Abstract
Among ascomycetous yeasts, the CTG clade is so-called because its constituent species translate CTG as serine instead of leucine. Though the biology of certain pathogenic species such as Candida albicans has been much studied, little is known about the life cycles of non-pathogen species of the CTG clade. Taking advantage of the recently obtained sequence of the biotechnological Millerozyma (Pichiasorbitophila) farinosa strain CBS 7064, we used MLST to better define phylogenic relationships between most of the Millerozyma farinosa strains available in public collections. This led to the constitution of four phylogenetic clades diverging from 8% to 15% at the DNA level and possibly constituting a species complex (M. farinosa) and to the proposal of two new species:Millerozyma miso sp. nov. CBS 2004T ( = CLIB 1230T) and Candida pseudofarinosa sp. nov.NCYC 386T( = CLIB 1231T).Further analysis showed that M. farinosa isolates exist as haploid and inter-clade hybrids. Despite the sequence divergence between the clades, secondary contacts after reproductive isolation were evidenced, as revealed by both introgression and mitochondria transfer between clades. We also showed that the inter-clade hybrids do sporulate to generate mainly viable vegetative diploid spores that are not the result of meiosis, and very rarely aneuploid spores possibly through the loss of heterozygosity during sporulation. Taken together, these results show that in this part of the CTG clade, non-Mendelian genetic exchanges occur at high rates through hybridization between divergent strainsfrom distinct clades and subsequent massive loss of heterozygosity. This combination of mechanisms could constitute an alternative sexuality leading to an unsuspected biodiversity.
Introduction
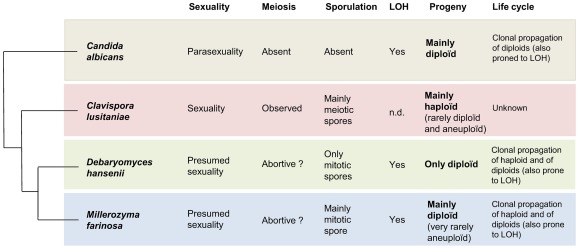
The life cycle of Saccharomyces species is well known. They propagate as haploid or diploids according to the environment, mate, sporulate and undergo meiosis to yield haploid spores [1]. Saccharomyces species are also known for forming inter-specific hybrids under the pressure of industrial fermentation conditions [2]. This genus is nevertheless far from being representative of the ascomycetous yeasts. Some Candida sp. pathogens belonging to the CTG clade were shown to have various life cycles. Candida albicans is a diploid that does mate and shows parasexuality, but does not perform sporulation or meiosis [3], [4]. Clavispora lusitaniae, on the other hand, can mate and sporulate. Its progeny is complex and comprises haploids resulting from meiosis, diploids and aneuploids [5]. Candida tropicalis was also recently shown to display parasexuality [6], suggesting that yeast pathogens are sexual species.
However, little is known about non-pathogenic species of this clade like the most studied Debaryomyces hansenii. This species is thought to have a rather clonal reproduction through fusion between mother and daughter cells followed by meiosis, whilemating between individual cells israrely observed [7]. Another CTG species, Pichia farinosa (Lindner) E.C. Hansen var. farinosa (teleomorph of Candida cacaoi) is a ubiquitous, halotolerant yeast found mainly in food (alcoholic beverages like beer and sake, soy sauce, miso, mash of rice vinegar etc.) but alsoon various substrates (cows with mastitis, sputum, giraffedung, laboratory media and highly concentrated sorbitol solutions) [8]. A reassessment of the phylogeny of Q9 enzyme-producing species changed the name of P. farinosa to Millerozyma farinosa [9]. Increased numbers of M. farinosa and other yeasts, mainly Candida albicans, were found in patients undergoing radiotherapy for cancer of the mouth [10] and an M. farinosa strain was found as an oral cavity colonizer [11]. Two further cases of human infections associated to M. farinosa were also described [12], [13].
In 1980, a related species, Pichia sorbitophila, was described [14]. The only member of this species, CBS 7064, is able to survive extremely high extracellular concentrations of salts (e.g., saturated solution of KCl) and other osmolytes (70% glucitol), although it has not been classified as halophilic or osmophilic [15], [16]. Both M. farinosa and P. sorbitophila can produce high yields of glycerol and xylitol, which makes them valuable in biotechnology. The placementof some strains in the M. farinosa species is a matter of debate [8]. However, the main controversy has concerned the taxonomy of M. farinosaand P. sorbitophila. On the basis of DNA relatedness, Debaryomyces halotolerans, P. sorbitophila, and Pichia petrophilum were considered to be conspecific with M. farinosa [17], whereas physiological properties and sequence analysis indicated they should be classified as separate species,in agreement with the divergent sequences of their respective mitochondrial DNAs [18], [19]. M. farinosa CBS 7064 carries two divergent URA3 genes, which were also different from that of the M. farinosa type strain [20]. The presence of the two divergent URA3 gene copies and the higher number of chromosomes in pulse field gel electrophoresis was in agreement with CBS 7064 being diploid.
From therecently sequenced CBS 7064 genome [21], it appears thatthis strain is a recent hybrid between two distant parents: the two copies of their homeologous chromosomes present more than 15% divergence at the DNA level. The parental contributorsremain unknown, but are certainly distinct from the M. farinosa type strain CBS 185T. CBS 7064 contains sevenpairs of chromosome. In three ofthese both parents contribute one chromosome; in two the chromosomes contain regions that are identical and regions that are diverged;two consist of identical chromosomes, indicative of massive loss of heterozygosity (LOH)[21]. Our analysis of the respective contribution of each parent to the hybrid reveals the major genetic changes that occurredduring the time that separated the last common ancestor of both parents and the hybridization event. These include one reciprocal translocation between two chromosomes, a number of gene losses and unequal mitochondrial DNA insertions (NUMT) in the nuclear genomes of the parents.
In order to define the parental contributors of CBS 7064, to resolve the taxonomy of the M. farinosa species and to gain insight into the life cycle of CTG species, we took advantage of the available complete genome sequence of the CBS 7064 strain [21] to analyzealmost all the M. farinosa strains available in public collections. In this paperwe use MLST to constitute phylogenetic groups. We demonstrate reproductive isolation of these groups. We show that M. farinosa is a species complex made of distinct taxa, and in which isolates exist as haploid organisms and as hybrids resulting from crosses between strains from these taxa. We also show that the hybrids can sporulate to generate viable diploid spores that are not the result of meiosis. In this species complex, an alternative sexuality allows the formation of hybrids subjected to LOH and of rare aneuploids.
Results
Structuring the Millerozyma farinosa populations in five clades
In a previous study, we have observed that Debaryomyces hansenii, a closely related species to M. farinose [22], consisted of distinct clades with haploid and diploid strains, the latter being hybrids from diverged strains belonging to these distinct clades [23]. We also demonstrated the efficacy of housekeeping gene intron sequence analysis in distinguishing between clades of D. hansenii. The most informative intron sequences were those of the ACT1 and RPL33 genes. In order to analyze the M. farinosa species, we therefore amplified and sequenced the introns of these two genes in 41 M. farinosa strains (Table 1). In a number of cases, PCR products were a mixture of two species which needed to be cloned before sequencing. In these cases, sequences were arbitrarily given the prefix A and B. We compared the obtained sequences and constructed gene trees using the neighbor-joining algorithm. Identical topologies were obtained when the trees were constructed with maximum-likelihood program (data not shown). The ACT1 gene intron tree is shown in Figure 1 and the RPL33 gene intron tree is shown in Figure S1. This analysis, both with ACT1 and RPL33, separated the sequences into the same five clades. Some strains displayed two different ACT1 alleles and one RPL33 allele or vice-versa, consistent with LOH. Using flow cytometry, we confirmed that the strains presenting ACT1 and/orRPL33 heterozygous alleles had a 2n DNA content. Table 2 gives the estimated genome size of all the strains presenting some heterozygosityfor ACT 1 and/or RPL33 together with ahaploid strain from each clade as a control. For CBS 185T and the haploidstrain of each clade, the estimated size of the genome varied from 9.2 to 10.8 Mb, similar to that of some species in theSaccharomycetaceae [24], but smaller than that of the closely related species D. hansenii (13.2 Mb) [25], [26]. The estimated values for the strains presenting sequence heterozygosityvaried between 18 and 22.9 Mb, approximatelytwice the value of CBS 185T. An example in Figure S2 shows a shift of the fluorescence intensity peaks in the strains CBS 7064 and CBS 5730 compared to CBS 185T and CBS 2001. These observations strongly suggest that the strains displaying heterozygosity result from crosses between strains belonging to different clades, as already observed for D. hansenii [23].
Table 1. List of strains used in this study.
| Species | Strain number | Substrate of isolation | Country of origin | Ploidy | Clade | Sporulation |
| Millerozyma farinosa | CBS 185T | Jopen beer | Poland | haploid | 1 | + |
| CBS 242 | katjang-bungil | unknown | haploid | 1 | nd | |
| CBS 2020 | fermenting cacao | Trinidad and Tobago | haploid | 1 | nd | |
| CBS 2597 | liquid medium, for tissue culture | UK | haploid | 1 | nd | |
| CBS 2768 | mash of rice vinegar | Japan | nd | 1 | nd | |
| CBS 4841 | miso | unknown | nd | 1 | nd | |
| CBS 11514 | human infection | Israel | nd | 1 | nd | |
| NCYC 3111 | food industry | UK | nd | 1 | nd | |
| NBRC 0602 | tamari koji | Japan | nd | 1 | nd | |
| PYCC 2633 | human | Israel | nd | 1 | nd | |
| VTT C-86127 | unknown | unknown | nd | 1 | nd | |
| CBS 2001T | sputum | Norway | haploid | 2 | − | |
| CBS 2101 | lung of hen | UK | haploid | 2 | + | |
| CBS 4548 | dung of giraffe | Japan | nd | 2 | − | |
| CBS 5031 | maize meal | South Africa | nd | 2 | + | |
| CBS 5116 | mastitis milk | Switzerland | nd | 2 | − | |
| DSMZ 70362 | mastitis of a cow | unknown | nd | 2 | + | |
| MUCL 38866 | soy sauce mash (moromi) | Indonesia | nd | 2 | nd | |
| MUCL 38867 | soy sauce mash (moromi) | Indonesia | nd | 2 | nd | |
| MUCL 38869 | soy sauce mash (moromi) | Indonesia | nd | 2 | nd | |
| MUCL 38870 | soy sauce mash (moromi) | Indonesia | nd | 2 | nd | |
| MUCL 38871 | soy sauce mash (moromi) | Indonesia | nd | 2 | + | |
| CBS 2007 | sake | unknown | haploid | 3 | − | |
| NBRC 10896 | unknown | unknown | nd | 3 | nd | |
| CBS 7064 | 70% sorbitol solution | Germany | diploid | 2–4 | + | |
| CBS 8045 | sorbitol stock | South Africa | diploid | 2–4 | + | |
| CBS 2006 | homare-miso | unknown | diploid | 2–3 | + | |
| CBS 4073 | tamari koji | unknown | diploid | 2–3 | − | |
| CBS 4710 | miso | Japan | diploid | 2–3 | + | |
| CBS 5949 | soya mash | Japan | diploid | 2–3 | no growth | |
| CBS 5730 | edo miso | Japan | diploid | 2–3 | + | |
| CBS 7911 | soy sauce | China | diploid | *−3 | + | |
| VTT C-86125 | unknown | unknown | diploid | 2–3 | ||
| MUCL 30824 | liquid sugar | Belgium | diploid | nd | + | |
| CLIB 1250 | Spore 28 from CBS 2006 | − | aneuploid | 2–3 | nd | |
| CLIB 1251 | Spore G6 from CBS 2006 | − | diploid | 2–3 | nd | |
| Millerozyma miso | CBS 2004T (CLIB 1230T) | sendai miso | unknown | haploid | 5 | + |
| CBS 4709 | miso | Japan | nd | 5 | nd | |
| CBS 4842 | unknown | unknown | nd | 5 | nd | |
| CBS 5013 | maize meal | South Africa | nd | 5 | nd | |
| NBRC 01003 | unknown | unknown | nd | 5** | nd | |
| M. farinosa x M. miso | CBS 2607 | liquid medium for tissue culture | UK | diploid | 3–5 | + |
| Candida pseudofarinosa | NCYC 386T (CLIB 1231T) | unknown | Japan | haploid | na | – |
| NBRC 0465 | unknown | unknown | nd | na | nd | |
| NBRC 0574 | unknown | unknown | nd | na | nd |
nd: not determined, na: not applicable,* 0.5% divergence in ACT1 with clade 1, ** 1.3% divergence in ACT1 with clade 4.
For a number of strains, ACT1 and RPL33 introns could not be PCR-amplified and the sequence of their ACT1 coding gene helped to place these strains into the different clades. This is the case for NBRC 0602 and NBRC 10896, CBS 2607(carrying a clade 2 allele and a clade 5 allele and being the only strains carrying this type of combination), NBRC 01003, (** 1.3% divergence with clade 4) and CBS 7911 (a clade 3 allele and *0.5 % divergence with clade 1 allele). MUCL 30824 presented a complex distribution of alleles. The strain CECT 10348 was not related to M. farinosa, but rather to the type strain NRRL YB-2798T of Sugiyamaella sp. [28] with 2 bp difference over the rDNA D1D2 585 bp long sequence.
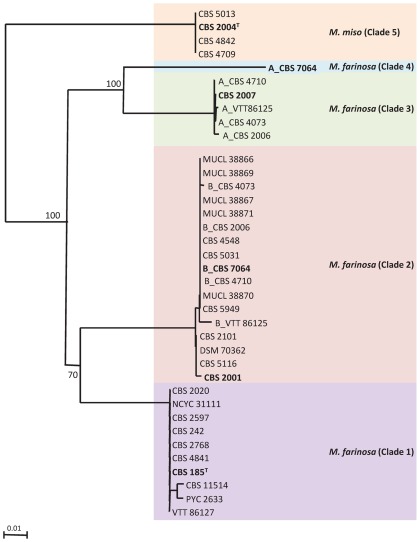
Figure 1. Neighbor-joining phylogram of the ACT1 gene intron of M. farinosa isolates.
Bootstrap values (%) based on 1000 replicates are indicated at the nodes for main groups and clades. All positions containing gaps and missing data were eliminated from the dataset. Clades are indicated.Typical strains are in bold characters. Heterozygous alleles in hybrids were arbitrarily given the prefix A or B. Bar, 0.01 substitutions per site.
Table 2. Genome size of various M. farinose strains estimated by flow-cytometry.
| Strains | Genome size (Mb) | Ratio strain/type strain |
| CBS 2004T | 9.20+/−0.2 | 0.85 |
| CBS 2001 | 10.6+/−0.2 | 0.98 |
| CBS 2007 | 10.6+/−0.5 | 0.98 |
| CBS 185T | 10.8+/−0.5 | 1.00 |
| CBS 5949 | 18.0+/−0.3 | 1.67 |
| CBS 7064 | 18.9+/−0.6 | 1.75 |
| CBS 5730 | 20.1+/−1.3 | 1.86 |
| CBS 8045 | 20.5+/−0.6 | 1.90 |
| MUCL 30824 | 21.1+/−0.9 | 1.95 |
| CBS 4073 | 21.3+/−0.3 | 1.97 |
| CBS 2006 | 21.9+/−1.5 | 2.02 |
| CBS 4710 | 22.4+/−0.5 | 2.07 |
| CBS 7911 | 22.9+/−1.4 | 2.12 |
| CLIB 1251 (spore G6) | 24.9+/−0.9 | 2.30 |
| CLIB 1250 (spore 28) | 13.1+/−0.2 | 1.23 |
The ratio of the genome size of each strain on that of CBS 185T is indicated.
The first clade (ascribed the number 1) was defined by the sequence of the ACT1 alleles of 10 strains including the type strain CBS 185T. As mentioned above, the results from the analysis of the RPL33 alleles gave essentially the same clades and are not further discussed here. Clade 2 contained 18sequencesincluding one of theACT1 alleles of CBS 7064. Sequence divergence in the ACT1 intron between these two clades was 8.5% (Table 3). We note that the five strains MUCL 38866, MUCL 38867, MUCL 38869, MUCL 38870 and MUCL 38871 all belong to clade 2 and have identical intronsequences. Since they were all isolated from the same substrate in Indonesia, they may well constitute asingle clone. The clade 3 is comprised of 5 isolates. Only one of these strains, CBS 2007, carried a unique copy of ACT1 and of RPL33; the four other strains, CBS 2006, CBS 4073, CBS 4710 and VTT 86125, each carried two alleles of ACT1, one grouping with clade 3 and the other with clade 2. The average sequence divergence in the ACT1 intronbetween clade 1 and clade 3 was 11.5% (Table 3). The fourth clade contains only one sequence, being that of the second ACT1 allele of M. farinosa CBS 7064, withasequencehighly divergent (13.7%) from that of clade 1 (Table 3). Finally, clade 5 consisted of the sequences of four strains that showed 14.5% divergence fromclade 1 ACT1 intron sequences and an even higher sequence divergence – above 15% with that of the other clades. Because intronic sequences could not be obtained for anumber of strains, weused coding sequences to place these strainsinto the defined clades (Table 1 and Table S2).
Table 3. Sequence divergence (%) within the ACT1 gene intron between M. farinosa clades.
| Clades | 1 | 2 | 3 | 4 | 5 | |
| Strains | CBS 185T | CBS 2001 | CBS 2007 | CBS 7064a | CBS 2004 T | |
| 1 | CBS 185T | − | 8.5 | 11.3 | 13.7 | 14.5 |
| 2 | CBS 2001 | − | 11.7 | 14.5 | 16 | |
| 3 | CBS 2007 | − | 11 | 17 | ||
| 4 | CBS 7064 a | − | 19.5 | |||
| 5 | CBS 2004 T | − |
Clade 4 allele.
As seen above, sequence divergence between the clades varied from 8.5 to 19.5% (Table 3), indicating that these clades are clearly distinct. We defined a typical strain for each clade as being, among haploids, the first strain described (see the ACT1 gene intron tree in Figure 1). The topology of the RPL33 gene intron tree (Figure S1) was very slightly different from that of the ACT1 gene tree. Comparison of the two gene trees indicated that the clades were composed of the same strains, suggesting the absence of marker exchange between clades, ACT1 and RPL33 being located on different chromosomes ([21]; our unpublished data).
Interestingly, one of the two RPL33 alleles of strain CBS 8045 (the defining and unique strain of clade 4) was different from all sequenced RPL33 alleles; the other allele belongs to clade 2 as does the unique ACT1 allele of this hybrid. Since CBS 7064 was homozygous for clade 2 RPL33 allele ([21]; our data), no “clade 4 RPL33 sequence” was available. To know whether CBS 8045 was also, like CBS 7064, a clade 2/clade 4 hybrid and effectively carried clade 4 sequences, three markers AB-r, CD-l and IJ-r(see Table S3 and Figure S3) known to existin two heterozygous copies in the genome of CBS 7064 were sequenced in CBS 8045.For each locus we found identical sequences in CBS 7064 and in CBS 8045, indicating that CBS 8045 was indeed another strain carrying both clade 2 and clade 4 sequences. Interestingly, both strains had been isolated as contaminants from highly concentrated sorbitol solutions [27].
Finally, two strains, CECT 10348 and NCYC 386T, previously assigned to the M. farinosa species did not fall in any of our five clades. CECT 10348 was unrelated to our test group and belonged in fact on the basis of rDNA D1/D2 sequence comparison to a species of the recently described genus Sugiyamaella [28]. The strain NCYC 386T, though not within the M. farinosa complex was the closest on the basis of the ACT1 coding sequence. Nevertheless, its ACT1 intron does not show any similarity with that of any strain from the five clades of M. farinosa (see later).
Origin of the CBS 7064chromosomes in relation with the M. farinosa clades
Using two housekeeping genes ACT1 and RPL33, we were able to define five distinct clades in the M. farinosa species (see above). This MLST analysisshowed unambiguously that strains belonging to clade 2 and clade 4 contributed to the hybrid CBS 7064. In the description of the genome of CBS 7064, a distinction was made between the two parental contributing genomes (designated Pγ and Pε?) on the basis of the unequal distribution of NUMT insertions and on the G+C percentage of each subgenome [21].The CBS 7064 genome contains seven pairs of chromosomes: three are heterozygous, two are homozygous and two are heterozygous along a proportion of their length, indicative of a massive loss of heterozygosity ([21], see Figure S3).Having defined the parental clades of CBS 7064 will allow us to determine which part of the CBS 7064 genome was contributed by which clade.In addition, it was shown that synteny was conserved between the two contributors of CBS 7064, except for a single reciprocal translocation between two chromosomes of one the CBS 7064 contributor [21]. Using the genome sequence of CBS 7064, it was therefore interesting to perform a wider MLST analysis on the typical, haploid strains of the four clades (CBS 185T, 2001, 2007 and 2004T from clades 1, 2, 3 and 5, respectively), using 16 markers located at the extremities of each of the chromosomes of CBS 7064 to define the origin of each of the chromosomes of CBS 7064 and to better define each of M. farinosa clades.
MLST analysis was obtained for one gene on each extremity of the seven chromosome pairs and on each side of the translocation breakpoints between chromosomes E/F and I/J (Figure S3 and Table S3). Out of the four typical strains tested, only the clade 2 CBS 2001 strain gave 100% sequence identity with at least one of the two alleles of CBS 7064. In the case of the EF-l marker,only 1 bp out of 478 bp differedbetween CBS 7064 and CBS 2001. The three other typical strains: CBS 185T for clade 1, CBS 2007 for clade 3 and CBS 2004T for clade 5 displayed 81 to 96% identity with that of CBS 7064. These results there fore confirmed what we had observed with the ACT1 and the RPL33 markers, i.e.a clade 2 strain is a parental contributor of CBS 7064,whileneither clade 1, clade 3 nor clade 5 strains contributed toCBS 7064.Interestingly, none of the strains tested gave 100% identity with the right end markers of chromosomes A and B of CBS 7064 (Figure S3), indicating that the right arm of chromosome A, which is homozygous to the right arm of chromosome B due to LOH [21], corresponds to an as-yet undescribed taxon. Here, we were able to confirm the distinction foundbetween the Pγ and the Pεsubgenomes [21]and to establish unequivocally the source of the parental subgenomes; a clade 2 strain is the contributor of the Pγ subgenome, whereas the Pεsubgenome comes from an as-yet undescribed taxon, which has no haploid representative in our M. farinose collection and which we have characterized as clade 4 (Figure 1).
In addition, pair-wise comparisons of the marker sequences in the four typical strains gave identities of 78 to 96%, further supporting the existence of distinct clades within M. farinosa based on the 16 chromosomal markers (Table S4). The existence of four divergent sequences for each marker of the typical haploid strains CBS 185T, 2001, 2007 and 2004T also indicated thatneither genetic exchange due to hybridization followed by LOH, nor introgression of the tested markers between strains had occurred at the chromosome extremities in representatives of clades 1, 2, 3 and 5.
Sporulation of the hybrids and characterization of progeny
As some isolates of M. farinosa were found to be inter-clade hybrids, it was interesting to examine their ability to sporulate and if they did, the content of the resulting spores. In the genus Saccharomycesinter-specific hybrids have been shown to sporulate but to produce unviable spores. In the CTG clade few studies exist on sporulation of pathogen species [5] and none on non-pathogen species, except for CBS 7064, which was described as a sporulating isolate [14].
All the M. farinosa inter-clade hybridstrains and a number of haploids listed in Table 1 were allowed to sporulate.All the hybrids sporulated well,exceptfor CBS 4073andhybrid CBS 5949 which did not grow on the sporulation medium used. On the other hand, some of the haploid strains showed little sporulation, if at all; in particular, the typical strains of clade 2 and 3, CBS 2001 and CBS 2007 respectively, did not sporulate, unlike the type strain CBS 185T.
The progeny of the clade 2/clade 4 hybrid CBS7064 and of the clade 2/clade 3 hybrid CBS 2006 were analyzed.Only four-spore asci were considered (Figure S4A and S4B). Asci were microdissected and spores were germinated. A total of 52 spores of CBS 7064 out of 163 grew on YPD (32% spore viability). This result seemed surprisingly high for progeny coming from a hybrid of highly diverged parents(15.3% between the two CBS 7064 subgenomes)[21]. Indeed, in Saccharomyces, the hybrids with over 10% divergence do not produce viable spores [29]. Using a heterozygous NUMT insertion as marker, we showed that the 52 viable spores had all inherited both parental alleles, suggesting that meiosis had not taken place. Flow cytometry confirmed that 10 of these spores chosen at random had a 2n DNA content (data not shown).
Sporulation of strain CBS 2006 yielded 132 spores, out of which 103 grew on YPD (78% spore viability). A total of 70 spores were analyzedfurther. As the NUMT content of CBS 2006 is not known, we used SNP in genomic markers to analyze the progeny. We amplified and sequenced the AB-r and CD-l markers (which are locatedin CBS 7064 on the right endof the A/B chromosome pair and onthe left endof the C/D chromosome pair, respectively; Figure S3, Table S3). Two spores (designated G6 and 28) carried only one type of CD-l allele, that of clade 3. The rest of the spores carried both clade 2 and clade 3 CD-l alleles. All the spores carried both of the AB-r alleles. This suggested that all the spores except G6 and 28 were diploid, which was confirmed by flow cytometry on 16 spores. Interestingly, the DNA content of spores G6 and 28 were estimated at24.9±0.9 Mb and 13.1±0.2 Mb respectively, the parental CBS 2006 genome size amounting to 21.9±1.5 Mb (Table 2). This suggests a diploid state (∼2.3n)for spore G6 and a haploid or aneuploid state (∼1.23n)for spore 28.
The spores G6 and 28 were further analyzed using theset of 16 markers located on each extremityof all chromosome pairs. The results are summarized in Table 4. Despite a slightly higher number of heterozygous markers than CBS 7064 (9/13 vs. 7/13), the hybrid CBS 2006 harbored also some level of LOH. When compared to its parent CBS 2006, spore G6 contained the same four heterozygous regions at the left and right ends of chromosome pair A/B, left end of E/F and right end of K/L, but homozygous markers were found in the place of the five other heterozygous regions of CBS 2006 (Table 4). Taken together, these results indicate that this spore is diploid and has undergone LOH, possibly during sporulation. On the other hand, spore 28 only displayed a single heterozygous region on the left end of chromosome pair A/B. This spore has a close to 1.23n DNA content with an estimated genome size of 13.1 Mb, which is slightly over the values between 9.2 and 10.8 Mb measured for haploid M. farinosa strains. Therefore, spore 28 is probably aneuploid, because it contains both clade 2 and clade 3 AB-r markersbut is homozygous for the other markers and it may have undergone meiosis (Table 4).
Table 4. Origin of left and right end side regions of spores G6 and 28 chromosomes.
| Markers * | Clades | |||
| CBS 2006 | spore G6 | spore 28 | CBS 7064 | |
| AB-l | 2/3 | 2/3 | 3 | 2/4 |
| AB-r | 2/3 | 2/3 | 2/3 | 4 |
| CD-l | 2/3 | 3 | 3 | 2/4 |
| CD-r | 2/3 | 3 | 3 | 2 |
| EF-l | 2/3 | 2/3 | 2 | 2/4 |
| EF-r | 3 | 3 | 3 | 2/4 |
| GH2-l | 2/3 | 2 | 2 | 2 |
| GH2-r | 2/3 | 3 | 3 | 2 |
| IJ-l | 2 | 2 | 2 | 2/4 |
| IJ-r | 2/3 | 3 | na | 2/4 |
| KL2-l | 3 | 3 | 3 | 2 |
| KL2-r | 2/3 | 2/3 | 3 | 2 |
| MN2-l | Na | na | na | 2/4 |
| MN2-r | 2 | 2 | 2 | 2/4 |
Analysis of mtDNA origin and NUMT insertions reveals secondary contacts after reproductive isolation
Within M. farinosa, we have described distinct clades whose sequence divergence is as high as that described for distinct species, as in the genus Saccharomycesfor instance. In addition, we have evidenced inter-clade hybridsthat sometimes yield aneuploids. This previously unobserved situation mimics horizontal gene transfer (HGT)and it was interesting to examine potential genetic exchanges in haploid strains. Mitochondria constitute good markers to follow genetic exchanges.
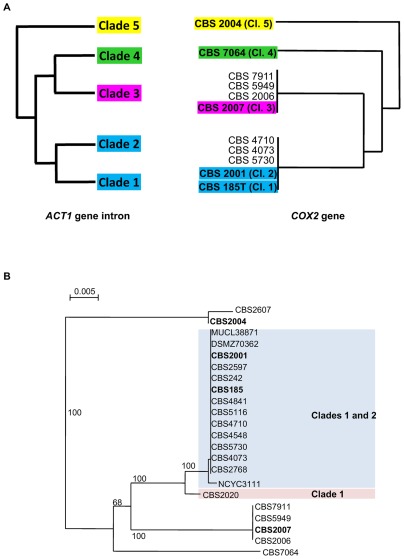
To address thequestion of the inheritance of mtDNA in the M.farinosa inter-clade hybrids, an MLST analysis of the COX2 and COX3 genes was first performed using sequence of the part of these two genes in the typical strains of each clade and in all the inter-clade hybrids. A schematic representation of the relationship between the mtDNA sequences in relation with the different clades is shown in Figure 2A. Clade 3 CBS 2007 and clade 5CBS 2004Tas expectedcarrieddivergentmtDNA alleles, while surprisinglystrains from clade 1 and clade 2 carried the same mtDNA alleles. All the tested hybrids,except CBS 7064 (clade 2/clade 4) and CBS 2167 (clade 3/clade 5), carried either the mtDNA alleles of clade 2 or that of clade 3, consistent with the idea that the hybridswere derived from crosses betweenthe clades. Our results show that, like in Saccharomyces [30]inter-specific hybrids [31] or Metschnikowia inter-specific hybrids [32], mtDNA can be transmitted by either of the parents in the M. farinosa species complex. This is in contrast with several reportson Saccharomycesfermentative hybrids involving S. cerevisiaeshowingthat mtDNA from the non-cerevisiaeparental contributor wasspecifically favored [33].
Figure 2. Origin of mtDNA in M. farinosa diploid strains.
(A) Relation between the clades according to the ACT1 gene intron tree generated with the sequences of the M. farinosa typical strains (left) and to the mitochondrial COX2 gene of theM. farinosa typical strains and of the inter-clade hybrids (right) are shown. Typical strains are in bold characters. Bar, 0.005 substitutions per site. (B) Concatenated COX2 (540 bp ) and COX3 (609 bp) gene tree form M. farinosa haploid strains of clades 1 and 2. Typical strains are in bold characters. Bar, 0.005 substitutions per site. Tree constructed with COX2 or COX3 were congruent.
Interestingly, the mtDNA carried CBS 7064 did not correspond to any of the alleles of clades 1, 2, 3 and 5. Comparison of the mitochondrial genomes of CBS 185T and CBS 7064 had already shown that they differed [18], [19]. From our results, we can deduce that mtDNA of CBS 7064 was transmitted by the parental strain for which we do not yet have a haploid representative strain.
The fact that both clade 1 CBS 185Tand clade 2 CBS 2001 typical strains carried the same mtDNA alleles prompted us to analyze the distal COX2 and COX3 geneson the mitochondrial genome of all haploid strains from clade 1 and 2 in order to try to identify the missing mtDNA type. As represented in Figure 2B, all the stains carried the same mtDNA alleles, except for clade 1 strain CBS 2020, which displayed threeSNP in COX2(0.5%) and five SNP in COX3(0.8%), demonstratingthat CBS 2020 carried another mtDNA type. These results suggest that mating events between clade 1 and clade 2 ancestors have occurred associated with unidirectional transfer of mtDNA. Unlike in Metschnikowia [32], we did not observe any recombination between the mtDNA from different clades.
NUMTinsertions can be found in most yeast genomes [34].Using NUMT insertions that we have identified in the genome of CBS 7064 [21] as markers, we could go further into the analysis of the relationships between the M. farinosa strains and confirm the existence of genetic exchanges between clades 1 and 2. By monitoring the presence /absence of NUMT in PCR products, we analyzed nine heterozygous and two homozygous NUMT loci present in the CBS 7064 genome in the three haploid strains CBS 185T, (clade 1), CBS 2001 (clade 2) and CBS 2007 (clade 3) as well as and two hybrids CBS 5730 (clade 2/clade 3) and CBS 8045 (clade 2/clade 4). Most insertions were absent in the typical strains from clades 1, 2 and 3, consistent with their presence on chromosomes E, J and M that are characteristic of the clade 4 parent of CBS 7064(Table 5). Conversely, the sole NUMT insertion of chromosome I of CBS 7064, contributed by clade 2 parent, was found in the clade 2 strain CBS 2001 and also in clade 1 strain CBS 185T.
Table 5. PCR analysis of 11 NUMT loci in six strains of M. farinosa.
| NUMT loci | |||||||||||||
| heterozygous loci | homozygous loci | ||||||||||||
| clade | strains | Es9* | Em3 | Es12 | Es13 | Ip | Js18 | Js19 | Jm4 | Ms24 | As1 | Am1* | |
| 1 | CBS 185T | − | − | − | − | + | − | − | − | − | − | na | |
| 2 | CBS 2001 | − | − | − | − | + | − | − | − | − | − | + | |
| 3 | CBS 2007 | − | − | − | − | na | − | − | − | − | − | + | |
| 2/4 | CBS8045 | −a | − | − | − | + | − | − | − | − | − | +b | |
| 2/3 | CBS 5730 | −c | − | − | − | na | − | − | − | − | − | +b | |
| 2/4 | CBS 7064 | +/− | +/− | +/− | +/− | +/− | +/− | +/− | +/− | +/− | +/+ | +/+ | |
The NUMT loci are called with their cognate chromosome letter (upper case) followed by the NUMT number in the sequenced reference strain CBS 7064 [21].
(+) indicates presence of the NUMT; (−) absence of the NUMT; (+/−) both alleles. In CBS 7064, the (+) loci are clade 4, except that on chromosome I which is clade 2.
indicate that the PCR products were sequenced; aclade 1 sequence; b clade 2 sequence; c clade 3 sequence.
na: no amplification.
During the analysis of a NUMT locus on chromosome E (NUMT s9) [21], we discovered that the sequence of the heterozygous allele on chromosome I of CBS 7064 was identical to that of clade 1 CBS 185T and displayed 9% divergence with the sequence of the parentCBS 2001(clade 2), which contributed the rest of the chromosome. Moreover,the clade 2/clade 4 hybrid CBS 8045 (see above) also displayed the clade 1 sequence for this locus. These observations suggest that some genetic exchange has occurred between clades 1 and 2 after reproductive isolation, in agreement with the presence of an identical mtDNA in most strains of clade 1 and clade 2.
Phylogeny of the M. farinosa complex in the CTG clade and taxonomic considerations
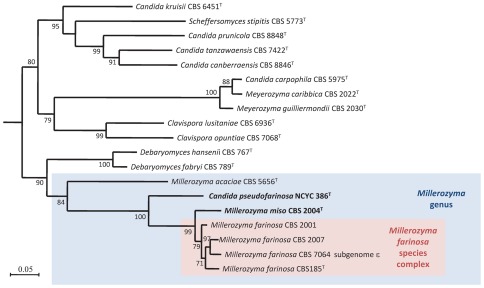
The M. farinosaspecies was shown to be made of related, but distant clades, and it was therefore necessary to establish a phylogenetic analysis of the species and clades constituting the genus Millerozyma based on a reliable multigenic analysis. Kurtzman et al [35]has circumscribed the genus Pichia, which now contains the type strain of the genus, Pichia membranifaciens, and various species from other previously described genera such as Issatchenkia. Kurtzman and Suzuki [9] have renamed the genus to which P. farinosa belongs as Millerozyma. We generated a phylogeny with the typical strain of each of the four clades, the clade 4 (the Pε subgenome) parent of CBS 7064, NCYC 386T,and the type strains of the previously decribed Millerozymaspecies using the RPB1, RPB2 and ACT1 exon2 sequences (Figure 3). All of the M. farinosa typical strains formed a monophyletic group with NCYC 386T having a basal position. We propose that the four closely related clades 1 to 4 that we have defined in this study constitute the species M. farinosa. NCYC 386T and CBS 2004T clearly defined two distinct taxa; the high sequence divergence that these two strains display from the other strains of the complex leads us to propose that they can be considered as a new species. We therefore propose to reinstate the species Zygopichia miso (type strain CBS 2004T = CLIB 1230T) previously used for CBS 2004T by Mogi in 1942 (but which had not been described) under the name Millerozyma miso and we would like to define a new species Candida pseudofarinosa (type strain NCYC 386T = CLIB 1231T).
Figure 3. Maximum likelihood tree of the concatenated ACT1, RPB1 and RPB2 coding genes of Millerozyma acaciae, Candida pseudofarinosa, Millerozyma miso type strains, of the typical strains of the M. farinosacomplex and of the most closely related species type strains or strains with a sequenced genome.
Candida fermenticarens CBS 704T was used as outgroup. Bootstrap values (%) based on 100 replicates are indicated at the nodes. Only bootstrap values over 70% are indicated. Most of the DNA sequences were retrieved from GenBank. Bar, 0.05 substitutions per site.
Latin diagnosis of Millerozyma miso Mogi exMallet, Weiss, Jacques etCasaregola
In agaro YPD post dies 2 ad 28°C cellulae vegetativae ovoidae aut oblongae, per gemmationem multipolarem reproducentes. Cultura est granula, albida vel cremea, margo glabro vel undulato.In agaro Zea mayidis pseudomycelia formatur.Asci non conjugate et non rumpuntur. Habentes 3–4 ascosporae globosae.
Glucosum, galactosum fermentatur. Maltosum, sucrosum, trehalosum, melibiosum, lactosum, cellobiosum, melezitosum et raffinosum non fermentatur.
Glucosum, galactosum, d-ribosum, l-rhamnosum, trehalosum, erythritolum, gluconatum et glycerolum assimilantur. Xylosum, sucrosum, maltosum, lactosum, cellobiosum, α-methylum-d-glucosidum, raffinosum,melezitosum, inositolum,acidum lacticum, sorbosum, d-glucosaminum, l-arabinosum, melibiosum, et sodium glucuronatum non assimilantur.Ethylaminum, lysinum et cadaverinum, assimilantur at non nitratum, nitritum creatinum etcreatininum.Vitamina externa crescentia non sunt necessaria. Crescere potest in 10% NaCl et 50% glucosum. 0.01% cycloheximidi non crescit.Maxima temperature crescentiae: 42°C. Materia amyloidea iodophila, acidum aceticum non formantur. Diazonium caeruleum B non respondens. Ureum non hydrolysatur.
Typus stirps CBS 2004T( = CLIB 1230T)
Description of Millerozyma miso Mogi exMallet, Weiss, Jacques &Casaregola
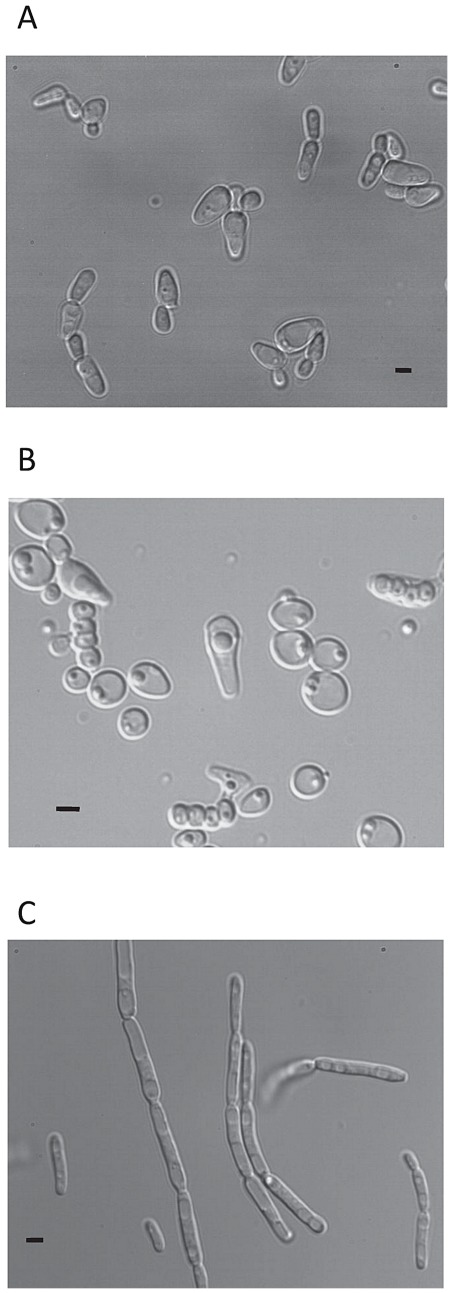
After 48 h at 28°C on YPD ovoid or oblong vegetative cells are formed (Figure 4A). Cell division is by multipolar budding. After two days on YPD agar the streak is rough, white to cream colored with a slightly undulating margin. Pseudomycelium wasobserved onculture corn meal agar.Formation of ascospores was observed. Asci are persistent and produce 3–4 spherical ascospores (Figure 4B).
Figure 4. Micrograph of cells.
(A): vegetative cells grown on YPD agar for three days at 28°C of Millerozyma miso CBS 2004T ; (B) sporulating cells after two weeks on Malt agar at room temperature ofCBS 2004T.and (C) vegetative cells grown on YPD agar for three days at 28°C of Candida pseudofarinosa sp.nov. NCYC 386T.Bar,5 µm.
Fermentation: d-glucose andd-galactose were fermented. d-maltose, saccharose, trehalose, melibiose, lactose, cellobiose, melezitose and raffinose are not fermented.
Assimilation reaction and other growth tests: d-glucose, d-galactose, d-ribose, l-rhamnose, trehalose, erythritol, 2-keto-d-gluconate, d-gluconate and glycerol are assimilated. d-xylose, sucrose, d-maltose, d-lactose, d-cellobiose, α-methyl-d-glucoside, d-raffinose, d-melezitose, inositol, d l-lactate, l-sorbose, d-glucosamine, l-arabinose, d-melibiose, sodium glucuronate, are not assimilated. Ethylamine, l-lysine and cadaverine are assimilated; potassium nitrate, sodium nitrite, creatine and creatinine are not assimilated.Growth in vitamin-free medium, 10% NaCl and 50% glucose are positive. 0.01% cycloheximide is negative. Maximum growth temperature is 42°C. Starch-like compounds and acetic acid are not produced. Diazonium blue B reaction is negative. Urease activity is negative.
Synonym: Zygopichia miso Mogi, J. Agric. Chem. Soc. Japan 18∶543-554, 629–641, 733–741, 940–944, 1942
Type: CBS 2004T( = CLIB 1230T)
Latin diagnosis of Candida pseudofarinosa Mallet, Weiss, Jacques et Casaregola sp. nov
In agaro YPD post dies 6 ad 28°C cellulae vel elongatae pseudohyphae formantes, per gemmationem multipolarem reproducentes. Cultura est butyrosa, sulcuma, centrum sublatum est, albida. In agaro Zea mayidis pseudomycelia formatur.Ascosporae nullae.
Glucosum, galactosum, maltosum, sucrosum, trehalosum, melibiosum, lactosum, cellobiosum, melezitosum et raffinosum non fermentatur.
Glucosum, gluconatum et galactosum assimilantur. D-ribosum, xylosum, L-rhamnosum, sucrosum, maltosum, lactosum, trehalosum, cellobiosum, alpha-methylum-D-glucosidum, raffinosum, melezitosum, glycerolum, erythritolum, inositolum, sorbitolum, D-gluconatum, acidum lacticum, sorbosum, D-glucosaminum, L-arabinosum, melibiosum, sodium glucuronatum et N-acetyl-glucosaminum non assimilantur.Nitratum, ethylaminum, lysinum, cadaverinum, creatinum et creatininum assimilantur at non nitritum et glucosaminum.Vitamina externa crescentia non sunt necessaria. Crescere potest in 10% NaCl et 50% glucosum. 0.01% cycloheximidi non crescit.Maxima temperature crescentiae: 40°C. Materia amyloidea iodophila, acidum aceticum non formantur. Diazonium caeruleum B non respondens. Ureum non hydrolysatur.
Typus stirps NCYC 386T( = CLIB 1231T)
Description of Candida pseudofarinosa Mallet, Weiss, Jacques &Casaregolasp. nov
After 6 days at 28°C on YPD, cells are elongated and mycelial pseudohyphae are formed (Figure 4C). Budding is multipolar. After two days on YPD agar the streak is butyrous, wrinkled, moderately raised and white-colored. Pseudomycelium was observed on culture corn meal agar. Formation of ascospores was not observed.
Fermentation: D-glucose, D-galactose, D-maltose, saccharose, trehalose, melibiose, lactose, cellobiose, melezitose and raffinose are not fermented.
Assimilation reaction and other growth tests: D-glucose, D-gluconate and D-galactose are assimilated. D-ribose, D-xylose, L-rhamnose, sucrose, D-maltose, D-lactose, trehalose, D-cellobiose, alpha-methyl-D-glucoside, D-raffinose, D-melezitose, glycerol, erythritol, inositol, D-sorbitol, 2-keto-D-gluconate, D L-lactate, L-sorbose, D-glucosamine, L-arabinose, D-melibiose, sodium glucuronate, N-acetyl-glucosamine are not assimilated. Potassium nitrate, ethylamine, L-lysine, cadaverine, creatine and creatinine are assimilated; sodium nitrite and glucosamine are not assimilated. Growth in vitamin-free medium, 10% NaCl and 50% glucose are positive. 0.01% cycloheximide is negative. Maximum growth temperature is 40°C. Starch-like compounds and acetic acid are not produced. Diazonium blue B reaction is negative. Urease activity is negative.
The type strain, NCYC 386T ( = CLIB 1231T), was isolated in Japan and was deposited at National Collection Yeast Culture by IFO collection during December 1953.
Discussion
Species whose taxonomy is debatable may reflect the possible existence of a species complex containing cryptic species. By evaluating the sequence divergence in regions little subjected to selective pressure, we have shown that M. farinosa was composed of at least four diverged taxa. In addition, two taxa that presented high sequence divergence were considered as new species: Candida pseudofarinosa and Millerozyma miso. We have previously shown that another CTG species, D. hansenii, whose taxonomy has been discussed over the years [36], [37] is a species complex with at least three distinct taxa [23]. Similarly, a third CTG species, Candida parapsilosis was also shown to be a complex made of three species [38].
Since the discovery of the inter-specific hybrid lager yeast S. pastorianus strains [39], a large number of hybrids have been detected in food [40] and in fermentation environments such as cider making [41], [42], wine making [43], and brewing [44]. As in the Saccharomyces, in which hybrids provide new technological potentialitiessuch as specific fermentations [45], hybrids of the CTG clade may form in constrained environmentsand generate new (and beneficial) properties to the cells. In this context, it isperhaps relevant that CBS 7064 and CBS 8045, the sole clade 2/clade 4 hybrids, were both isolated in very high sugar concentrations; the association of genetic material from these two clades may increase resistance to osmotic pressure. However, our test in the laboratory for the growth in high salt concentration (4 M NaCl) did not differentiate these two strains from the other M. farinosa isolates (data not shown), in contrast to what was described by [15]; thus new phenotypes associated with hybrid formation in M. farinosa remains to be found.
In the M. farinosa hybrids, sequence divergence in the ACT1 intron varied between 8.5 and 14.5%. The overall average genomic divergence between the contributors of CBS 7064 is 15.3% [21], consistent with the value of 15% for the introns of ACT1 and RPL33. Introns are therefore good indicators of overall sequence divergence in yeast genomes, and are superior to sequences like the rDNA D1/D2 [8] or the ACT1coding gene [46].
In the M. farinosahybrids, the overall divergence may be reduced by LOH. Indeed, massive LOH was observed in M. farinosa CBS 7064 to the point where some of the chromosome pairs became entirely homozygous [21]. We confirmed that LOH also occurred at high frequency in other M. farinosa hybrids.The distribution of markers in the diploid spore G6showed that the genetic information from one of the parents became predominant. This processseems to vary according to the strain analyzed, as seen from the comparison betweenCBS 7064and spore G6 of CBS 2006. In our study of LOH in a large number of diploids inD. hansenii,we observed a tendency, which favored only one parental contributor [47]. The present study demonstrates that M. farinosa shares common featureswith other described CTG species such as C. albicans [48], Candidadubliniensis [49], and D. hansenii, for the existence of hybrids and LOH. In addition, natural populations of both haploids and hybrids exist in M. farinosa and D. hansenii.
In the case of hybrids, LOH would certainly alleviate the heterozygosity-driven inhibition of meiotic recombination. Liti et al [29] have observed a threshold of sequence divergence (around 5%), above which the yield of viable spores in S. cerevisiaeis greatly diminished. Sequence divergence(as low as 3.6% in populations of S. paradoxus) was shown to drastically lower spore viability [50]. The sequence divergence between the M. farinosa clades (from 8.5 to 14%) is well above these thresholds and would almost certainly prevent recombination at meiosis if meiosis occurred. Consistent with this observation, our study has shown that there is no shuffling of alleles between analyzed strains using two markers. This suggests that M. farinosa has mainly a clonal propagation. Hence, it is possible that the concept of species defined by fecundity as described by Liti et al [29] for the Saccharomyces does not apply to yeasts of the CTG clade.
Although most of the M. farinosa hybrids did sporulate, one can predict that they do not undergo meiosis. In Zygosaccharomyces bailii, a species that is closer to the Saccharomyces and does not belong to the CTG clade, a lack of meiotic spores was observed. This was explained by the absence of nuclear fusion after mating [51]. In our study, none of the 52 spores of CBS 7064 and none of the 70 spores of CBS 2006 underwent meiosis, with the possible exception of one spore from CBS 2006 (spore 28). The reason why these yeasts undergo sporulation, but do not perform meiosis is unknown. This is reminiscent of C. lusitaniae, whose progeny from crosses was complexand contained 70% haploid spores, 21% diploid spores with the rest being aneuploid [5]. Obviously, the rarity of haploid spores in M. farinosa may be due to the important sequence divergence between the parents, which was not the case in C. lusitaniae. On the other hand, all the spores from D. hanseniiinter-clade hybrids are also diploid (our unpublished observation), though in this case the sequence divergence was lower (at most, 3%) than that of M. farinosa clades. This observationsuggests that sequence divergence may not be the only reason for the lack of meiosis in these species. Alternatively, in these species, a 3% sequence divergence may still be too high to allow meiosis to occur. It has been proposed that errors in meiosis such as meiosis I non-disjunction or precocious sister segregationor a possibleabsence of synaptonemal complex could lead to aneuploid and diploid sporesin C. lusitaniae [5]. Indeed, sporulation of inter-specific hybrids generates aneuploids in Saccharomyces [52], because the reduction of meiotic recombination causes a non-disjunction of chromosomes [53]. Therefore, the lack of recombination at meiosis couldexplain the death of aneuploids in M. farinosa.
After sporulation, M. farinosa could undergo LOH leading to a reduced heterozygosity allowing meiosis in the resulting organism. The study of the spore G6 confirmed that LOH could be massive in M. farinosa, as seen in M. farinosa CBS 7064. As in C. albicansin which stress affects LOH [54], stress caused by sporulation could increase LOH in M. farinosa. Figure 5 summarizes the various situations with regard to meiosis and sexual reproduction in four studied species from the CTG clade. The production of aneuploids by hybrids may be a common feature in the CTG clade, since aneuploids were also described in the species complex C. parapsilosis [55]. Interestingly, these authors also observed genetic exchanges in this species that are similar to that which we describe between M. farinosaclade 1 and 2.
Figure 5. Diversity of the mode of propagation in some yeasts of the CTG clade.nd: non determined.
The progeny of some of the CTG yeast species can therefore be complex, contributing to the diversity in this clade. M. farinosa is a mix of haploid clones belonging to different clades, hybrids resulting from mating between these clades and subjected to LOH and aneuploids having variable DNA content resulting from the sporulation of the hybrids through abortive meiosis or possible other mechanisms. The haploid or close to haploid spores resulting from the hybrids therefore display a non-Mendelian marker inheritance. For instance, these haploids may be the ancestor of strains of M. farinosa clade 1 or clade 2, which have exchanged material, as deduced from the transmission of mtDNA and from the introgression of unknown extent that we have evidenced from the NUMT analysis. Together, these results indicate that hybridization followedbyLOH is an alternative sexuality for these very divergent strains, although arare occurrence of meiosis isnot excluded. In conclusion, M. farinosa constitutes a species complex made of diverged cryptic species, whose genomes are affected by secondary contacts and LOH. We hypothesizethat this situation could be the case for a number of species of the CTG clade. This ensemble of observations invites reflection on thespecies concept in this clade.
Materials and Methods
Yeast strains and cultivation
The yeast strains used in this study are from CBS (http://www.CBS.knaw.nl), CECT (http://www.cect.org/), CIRM-Levures (http://www.inra.fr/cirmlevures), DSMZ (http://www.dsmz.de/), MUCL (http://bccm.belspo.be/about/mucl.php), NBRC (http://www.nbrc.nite.go.jp/e/), NCYC (http://www.ncyc.co.uk/), PYCC (http://www.crem.fct.unl.pt/) and VTT (http://culturecollection.vtt.fi/). They are listed in Table 1. Cells were routinely grown in YPD medium (1% yeast extract, 1% peptone, 1% glucose) at 28°C.For sporulation, cells were streaked on sporulation Malt Agar (Difco) plates and were grown for two days at 28°C, then left at room temperature for two weeks. The resulting cells were suspended in 50 µl of zymolyase 100T (1 mg/ml in a 1 M sorbitol solution) for 15 min at 37°C. The 4-spore tetrads were dissected with a Singer MCM System series 200 micromanipulator. Each spore was transplanted on YPD agar plates and incubated at 28°C overnight.
PCR amplification
The sequences of the nucleotide primersused in this study are listed in Table S1. Primers weredesigned with Primer3 (http://fokker.wi.mit.edu/primer3) on the alignments of genes from complete sequence genomes (http://www.genolevures.org/). Some of the primers were already described [23].Genomic DNA was extracted as previously described [23].
NUMT analysis
Primers were designed on the complete sequence of M. (P. sorbitophila) farinosa CBS7064 around each NUMT locus. For the nine heterozygous loci (with the insertion being present on only one parental allele), we chose primer sequences that were identical on both alleles. Two pairs of primers were designed for each of the two homozygous NUMT loci to increase the chance to amplify these regions in all strains.
Cloning
PCR products were purified using Nucleospin extract II purification kit (Macherey-Nagel, Germany). They were cloned intoa pGemT Easy (Promega, France) using T4 DNA ligase (Promega). DH10B E. coli cells were transformed by electroporation with pGemT easy recombinant plasmids. Cells were selected for their ability to grow on 50 µg/ml ampicillin. Positive recombinant clones were chosen after PCR screening for the presence of insert by restriction analysis. Inserts were then sequenced using universal primers (T7 and SP6) or the primers used for the PCR amplification.
DNA sequence determination and phylogenetic analysis
PCR fragments were sequenced on both strands by Cogenics (Takeley, U.K.) and Eurofins MWG Operon (Ebersberg, Germany). Sequences were assembled with the phred/phrap/consed package. Sequences were analysed with various programs including BLAST implemented at NCBI (http://www.ncbi.nlm.nih.gov). Sequence alignments were generated by using ClustalX [56], MAFFT [57] or Muscle [58] and were manually adjusted with Genedoc (http://www.psc.edu/biomed/genedoc). Phylogenetic trees were reconstructed with Neighbor-Joining program implemented in ClustalX and in MAFFT or with Phyml [59]. Phylogenetic trees were visualized with NJplot [60].
Flow cytometry
Flow cytometry was performed as described in [47].
Nucleotide sequence and Mycobank accession numbers
The GenBank/EMBL/DDBJ accession numbers for sequences described in this article are listed in Table S2.The Mycobank (http://www.mycobank.org) accession numbers for Millerozyma miso sp. nov. and Candida pseudofarinosasp. nov. are MB 563405 and MB 563406, respectively.
Supporting Information
Neighbor-joining phylogram of the RPL33 gene intron of M. farinosa isolates. Bootstrap values (%) based on 1000 replicates are indicated at the nodes for main groups and clades. All positions containing gaps and missing data were eliminated from the dataset. Clades are indicated.Typical strains are in bold characters. Heterozygous alleles in hybrids were arbitrarily given the prefix A or B. Bar, 0.02 substitutions per site.
(TIF)
Fluorescence histograms of various M. farinosa isolates after propidium iodide staining.
(TIF)
Schematic representation of the localization of the studied markers on the CBS 7064 chromosomes. The representation of the CBS 7064 chromosomes is taken from [21]. Letters on the right indicate the chromosome name. The markers selected for PCR amplification and sequencing are represented by their gene names (Genolevures nomenclature). Simple name used in this study areindicated in red.Orange:clade 2 (Pγ subgenome). Green:divergent sequence from any of the sequences of clades 1, 2, 3 or 5, i.e. clade 4 (Pε subgenome).
(TIF)
Micrograph of sporulating cells after two weeks on Malt agar at room temperature of (A) CBS 7064 and (B) CBS 2006.Bar,5 µm.
(TIF)
Primers used in this study.
(DOCX)
Accession numbers.
(XLSX)
CBS 7064 markers used in this study.
(XLSX)
Sequence identity(%) of various markers of the M. farinosa typical strains (CBS 185T, CBS 2001, CBS 2007) and M. miso (CBS 2004T) with M. (P. sorbitophila) farinosa CBS 7064 and with each other.
(XLSX)
Acknowledgments
We thank Christelle Louis-Mondésir for expert technical assistance. We are grateful to the Génolevures consortium for the access to the genome sequence of CBS 7064. We thank our colleagues of the Génolevures consortium for helpful discussion.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by an AIP Bioresources INRA grant. This work has received funding from the European Community's Seventh Framework Programme (FP7, 2007–2013), Research Infrastructures action, under grant agreement No. FP7-228310 (EMbaRC project). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. . Microbiol Rev. 1988;52:536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sicard D, Legras JL. Bread, beer and wine: yeast domestication in the Saccharomyces sensu stricto complex. C R Biol. 2011;334:229–236. doi: 10.1016/j.crvi.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 3.Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Annu Rev Microbiol. 2005;59:233–255. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- 4.Heitman J. Microbial pathogens in the fungal kingdom. Fungal Biol Rev. 2011;25:48–60. doi: 10.1016/j.fbr.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reedy JL, Floyd AM, Heitman J. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol. 2009;19:891–899. doi: 10.1016/j.cub.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porman AM, Alby K, Hirakawa MP, Bennett RJ. Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc Natl Acad Sci U S A. 2011;108:21158–21163. doi: 10.1073/pnas.1112076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Walt JP, Taylor MB, Liebenberg NV. Ploidy, ascus formation and recombination in Torulaspora (Debaryomyces) hansenii. Antonie van Leeuwenhoek. 1977;43:205–218. doi: 10.1007/BF00395675. [DOI] [PubMed] [Google Scholar]
- 8.Kurtzman CP. Pichia E. C. Hansen amend. Kurtzman. In: Kurtzman CP, Fell JW, editors. The yeasts, a taxonomic study. Amsterdam: Elsevier; 1998. pp. 273–352. [Google Scholar]
- 9.Kurtzman CP, Suzuki M. Phylogenetic analysis of ascomycete yeasts that form coenzyme Q-9 and the proposal of the new genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces and Scheffersomyces. Mycoscience. 2010;51 [Google Scholar]
- 10.Paula CR, Sampaio MC, Birman EG, Siqueira AM. Oral yeasts in patients with cancer of the mouth, before and during radiotherapy. Mycopathologia. 1990;112:119–124. doi: 10.1007/BF00436507. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Martos P, Mira J, Galan F, Hernandez JM. Sexual forms of yeasts in clinical samples. Mycopathologia. 1996;136:67–70. doi: 10.1007/BF00437497. [DOI] [PubMed] [Google Scholar]
- 12.Anaissie E, Bodey GP, Kantarjian H, Ro J, Vartivarian SE, et al. New spectrum of fungal infections in patients with cancer. Rev Infect Dis. 1989;11:369–378. doi: 10.1093/clinids/11.3.369. [DOI] [PubMed] [Google Scholar]
- 13.Adler A, Hidalgo-Grass C, Boekhout T, Theelen B, Sionov E, et al. Pichia farinosa bloodstream infection in a lymphoma patient. J Clin Microbiol. 2007;45:3456–3458. doi: 10.1128/JCM.00788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriques de Miranda L, Appel KR, Seyfarth H. Pichia sorbitophila sp nov. Antonie Van Leeuwenhoek. 1980;46:157–159. doi: 10.1007/BF00444070. [DOI] [PubMed] [Google Scholar]
- 15.Lages F, Lucas C. Characterization of a glycerol/H+ symport in the halotolerant yeast Pichia sorbitophila. Yeast. 1995;11:111–119. doi: 10.1002/yea.320110203. [DOI] [PubMed] [Google Scholar]
- 16.Maresova L, Sychrova H. Physiological characterization of osmotolerant yeast Pichia sorbitophila and comparison with a putative synonym Pichia farinosa. Folia Microbiol (Praha) 2003;48:211–217. doi: 10.1007/BF02930958. [DOI] [PubMed] [Google Scholar]
- 17.Kurtzman CP, Smiley MJ, Baker FL. Scanning electron microscopy of ascospores of Debaryomyces and Saccharomyces. Mycopathologia. 1975;55:29–34. doi: 10.1007/BF00467088. [DOI] [PubMed] [Google Scholar]
- 18.Jung PP, Schacherer J, Souciet JL, Potier S, Wincker P, et al. The complete mitochondrial genome of the yeast Pichia sorbitophila. FEMS Yeast Res. 2009;9:903–910. doi: 10.1111/j.1567-1364.2009.00540.x. [DOI] [PubMed] [Google Scholar]
- 19.Jung PP, Friedrich A, Souciet JL, Louis V, Potier S, et al. Complete mitochondrial genome sequence of the yeast Pichia farinosa and comparative analysis of closely related species. Curr Genet. 2010;56:507–515. doi: 10.1007/s00294-010-0318-y. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki C, Yoshida N, Okano E, Kawasumi T, Kashiwagi Y. Cloning and chromosomal mapping of URA3 genes of Pichia farinosa and P. sorbitophila encoding orotidine-5′-phosphate decarboxylase. Yeast. 2003;20:905–912. doi: 10.1002/yea.1017. [DOI] [PubMed] [Google Scholar]
- 21.Leh Louis V, Despons L, Friedrich A, Martin T, Durrens P, et al. Pichia sorbitophila, an interspecies yeast hybrid, reveals early steps of genome resolution following polyploidization. G3. 2012;2:299–311. doi: 10.1534/g3.111.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsui CK, Daniel HM, Robert V, Meyer W. Re-examining the phylogeny of clinically relevant Candida species and allied genera based on multigene analyses. FEMS Yeast Res. 2008;8:651–659. doi: 10.1111/j.1567-1364.2007.00342.x. [DOI] [PubMed] [Google Scholar]
- 23.Jacques N, Mallet S, Casaregola S. Delimitation of the species of the Debaryomyces hansenii complex by intron sequence analysis. Int J Syst Evol Microbiol. 2009;59:1242–1251. doi: 10.1099/ijs.0.004325-0. [DOI] [PubMed] [Google Scholar]
- 24.Fritsch ES, Schacherer J, Bleykasten-Grosshans C, Souciet JL, Potier S, et al. Influence of genetic background on the occurrence of chromosomal rearrangements in Saccharomyces cerevisiae. BMC Genomics. 2009;10:99. doi: 10.1186/1471-2164-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 26.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee F-L, Lee C-F, Okada S, Uchimura T, Kozaki M. Chemotaxonomic comparison of Pichia farinosa, Pichia sorbitophila and Candida cacaoi Bull Jpn Fed Cult Collect. 1992;8:71–78. [Google Scholar]
- 28.Kurtzman CP, Robnett CJ. Multigene phylogenetic analysis of the Trichomonascus, Wickerhamiella and Zygoascus yeast clades, and the proposal of Sugiyamaella gen. nov. and 14 new species combinations. FEMS Yeast Res. 2007;7:141–151. doi: 10.1111/j.1567-1364.2006.00157.x. [DOI] [PubMed] [Google Scholar]
- 29.Liti G, Barton DB, Louis EJ. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics. 2006;174:839–850. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger KH, Yaffe MP. Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol. 2000;8:508–513. doi: 10.1016/s0966-842x(00)01862-x. [DOI] [PubMed] [Google Scholar]
- 31.Rainieri S, Kodama Y, Nakao Y, Pulvirenti A, Giudici P. The inheritance of mtDNA in lager brewing strains. FEMS Yeast Res. 2008;8:586–596. doi: 10.1111/j.1567-1364.2008.00363.x. [DOI] [PubMed] [Google Scholar]
- 32.Marinoni G, Lachance MA. Speciation in the large-spored Metschnikowia clade and establishment of a new species, Metschnikowia borealis comb. nov. FEMS Yeast Research. 2004;4:587–596. doi: 10.1016/j.femsyr.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Solieri L. Mitochondrial inheritance in budding yeasts: towards an integrated understanding. Trends Microbiol. 2010;18:521–530. doi: 10.1016/j.tim.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Sacerdot C, Casaregola S, Lafontaine I, Tekaia F, Dujon B, et al. Promiscuous DNA in the nuclear genomes of hemiascomycetous yeasts. FEMS Yeast Res. 2008;8:846–857. doi: 10.1111/j.1567-1364.2008.00409.x. [DOI] [PubMed] [Google Scholar]
- 35.Kurtzman CP, Robnett CJ, Basehoar-Powers E. Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Res. 2008;8:939–954. doi: 10.1111/j.1567-1364.2008.00419.x. [DOI] [PubMed] [Google Scholar]
- 36.Prillinger H, Molnar O, Eliskases-Lechner F, Lopandic K. Phenotypic and genotypic identification of yeasts from cheese. Antonie Van Leeuwenhoek. 1999;75:267–283. doi: 10.1023/a:1001889917533. [DOI] [PubMed] [Google Scholar]
- 37.Corredor M, Davila AM, Casaregola S, Gaillardin C. Chromosomal polymorphism in the yeast species Debaryomyces hansenii. Antonie Van Leeuwenhoek. 2003;83:215–222. doi: 10.1023/a:1023322301634. [DOI] [PubMed] [Google Scholar]
- 38.Tavanti A, Davidson AD, Gow NA, Maiden MC, Odds FC. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J Clin Microbiol. 2005;43:284–292. doi: 10.1128/JCM.43.1.284-292.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaughan-Martini A, Kurtzman CP. Deoxyribonucleic acid relatedness among species of the genus Saccharomyces Sensu Stricto. Int J Syst Bacteriol. 1985;35:508–511. [Google Scholar]
- 40.James SA, Bond CJ, Stratford M, Roberts IN. Molecular evidence for the existence of natural hybrids in the genus Zygosaccharomyces. FEMS Yeast Research. 2005;5:747–755. doi: 10.1016/j.femsyr.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Masneuf I, Hansen J, Groth C, Piskur J, Dubourdieu D. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl Environ Microbiol. 1998;64:3887–3892. doi: 10.1128/aem.64.10.3887-3892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Groth C, Hansen J, Piskur J. A natural chimeric yeast containing genetic material from three species. Int J Syst Bacteriol 49 Pt. 1999;4:1933–1938. doi: 10.1099/00207713-49-4-1933. [DOI] [PubMed] [Google Scholar]
- 43.Gonzalez SS, Barrio E, Gafner J, Querol A. Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res. 2006;6:1221–1234. doi: 10.1111/j.1567-1364.2006.00126.x. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez SS, Barrio E, Querol A. Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing. Appl Environ Microbiol. 2008;74:2314–2320. doi: 10.1128/AEM.01867-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gangl H, Batusic M, Tscheik G, Tiefenbrunner W, Hack C, et al. Exceptional fermentation characteristics of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. N Biotechnol. 2009;25:244–251. doi: 10.1016/j.nbt.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Daniel HM, Sorrell TC, Meyer W. Partial sequence analysis of the actin gene and its potential for studying the phylogeny of Candida species and their teleomorphs. Int J Syst Evol Microbiol. 2001;51:1593–1606. doi: 10.1099/00207713-51-4-1593. [DOI] [PubMed] [Google Scholar]
- 47.Jacques N, Sacerdot C, Derkaoui M, Dujon B, Ozier-Kalogeropoulos O, et al. Population polymorphism of nuclear mitochondrial DNA insertions reveals widespread diploidy associated with loss of heterozygosity in Debaryomyces hansenii. Eukaryot Cell. 2010;9:449–459. doi: 10.1128/EC.00263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forche A, May G, Magee PT. Demonstration of loss of heterozygosity by single-nucleotide polymorphism microarray analysis and alterations in strain morphology in Candida albicans strains during infection. Eukaryot Cell. 2005;4:156–165. doi: 10.1128/EC.4.1.156-165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson AP, Gamble JA, Yeomans T, Moran GP, Saunders D, et al. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res. 2009;19:2231–2244. doi: 10.1101/gr.097501.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuehne HA, Murphy HA, Francis CA, Sniegowski PD. Allopatric divergence, secondary contact, and genetic isolation in wild yeast populations. Curr Biol. 2007;17:407–411. doi: 10.1016/j.cub.2006.12.047. [DOI] [PubMed] [Google Scholar]
- 51.Rodrigues F, Ludovico P, Sousa MJ, Steensma HY, Corte-Real M, et al. The spoilage yeast Zygosaccharomyces bailii forms mitotic spores: a screening method for haploidization. Appl Environ Microbiol. 2003;69:649–653. doi: 10.1128/AEM.69.1.649-653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roeder GS. Chromosome synapsis and genetic recombination: their roles in meiotic chromosome segregation. Trends Genet. 1990;6:385–389. doi: 10.1016/0168-9525(90)90297-j. [DOI] [PubMed] [Google Scholar]
- 53.Szekvolgyi L, Nicolas A. From meiosis to postmeiotic events: homologous recombination is obligatory but flexible. FEBS J. 2010;277:571–589. doi: 10.1111/j.1742-4658.2009.07502.x. [DOI] [PubMed] [Google Scholar]
- 54.Forche A, Abbey D, Pisithkul T, Weinzierl MA, Ringstrom T, et al. Stress alters rates and types of loss of heterozygosity in Candida albicans. MBio. 2011;2 doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fundyga RE, Kuykendall RJ, Lee-Yang W, Lott TJ. Evidence for aneuploidy and recombination in the human commensal yeast Candida parapsilosis. Infect Genet Evol. 2004;4:37–43. doi: 10.1016/j.meegid.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 56.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 58.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 60.Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neighbor-joining phylogram of the RPL33 gene intron of M. farinosa isolates. Bootstrap values (%) based on 1000 replicates are indicated at the nodes for main groups and clades. All positions containing gaps and missing data were eliminated from the dataset. Clades are indicated.Typical strains are in bold characters. Heterozygous alleles in hybrids were arbitrarily given the prefix A or B. Bar, 0.02 substitutions per site.
(TIF)
Fluorescence histograms of various M. farinosa isolates after propidium iodide staining.
(TIF)
Schematic representation of the localization of the studied markers on the CBS 7064 chromosomes. The representation of the CBS 7064 chromosomes is taken from [21]. Letters on the right indicate the chromosome name. The markers selected for PCR amplification and sequencing are represented by their gene names (Genolevures nomenclature). Simple name used in this study areindicated in red.Orange:clade 2 (Pγ subgenome). Green:divergent sequence from any of the sequences of clades 1, 2, 3 or 5, i.e. clade 4 (Pε subgenome).
(TIF)
Micrograph of sporulating cells after two weeks on Malt agar at room temperature of (A) CBS 7064 and (B) CBS 2006.Bar,5 µm.
(TIF)
Primers used in this study.
(DOCX)
Accession numbers.
(XLSX)
CBS 7064 markers used in this study.
(XLSX)
Sequence identity(%) of various markers of the M. farinosa typical strains (CBS 185T, CBS 2001, CBS 2007) and M. miso (CBS 2004T) with M. (P. sorbitophila) farinosa CBS 7064 and with each other.
(XLSX)