Abstract
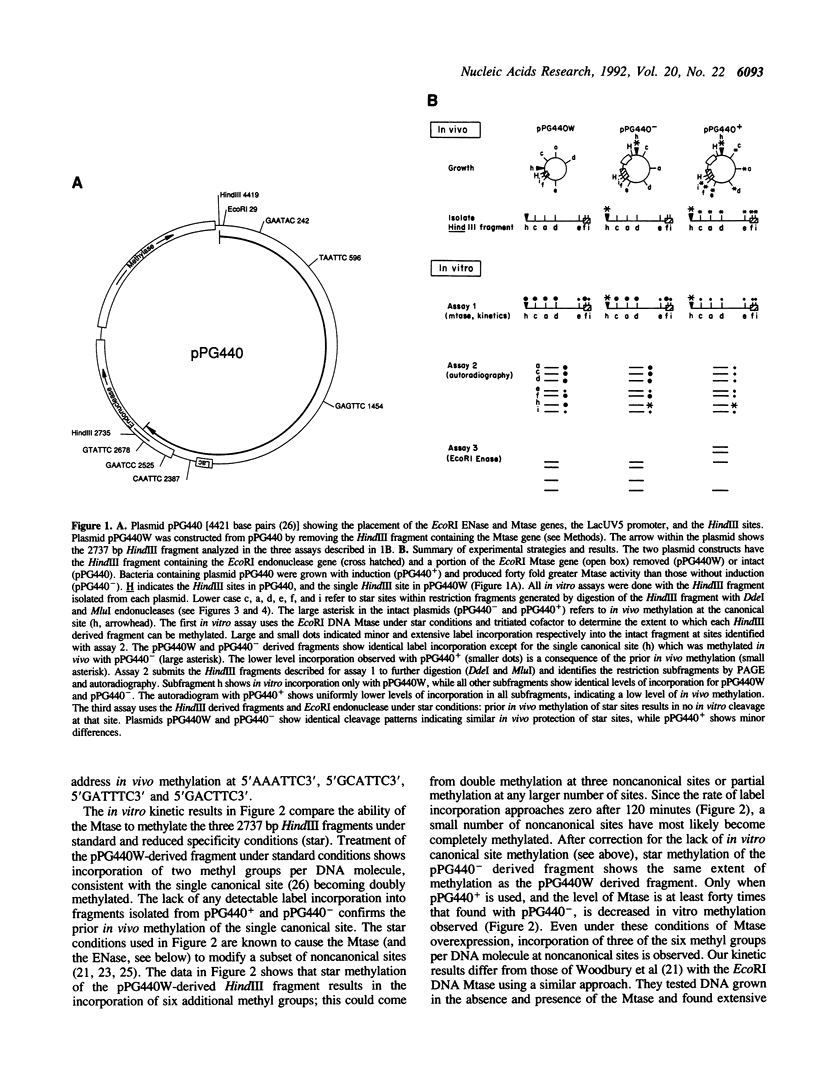
The EcoRI adenine DNA methyltransferase forms part of a bacterial restriction/modification system; the methyltransferase modifies the second adenine within the canonical site GAATTC, thereby preventing the EcoRI endonuclease from cleaving this site. We show that five noncanonical EcoRI sites (TAATTC, CAATTC, GTATTC, GGATTC and GAGTTC) are not methylated in vivo under conditions when the canonical site is methylated. Only when the methyltransferase is overexpressed is partial in vivo methylation of the five sites detected. Our results suggest that the methyltransferase does not protect host DNA against potential endonuclease-mediated cleavage at noncanonical sites. Our related in vitro analysis of the methyltransferase reveals a low level of sequence-discrimination. We propose that the high in vivo specificity may be due to the active removal of methylated sequences by DNA repair enzymes (J. Bacteriology (1987), 169 3243-3250).
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alves J., Rüter T., Geiger R., Fliess A., Maass G., Pingoud A. Changing the hydrogen-bonding potential in the DNA binding site of EcoRI by site-directed mutagenesis drastically reduces the enzymatic activity, not, however, the preference of this restriction endonuclease for cleavage within the site-GAATTC-. Biochemistry. 1989 Mar 21;28(6):2678–2684. doi: 10.1021/bi00432a047. [DOI] [PubMed] [Google Scholar]
- Corey D. R., Schultz P. G. Generation of a hybrid sequence-specific single-stranded deoxyribonuclease. Science. 1987 Dec 4;238(4832):1401–1403. doi: 10.1126/science.3685986. [DOI] [PubMed] [Google Scholar]
- Hager P. W., Reich N. O., Day J. P., Coche T. G., Boyer H. W., Rosenberg J. M., Greene P. J. Probing the role of glutamic acid 144 in the EcoRI endonuclease using aspartic acid and glutamine replacements. J Biol Chem. 1990 Dec 15;265(35):21520–21526. [PubMed] [Google Scholar]
- Heitman J., Model P. Mutants of the EcoRI endonuclease with promiscuous substrate specificity implicate residues involved in substrate recognition. EMBO J. 1990 Oct;9(10):3369–3378. doi: 10.1002/j.1460-2075.1990.tb07538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Model P. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J Bacteriol. 1987 Jul;169(7):3243–3250. doi: 10.1128/jb.169.7.3243-3250.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. C., Grable J. C., Love R., Greene P. J., Rosenberg J. M. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990 Sep 14;249(4974):1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- Koob M., Grimes E., Szybalski W. Conferring operator specificity on restriction endonucleases. Science. 1988 Aug 26;241(4869):1084–1086. doi: 10.1126/science.2842862. [DOI] [PubMed] [Google Scholar]
- Kretz P. L., Kohler S. W., Short J. M. Identification and characterization of a gene responsible for inhibiting propagation of methylated DNA sequences in mcrA mcrB1 Escherichia coli strains. J Bacteriol. 1991 Aug;173(15):4707–4716. doi: 10.1128/jb.173.15.4707-4716.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn I., Stephenson F. H., Boyer H. W., Greene P. J. Positive-selection vectors utilizing lethality of the EcoRI endonuclease. Gene. 1986;42(3):253–263. doi: 10.1016/0378-1119(86)90229-5. [DOI] [PubMed] [Google Scholar]
- Lauster R. Evolution of type II DNA methyltransferases. A gene duplication model. J Mol Biol. 1989 Mar 20;206(2):313–321. doi: 10.1016/0022-2836(89)90481-6. [DOI] [PubMed] [Google Scholar]
- Lesser D. R., Kurpiewski M. R., Jen-Jacobson L. The energetic basis of specificity in the Eco RI endonuclease--DNA interaction. Science. 1990 Nov 9;250(4982):776–786. doi: 10.1126/science.2237428. [DOI] [PubMed] [Google Scholar]
- Needels M. C., Fried S. R., Love R., Rosenberg J. M., Boyer H. W., Greene P. J. Determinants of EcoRI endonuclease sequence discrimination. Proc Natl Acad Sci U S A. 1989 May;86(10):3579–3583. doi: 10.1073/pnas.86.10.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. K., Rubin R. A., Kim S. H., Modrich P. DNA sequences of structural genes for Eco RI DNA restriction and modification enzymes. J Biol Chem. 1981 Mar 10;256(5):2131–2139. [PubMed] [Google Scholar]
- Pogolotti A. L., Jr, Ono A., Subramaniam R., Santi D. V. On the mechanism of DNA-adenine methylase. J Biol Chem. 1988 Jun 5;263(16):7461–7464. [PubMed] [Google Scholar]
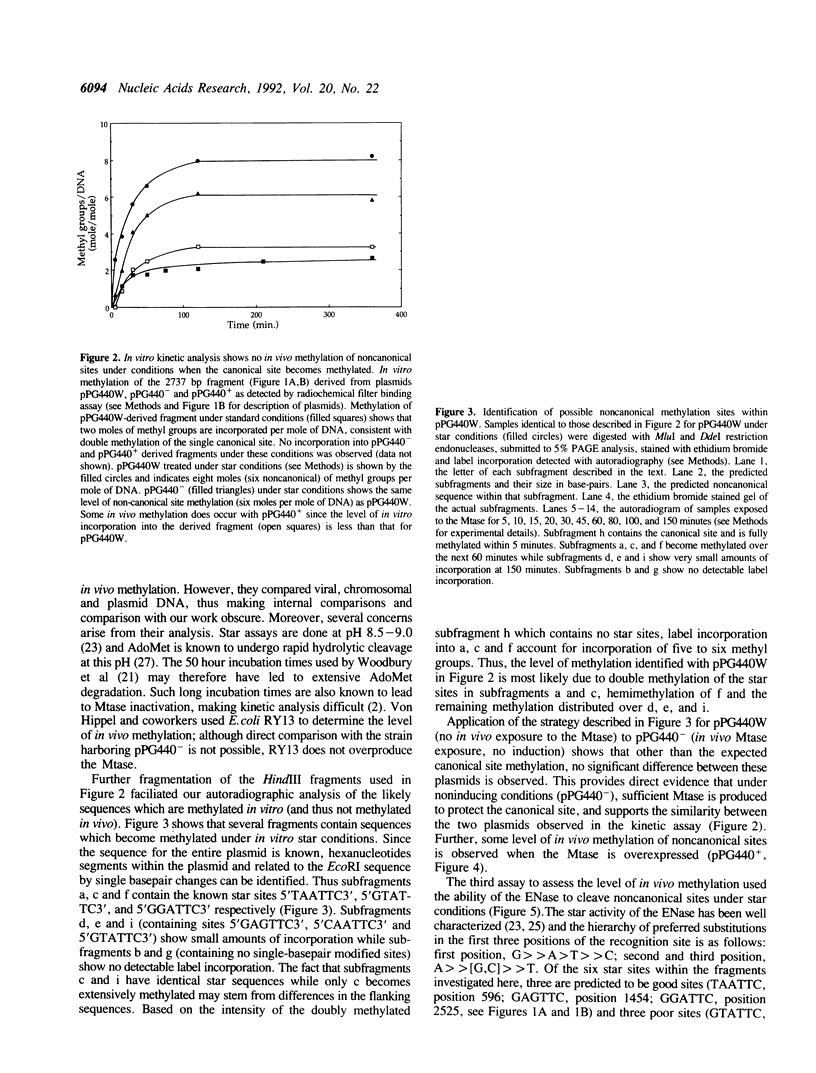
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich N. O., Mashhoon N. Kinetic mechanism of the EcoRI DNA methyltransferase. Biochemistry. 1991 Mar 19;30(11):2933–2939. doi: 10.1021/bi00225a029. [DOI] [PubMed] [Google Scholar]
- Reich N. O., Olsen C., Osti F., Murphy J. In vitro specificity of EcoRI DNA methyltransferase. J Biol Chem. 1992 Aug 5;267(22):15802–15807. [PubMed] [Google Scholar]
- Rosenberg J. M., Greene P. Eco RI* specificity and hydrogen bonding. DNA. 1982;1(2):117–124. doi: 10.1089/dna.1.1982.1.117. [DOI] [PubMed] [Google Scholar]
- Rubin R. A., Modrich P. EcoRI methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1977 Oct 25;252(20):7265–7272. [PubMed] [Google Scholar]
- Strobel S. A., Dervan P. B. Single-site enzymatic cleavage of yeast genomic DNA mediated by triple helix formation. Nature. 1991 Mar 14;350(6314):172–174. doi: 10.1038/350172a0. [DOI] [PubMed] [Google Scholar]
- Taylor J. D., Goodall A. J., Vermote C. L., Halford S. E. Fidelity of DNA recognition by the EcoRV restriction/modification system in vivo. Biochemistry. 1990 Dec 4;29(48):10727–10733. doi: 10.1021/bi00500a003. [DOI] [PubMed] [Google Scholar]
- Taylor J. D., Halford S. E. Discrimination between DNA sequences by the EcoRV restriction endonuclease. Biochemistry. 1989 Jul 25;28(15):6198–6207. doi: 10.1021/bi00441a011. [DOI] [PubMed] [Google Scholar]
- Wilson G. G. Organization of restriction-modification systems. Nucleic Acids Res. 1991 May 25;19(10):2539–2566. doi: 10.1093/nar/19.10.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfes H., Alves J., Fliess A., Geiger R., Pingoud A. Site directed mutagenesis experiments suggest that Glu 111, Glu 144 and Arg 145 are essential for endonucleolytic activity of EcoRI. Nucleic Acids Res. 1986 Nov 25;14(22):9063–9080. doi: 10.1093/nar/14.22.9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury C. P., Jr, Downey R. L., von Hippel P. H. DNA site recognition and overmethylation by the Eco RI methylase. J Biol Chem. 1980 Dec 10;255(23):11526–11533. [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]