1.338 (4) |
| C3A—H3A | 0.9300 | C3B—H3B | 0.9300 |
| C4A—H4A | 0.9300 | C4B—H4B | 0.9300 |
| C5A—C6A | 1.345 (3) | C5B—C6B | 1.340 (3) |
| C5A—H5A | 0.9300 | C5B—H5B | 0.9300 |
| C6A—C10A | 1.429 (4) | C6B—C10B | 1.430 (3) |
| C6A—C7A | 1.490 (3) | C6B—C7B | 1.478 (4) |
| C8A—C9A | 1.457 (4) | C8B—C9B | 1.385 (5) |
| C8A—H8A1 | 0.9700 | C8B—H8B1 | 0.9700 |
| C8A—H8A2 | 0.9700 | C8B—H8B2 | 0.9700 |
| C9A—H9A1 | 0.9600 | C9B—H9B1 | 0.9600 |
| C9A—H9A2 | 0.9600 | C9B—H9B2 | 0.9600 |
| C9A—H9A3 | 0.9600 | C9B—H9B3 | 0.9600 |
| C4A—O1A—C1A | 105.8 (2) | C4B—O1B—C1B | 105.5 (2) |
| C7A—O3A—C8A | 115.1 (2) | C7B—O3B—C8B | 117.1 (2) |
| C2A—C1A—O1A | 109.7 (2) | C2B—C1B—O1B | 109.8 (2) |
| C2A—C1A—C5A | 130.0 (3) | C2B—C1B—C5B | 130.0 (3) |
| O1A—C1A—C5A | 120.3 (2) | O1B—C1B—C5B | 120.3 (2) |
| C1A—C2A—C3A | 107.0 (3) | C1B—C2B—C3B | 107.3 (3) |
| C1A—C2A—H2A | 126.5 | C1B—C2B—H2B | 126.4 |
| C3A—C2A—H2A | 126.5 | C3B—C2B—H2B | 126.4 |
| C4A—C3A—C2A | 106.0 (3) | C4B—C3B—C2B | 105.9 (3) |
| C4A—C3A—H3A | 127.0 | C4B—C3B—H3B | 127.1 |
| C2A—C3A—H3A | 127.0 | C2B—C3B—H3B | 127.1 |
| C3A—C4A—O1A | 111.5 (3) | C3B—C4B—O1B | 111.5 (3) |
| C3A—C4A—H4A | 124.3 | C3B—C4B—H4B | 124.2 |
| O1A—C4A—H4A | 124.3 | O1B—C4B—H4B | 124.2 |
| C6A—C5A—C1A | 129.9 (2) | C6B—C5B—C1B | 130.5 (2) |
| C6A—C5A—H5A | 115.1 | C6B—C5B—H5B | 114.7 |
| C1A—C5A—H5A | 115.1 | C1B—C5B—H5B | 114.7 |
| C5A—C6A—C10A | 123.8 (2) | C5B—C6B—C10B | 123.7 (2) |
| C5A—C6A—C7A | 118.2 (2) | C5B—C6B—C7B | 118.5 (2) |
| C10A—C6A—C7A | 118.0 (2) | C10B—C6B—C7B | 117.9 (2) |
| O2A—C7A—O3A | 124.5 (2) | O2B—C7B—O3B | 123.8 (3) |
| O2A—C7A—C6A | 123.2 (2) | O2B—C7B—C6B | 124.1 (3) |
| O3A—C7A—C6A | 112.2 (2) | O3B—C7B—C6B | 112.1 (2) |
| C9A—C8A—O3A | 108.3 (3) | C9B—C8B—O3B | 111.6 (3) |
| C9A—C8A—H8A1 | 110.0 | C9B—C8B—H8B1 | 109.3 |
| O3A—C8A—H8A1 | 110.0 | O3B—C8B—H8B1 | 109.3 |
| C9A—C8A—H8A2 | 110.0 | C9B—C8B—H8B2 | 109.3 |
| O3A—C8A—H8A2 | 110.0 | O3B—C8B—H8B2 | 109.3 |
| H8A1—C8A—H8A2 | 108.4 | H8B1—C8B—H8B2 | 108.0 |
| C8A—C9A—H9A1 | 109.5 | C8B—C9B—H9B1 | 109.5 |
| C8A—C9A—H9A2 | 109.5 | C8B—C9B—H9B2 | 109.5 |
| H9A1—C9A—H9A2 | 109.5 | H9B1—C9B—H9B2 | 109.5 |
| C8A—C9A—H9A3 | 109.5 | C8B—C9B—H9B3 | 109.5 |
| H9A1—C9A—H9A3 | 109.5 | H9B1—C9B—H9B3 | 109.5 |
| H9A2—C9A—H9A3 | 109.5 | H9B2—C9B—H9B3 | 109.5 |
| N1A—C10A—C6A | 178.8 (3) | N1B—C10B—C6B | 177.9 (3) |
| C4A—O1A—C1A—C2A | 0.1 (3) | C4B—O1B—C1B—C2B | 0.1 (3) |
| C4A—O1A—C1A—C5A | −179.9 (2) | C4B—O1B—C1B—C5B | 179.8 (2) |
| O1A—C1A—C2A—C3A | 0.1 (3) | O1B—C1B—C2B—C3B | 0.0 (3) |
| C5A—C1A—C2A—C3A | −179.9 (3) | C5B—C1B—C2B—C3B | −179.7 (3) |
| C1A—C2A—C3A—C4A | −0.3 (3) | C1B—C2B—C3B—C4B | −0.1 (3) |
| C2A—C3A—C4A—O1A | 0.4 (4) | C2B—C3B—C4B—O1B | 0.2 (4) |
| C1A—O1A—C4A—C3A | −0.3 (4) | C1B—O1B—C4B—C3B | −0.2 (3) |
| C2A—C1A—C5A—C6A | −178.9 (3) | C2B—C1B—C5B—C6B | 177.4 (3) |
| O1A—C1A—C5A—C6A | 1.1 (4) | O1B—C1B—C5B—C6B | −2.2 (4) |
| C1A—C5A—C6A—C10A | −2.1 (4) | C1B—C5B—C6B—C10B | 0.5 (4) |
| C1A—C5A—C6A—C7A | 177.5 (2) | C1B—C5B—C6B—C7B | −179.0 (2) |
| C8A—O3A—C7A—O2A | −1.0 (4) | C8B—O3B—C7B—O2B | −0.2 (4) |
| C8A—O3A—C7A—C6A | 177.8 (2) | C8B—O3B—C7B—C6B | 179.4 (3) |
| C5A—C6A—C7A—O2A | 0.8 (4) | C5B—C6B—C7B—O2B | 1.7 (4) |
| C10A—C6A—C7A—O2A | −179.5 (2) | C10B—C6B—C7B—O2B | −177.8 (3) |
| C5A—C6A—C7A—O3A | −178.0 (2) | C5B—C6B—C7B—O3B | −177.9 (2) |
| C10A—C6A—C7A—O3A | 1.6 (3) | C10B—C6B—C7B—O3B | 2.6 (3) |
| C7A—O3A—C8A—C9A | −167.4 (3) | C7B—O3B—C8B—C9B | 125.3 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5A—H5A···O2Ai | 0.93 | 2.40 | 3.242 (3) | 151 |
| C5B—H5B···O2Bii | 0.93 | 2.46 | 3.320 (3) | 153 |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) −x+1, −y, −z+2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS5119).
References
- Oxford Diffraction (2010). CrysAlis PRO and CrysAlis RED Oxford Diffraction Ltd, Yarnton, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Wang, J.-G. & Jian, F.-F. (2008). Acta Cryst. E64, o2145. [DOI] [PMC free article] [PubMed]
- Yadav, J. S., Subba Reddy, B. V., Basak, A. K., Visali, B., Narsaiah, A. V. & Nagaiah, K. (2004). Eur. J. Org. Chem. pp. 546–551.
- Ye, Y., Shen, W.-L. & Wei, X.-W. (2009). Acta Cryst. E65, o2636. [DOI] [PMC free article] [PubMed]
- Yuvaraj, H., Gayathri, D., Kalkhambkar, R. G., Gupta, V. K. & Rajnikant, (2011). Acta Cryst. E67, o2135. [DOI] [PMC free article] [PubMed]
- Zhang, S.-J., Zheng, X.-M. & Hu, W.-X. (2009). Acta Cryst. E65, o2351. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812016510/is5119sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812016510/is5119Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812016510/is5119Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report