Abstract
Images of multiply-labeled fluorescent samples provide unique insights into the localization of molecules, cells, and tissues. The ability to image multiple channels simultaneously at high speed without cross talk is limited to a few colors and requires dedicated multi-channel or multi-spectral detection procedures. Simpler microscopes, in which each color is imaged sequentially, produce a much lower frame rate. Here, we describe a technique to image, at high frame rate, multiply-labeled samples that have a repeating motion. We capture images in a single channel at a time over one full occurrence of the motion then repeat acquisition for other channels over subsequent occurrences. We finally build a high-speed multi-channel image sequence by combining the images after applying a normalized mutual information-based time registration procedure.
We show that this technique is amenable to image the beating heart of a double-labeled embryonic quail in three channels (brightfield, yellow and mCherry fluorescent proteins) using a fluorescence wide-field microscope equipped with a single monochrome camera and without fast channel switching optics. We experimentally evaluate the accuracy of our method on image series from a two-channel confocal microscope.
Keywords: Cardiac imaging, quail, fluorescence imaging, multi-color imaging, heart development, registration, fast imaging, mutual information
1 Introduction
There are multiple modalities available for imaging biological samples, each mapping local optical or biochemical properties onto images of variable contrasts. Modalites such as brightfield, phase-contrast, or darkfield microscopy can reveal intrinsic optical properties of organelles, cells, or tissues, including local indices of absorption, refraction, or scattering. Vital fluorescent dyes, probes, and fluorescent proteins can be used to specifically label proteins, organelles, cells, and tissues or to report about cell activity. Bright fluorescent molecules have been developed over the entire visible spectrum and it is increasingly common to find samples that carry multiple fluorescent labels of different colors.
Many microscopes allow imaging samples in more than one modality or color. Switching from imaging in one modality to another or from imaging one fluorophore to another is done by inserting additional optical elements in the light path to adjust illumination and light collection.
Imaging fluorescent samples is achieved by illuminating the fluorophores to excite them, then collect the emitted fluorescence signal while rejecting illumination light.
Several strategies exist to image samples that carry fluorescent molecules of different colors. We can broadly categorize them into two classes depending on whether all channels are measured synchronously or sequentially.
Synchronous imaging requires that all fluorophore be excited simultaneously (using one or multiple laser lines or a multi-band excitation filter). Fluorescence emission from each fluorescent species can then be directed to multiple detectors (cameras or point detectors, in the case of scanning microscopes (Garini et al., 2006; Wolleschensky et al., 2006)) after successively selecting appropriate spectral bands, for example by applying a cascade of two or more dichroic mirrors and emission filters. Alternatively, several approaches to direct the emitted light to multi-spectral detectors have been proposed for scanning microscopes (Dickinson et al., 2001).
During sequential imaging, excitation and collection from only one fluorophore is desired. Illumination is limited to a single spectral band or laser line and the collection optics, typically dichroics and emission filters, are adjusted to collect fluorescence emission from the fluorephore of interest.
Both approaches have advantages and disadvantages. Synchronous imaging requires more sophisticated setups that can accommodate multiple illumination sources, requires filter sets that can selectively illuminate in multiple distinct light bands, and, for collection, requires that light from the different fluorophores is separated and routed to multiple detectors or cameras. The cost of setting up a system with multiple cameras can be particularly prohibitive. It is difficult to avoid cross-talk when imaging all fluorophores simultaneously since fluorophores have broad (and often overlapping) excitation and emission bands. A fluorescent species may get excited by light dedicated to another species and fluorescence light from one species can reach a detector dedicated to another species (spectral unmixing post processing can, in some cases, alleviate that problem (Dickinson et al., 2001)). Finally, cameras must be carefully aligned to produce co-registered images of the sample, with magnification and focus matching at each wavelength. The main advantage of synchronous imaging is that it is fast: all channels are acquired simultaneously (Fig 1a and b).
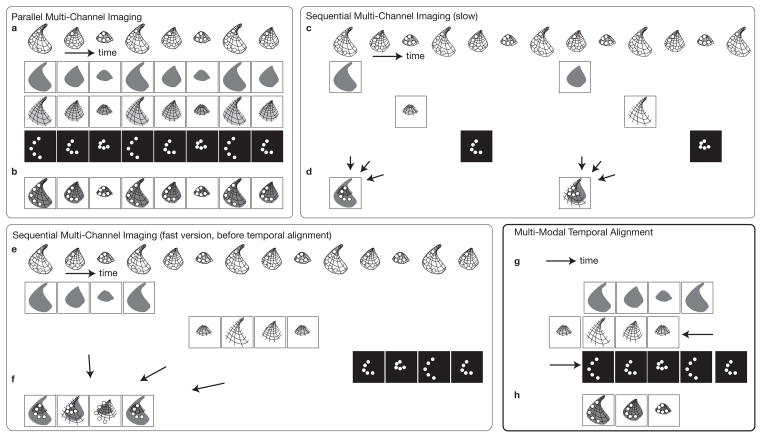
Figure 1.
Multi-channel microscopy procedure for imaging dynamic samples. a–b All channels are captured simultaneously on microscopes equipped for parallel imaging a Sample undergoing a periodic motion pattern imaged in three channels b Composite image of all channels. c–d Channels are captured sequentially on microscopes equipped with a single detector/camera c Sample undergoing a periodic motion pattern imaged alternatively in three channels d inaccurate composite image due to asynchronous imaging of the channels. e–f Channels captured sequentially at high frame rate over one ore more occurences of the motion pattern e Each channel is captured at full camera frame rate with short delays in between channels f random triggering leads to sequences that are temporally unregistered. g–h Multi-modal registration allows for high-speed multi-modal imaging g Temporal registration procedure of the sequences acquired in panel e. f Aligned images yield composite images at full frame rate.
Imaging each fluorescent species separately has the advantage that it can be carried out with simpler optical paths, each channel being acquired after inter-changing the filter set. Only a single camera or detector is required, which can significantly bring down the cost of the system. Also, since each species is illuminated and imaged separately, more choices are available to optimally illuminate the fluorophores and collect light with appropriate wavelength bands to minimize cross-talk from other species (images are also amenable to spectral unmixing). Interchangeable filter sets, often mounted on motorized turrets, are available for most commercial microscopes. Imaging each channel sequentially also allows imaging with modalities that may be irreconcilable with synchronous imaging, such as bright field imaging in a spectral band that overlaps with the excitation or emission of one fluorescent species. When imaging dynamic samples, the standard approach is to (rapidly) cycle through channels, switching filters in between snapshots. Unless the sample is static, images acquired sequentially are not synchronous (Fig 1c) leading to the main disadvantage of this technique: it is limited to imaging samples that undergo no or only slow motions. For samples that undergo fast motions, the resulting composite images are spatially mismatched (Fig 1d).
The integration time of each frame (the interval of time over which photons are collected to produce an image) is an essential parameter for determining image quality. The spatial and temporal resolutions of an image, as well as its signal-to-noise ratio are strongly inter-dependent. A moving structure produces motion-related blur. Biological processes occur over a wide range of velocities and scales. At high magnification, even seemingly slow processes can produce motion artifacts that rapidly outweigh the blur introduced by otherwise well designed, diffraction limited, optical elements (Vermot et al., 2008). For example, imaging cardiac dynamics in embryos require upwards from 30 frames per second to resolve moving cell on the surface of the beating heart wall (Vermot et al., 2008), while resolving red blood cells in the blood flow of the heart may require up to several thousands of frames per second (Ohn et al., 2009). Such processes can therefore not be imaged in multiple channels using a sequential imaging approach, as the time required to switch between channels (an operation that may involve mechanically turning a turret or opening a shutter) slows down the achievable frame rate. The number of channels that can be imaged at high speed therefore depends on the microscope, ranging from one for the simplest single detector microscopes to a handful for more sophisticated multi-path or multi-spectral microscopes.
Here, we show that for a class of samples that undergo motions that are repeatable, multi-channel imaging can be achieved even on microscopes that are not amenable to do synchronous imaging of all channels. The central assumption of repeatability is that the sample motion is spatially stereotypical, which allows to image each channel individually, each over a different occurrence of the motion. Repeating dynamic processes in biology include various organ and tissue contraction processes such as in the heart and other muscle tissues, as well as cyclically occurring calcium waves and beating mono cilia.
The key idea is that since imaging each channel individually is fast and minimizes cross-talk between channels it would allow for fast imaging on even simple microscopes provided a single channel can be imaged at the desired frame rate. Similarly to sequential imaging, in which microscope settings are adjusted in between imaging of the individual snapshots, we propose to measure the contributions in each channel sequentially, but rather than attempting to cycle through channels after each snapshot of a single channel, one or several occurrence of the process are imaged without changing settings. The other channels are then similarly imaged over subsequent occurrences of the process (Fig 1e).
Unless acquisition of each channel is triggered at a set instant in the dynamic process, the movies in each channel are not synchronized (Fig 1f). Given the scale at which microscopy operates, measuring a reliable gating signal might not be possible and we therefore rely on the measured signals themselves to build a synchronized, multi-channel sequence. Temporally aligning several series of images is an instance of image registration (Brown, 1992) an image processing technique that aims at finding a geometrical transformation (such as a temporal offset of the image series in the simplest synchronization case) so that the transformed image matches a reference image. To achieve this task automatically, one defines an image similarity measure. A candidate transformation is applied to the image whose similarity to a the reference is then evaluated. The transformation that yields the highest similarity is taken to be the solution to the registration problem. When aligning images that were generated through the same contrast mechanism, an intensity-based criterion is appropriate (Thévenaz et al., 1998): two images to be compared are subtracted from each other (pixel-by-pixel) and the sum of the absolute value of all pixels in the difference image produces a score, which is minimum when both the transformed and reference images are identical (in the latter case their difference image is then zero, compare difference images in Fig. 3a and c). When images have been acquired using different modalities, intensity-based similarity measures are inappropriate, since difference images, even for perfectly aligned images are not zero due to the differing contrast mechanisms that produced the images (see Fig. 3e and g). We therefore follow an approach that has been shown successful for multi-modal imaging in the medical field to register, for example, computed X-ray tomography images to magnetic resonance images or positron emission tomography images (Wells et al., 1996; Maes et al., 1997; Pluim et al., 2003). The key idea is to concentrate, rather than on the difference image of the candidate transformed and reference image, on the joint histogram of the two images. A joint histogram of two images (for example two channels of a multi-color image) reports how frequently an intensity pair occurs in an image (a pair consists of a pixel in one channel and the same pixel in the other channel). In fluorescence microscopy, joint histograms are used to analyze fluorophore coloalization (Manders et al., 1993). Identical images lead to a joint histogram whose counts are concentrated along the identity line (main diagonal, Fig. 3d) since there are only pairs of identical intensites [(0,0),(1,1), etc.]. Two similar, yet not identical, images yield joint histograms that have a wider spread (Fig. 3a). When comparing images produced with different contrast mechanisms, histograms may wary, yet aligned images will produce joint histograms that have a lesser spread (Fig 3f and h). The normalized mutual information (Studholme et al., 1999) is a metric that effectively captures similarity between two images of different modalities and can be estimated from the joint and marginal histograms (the latter are the grayscale histograms of each channel taken on their own). We have used this metric as an alternative to an intensity based metric we previously used for temporally registering images of the same modality (Liebling et al., 2005, 2006; Liebling and Ranganathan, 2009). Here we demonstrate that this technique is applicable to effectively image cardiac contractions in a quail embryo labeled with two fluorophores, yellow fluorescent protein (YFP) and (red) mCherry fluorescent protein.
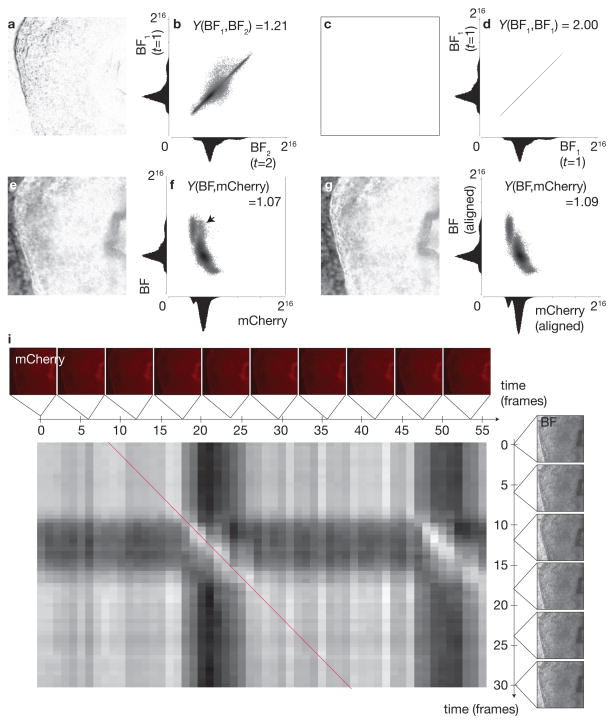
Figure 3.
Temporal registration metric is based on normalized mutual information rather than intensity comparison. a–b Comparing two similar but non-identical frames in the brightfield image sequence (Fig. 2 and e) leads to a non-zero absolute difference image (a) and a spread in their joint histogram (b). c–d Comparing two identical frames in a brightfield image sequence (Fig. 2 with itself) leads to a zero absolute difference image (c) and a joint histogram with values concentrated along the main diagonal (d). e–f Comparing two non-matching frames (brightfield image sequence, Fig. 2 and mCherry, Fig. 2) leads to a non-zero absolute difference image (e) and a spread (marked by arrow) in their joint histogram (f). g–h Comparing two matching frames (brightfield image sequence, Fig. 2 and mCherry, Fig. 2) still leads to a nonzero absolute difference image (g; making absolute difference unsuitable as a base criterion for measuring accurately matched images) but joint histogram exhibits less spread (h, compare in region with arrow marked in f). Normalized mutual information Y, (Studholme et al., 1999), increases for matching frames. (i) Normalized mutual information matrix; bright entries correspond to matching frames. Synchronization is achieved by finding a maximum merit path through the matrix to match the test and reference sequences (red line).
2 Results
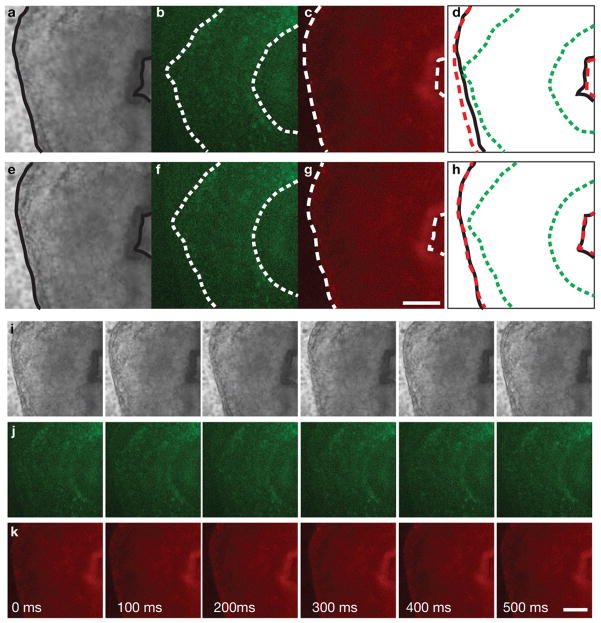
Fig. 2 shows the respective first frames of sequences of a beating embryonic quail heart acquired using brightfield (Fig. 2a) and epi-fluorescence wide-field (with yellow and red fluorescence filter cubes, Fig. 2b and c, respectively) contrasts. Since acquisition of each channel was carried out sequentially, the images correspond to different instants in the cardiac cycle and do not coincide (Fig. 2d) as indicated by anatomical landmarks such as the outer heart wall (myocardium) and clearly visible in Movies 1 and 2. Temporal registration of each fluorescence channel to the brightfield channel yields new sequences where the anatomical features coincide (Fig. 2e–h, Movies 1 and 3).
Figure 2.
a–d Anatomical features do not coincide in corresponding frames of three sequentially acquired image sequences showing the looping heart tube of a double-labeled embryonic quail. a Brightfield with outlined myocardium b YFP channels with outlined endocardium c mCherry with outlined myocardium d before temporal registration frames capture different heart contraction phases e–h Anatomical features coincide in corresponding frames after image sequences were temporally registered. e Brightfield with outlined myocardium f YFP channels with outlined endocardium g mCherry with outlined myocardium h after temporal registration frames capture the same heart contraction phase i–k Time-course of the dynamic part of heart beat after temporal registration. All scale bars are 100 μm. See also Movies 1–3.
To illustrate the necessity of using a similarity criterion other than direct intensity comparison between images (which is not appropriate for images of different modalities or colors) we show the absolute difference image and joint histograms for two neighboring frames in the brightfield image sequence (Fig. 3a and b, respectively) and, by comparison, the difference images for brightfield and red fluorescence channel frame pairs before and after the two sequences are temporally registered (Fig. 3e and g, respectively). The difference image of two identical frames yields an identically zero absolute difference image (Fig. 3c) and a joint histogram with values concentrated along the mid-diagonal (Fig. 3d). When the modalities are different, the absolute difference image is non-zero whether the images are matching or not and is therefore not appropriate to estimate successful registration. When examining the joint histograms for non-matching and matching frames (Fig. 3f and h, respectively) we notice that in the case of non matching frames, the histogram has a wider spread (as indicated by the arrow in Fig. 3f). Both in single modality and multi-modality images, the normalized mutual information, which quantifies the spread in the joint histogram (a lower spread yields a higher normalized mutual information), is higher for matching frames than for non-matching frames.
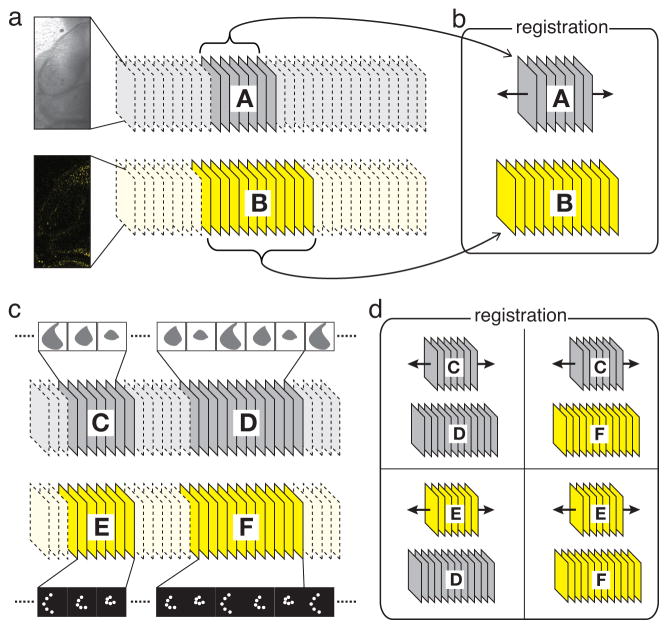
We estimated the accuracy of our algorithm by using a two-channel image sequence acquired on a fast confocal microscope. In this sequence, YFP fluorescence light (FL) and transmitted light (TL) were acquired simultaneously, providing a ground truth for evaluating synchronization accuracy. We first selected a one-heartbeat long sequence with arbitrary starting point from the TL channel as well as a longer sequence in the fluorescence channel that contained the sequence corresponding to the TL subset (Fig. 4a). Our registration algorithm lead to exact synchronization (Fig. 4b and Movie 4). To verify that our algorithm gives accurate results even when registering sequences from different heartbeats (which is the case for sequential acquisition), we extracted frames covering a full heartbeat in both the TL and FL channels (acting as reference sequences), as well as frames in the TL and FL channels covering two periods of subsequent heartbeats and acting as test sequences (Fig. 4c and Movie 4). The TL and FL reference sequences were then matched to the TL or FL test sequences, respectively. When compared to the registered TL-TL sequence, the registered FL-TL and TL-FL sequences were delayed or advanced by a half frame, respectively, and the FL-FL synchronization timestamps were identical to those resulting from the TL-TL synchronization (Fig. 4d and Movie 4). Taken together, these results suggest that inter- and intra-modal synchronizations yield results consistent and accurate, with only 0.5 frames discrepancy.
Figure 4.
Validation procedure for multi-modal synchronization of microscopy images. a Synchronously-acquired transmission and fluorescence channels. b a reference sequence A and a test sequence B are extracted; synchronization matches reference sequence A to its correct temporal position by maximizing mutual information with sequence B. c two reference sequences, C and E, and two test sequences, D and F are extracted from the synchronously acquired data; d intra-modal and inter-modal synchronization of the reference sequences to the test sequences yield consistent (within 0.5 frames) synchronization to a subsequent cardiac cycle in the test sequences. See also Movie 4.
3 Discussion
Our data show that rapid acquisition of each individual channel, followed by temporal alignment is an effective technique for retrieving multi-color or multi-modality images at high frame rates. We achieve full frame rate (30 frames per second) for frames of size 512 by 512 pixels. Since our technique does not require that the channels be acquired simultaneously, it can be carried out on standard microscopes equipped with a single collection light path and a monochrome camera. Switching between channel settings does not have to be fast nor does it have to be automated. Sequential acquisition allows us to use filter sets that are optimally adapted to each fluorophore, thereby minimizing the risk for cross-talk between channels. The camera and the brightness of the samples dictate the frame rate. Our technique is not limited in the number of channels that can be imaged. The latter number is only limited by how many different fluorescent labels the sample can viably carry and could be imaged sequentially, were the sample static. Image quality is affected by the accuracy with which images can be temporally registered and the intrinsic reproducibility of the motion by the sample itself. Use of mutual information based metrics requires that the different channels bear a sufficient level of co-localization at the resolution intended for imaging. We found that alignment of fluorescence channels against a modality that reveals anatomical features, such as brightfield microscopy, was most effective. In the future, we expect our technique to be applicable to other multi-modality (cardiac) imaging protocols, such as imaging with confocal microscopy and optical coherence tomography (Yelin et al., 2007).
4 Methods
Sample Preparation
Double-labeled (YFP, mCherry) quail embryos were incubated at 37 °C. The embryos were dissected from the egg and mounted for ex ovo culture on filter papers with a central aperture (Chapman et al., 2001) on an agar-albumen substrate on glass-bottom Petri dishes for microscopic observation.
Imaging
We acquired images on a Leica DMI6000B inverted widefield microscope with a Leica PL S-APO 10×/0.3 dry objective (Leica Microsystems, Wetzlar, Germany). Images were captured with an electron multiplying charge coupled device (EM-CCD)-equipped camera (ImageEM C9100-13, Hamamatsu, Japan). During imaging, the temperature was maintained at 37 °C inside the incubation chamber (PeCon, Erbach, Germany). We acquired consecutive 5-second bursts (30 frames per second, 512×512-pixels per frame) in bright-field and fluorescence microscopy (YFP, then mCherry). For validation experiments, we acquired images on a Leica TCS SP5 confocal microscope with resonant scanner for dynamic imaging at high speed using a 10×/0.4NA objective lens. To achieve a frame rate of approximately 40 fps, the scanned image size was set to 512×256 pixels. The pinhole and gain were adjusted for optimal viewing at high frame rate, and the optical slice contained 5 seconds or approximately 200 frames to capture a minimum of two heartbeats. Epi-fluorescence and transmitted light were collected simultaneously.
Temporal aligment of multi-color image sequences
Sequences were synchronized using a custom written Matlab (The Mathworks, Natick, MA) implementation of a non-uniform temporal aligment algorithm (Liebling et al., 2006). One period of the heartbeat was cut in the brightfield image series (acting as the reference sequence) and the the YFP and mCherry sequences were automatically aligned to it. Briefly, the normalized mutual information (Studholme et al., 1999; Liebling and Ranganathan, 2009) is computed between all frame-pairs from the two sequences to be aligned (Fig 3i) and the diagonal path of maximal merit (high normalized mutual information and low temporal distortion), indicating optimal match, is determined using a dynamic programming algorithm (Liebling et al., 2006). Image sequences were rendered using ImageJ and Imaris (Bitplane, Zurich, Switzerland).
Supplementary Material
References
- Brown LG. A survey of image registration techniques. Computing Surveys. 1992;24:325–376. [Google Scholar]
- Chapman S, Collignon J, Schoenwolf G, Lumsden A. Improved method for chick whole-embryo culture using a filter paper carrier. Dev Dyn. 2001;220:284–289. doi: 10.1002/1097-0177(20010301)220:3<284::AID-DVDY1102>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Dickinson ME, Bearman G, Tille S, Lansford R, Fraser SE. Multi-spectral imaging and linear unmixing add a whole new dimension to laser scanning fluorescence microscopy. BioTechniques. 2001;31:1272–6. 1278. doi: 10.2144/01316bt01. [DOI] [PubMed] [Google Scholar]
- Garini Y, Young I, McNamara G. Spectral imaging: principles and applications. Cytometry A. 2006;69:735–47. doi: 10.1002/cyto.a.20311. [DOI] [PubMed] [Google Scholar]
- Liebling M, Forouhar AS, Gharib M, Fraser SE, Dickinson ME. Four-dimensional cardiac imaging in living embryos via postacquisition synchronization of nongated slice sequences. J Biomed Opt. 2005;10:054001. doi: 10.1117/1.2061567. [DOI] [PubMed] [Google Scholar]
- Liebling M, Ranganathan H. Wavelet domain mutual information synchronization of multi-modal cardiac microscopy image sequences. In: Goyal V, Van DeVille D, Papadakis M, editors. Wavelets: Applications in Signal and Image Processing XIII; Proc. SPIE; San Diego CA, USA: 2009. p. 744602. [Google Scholar]
- Liebling M, Vermot J, Forouhar AS, Gharib M, Dickinson ME, Fraser SE. Nonuniform temporal alignment of slice sequences for four-dimensional imaging of cyclically deforming embryonic structures. Proceedings of the 3rd IEEE International Symposium on Biomedical Imaging: Macro to Nano (ISBI’06); Arlington, VA, USA. 2006. pp. 1156–1159. [Google Scholar]
- Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE transactions on medical imaging. 1997;16:187–98. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- Manders E, Verbeek F, Aten J. Measurement of colocalization of objects in dual-color confocal images. Journal of Microscopy. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- Ohn J, Tsai HJ, Liebling M. Joint dynamic imaging of morphogenesis and function in the developing heart. Organogenesis. 2009;5:1–8. doi: 10.4161/org.5.4.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluim JPW, Maintz JBA, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE transactions on medical imaging. 2003;22:986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- Studholme C, Hill D, Hawkes D. An overlap invariant entropy measure of 3d medical image alignment. Pattern Recogn. 1999;32:71–86. [Google Scholar]
- Thévenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Transactions on Image Processing. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- Vermot J, Fraser SE, Liebling M. Fast fluorescence microscopy for imaging the dynamics of embryonic development. HFSP Journal. 2008;2:143–155. doi: 10.2976/1.2907579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells WM, Viola P, Atsumi H, Nakajima S, Kikinis R. Multi-modal volume registration by maximization of mutual information. Medical image analysis. 1996;1:35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- Wolleschensky R, Zimmermann B, Kempe M. High speed confocal fluorescence imaging with a novel line scanning microscope. J Biomed Opt. 2006;11(1–14):064 011. doi: 10.1117/1.2402110. [DOI] [PubMed] [Google Scholar]
- Yelin R, Yelin D, Oh WY, Yun SH, Boudoux C, Vakoc BJ, Bouma BE, Tearney GJ. Multimodality optical imaging of embryonic heart microstructure. J Biomed Opt. 2007;12:064 021. doi: 10.1117/1.2822904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.