Abstract
Background
Various Pseudomonas strains can use l-lactate as their sole carbon source for growth. However, the l-lactate-utilizing enzymes in Pseudomonas have never been identified and further studied.
Methodology/Principal Findings
An NAD-independent l-lactate dehydrogenase (l-iLDH) was purified from the membrane fraction of Pseudomonas stutzeri SDM. The enzyme catalyzes the oxidation of l-lactate to pyruvate by using FMN as cofactor. After cloning its encoding gene (lldD), l-iLDH was successfully expressed, purified from a recombinant Escherichia coli strain, and characterized. An lldD mutant of P. stutzeri SDM was constructed by gene knockout technology. This mutant was unable to grow on l-lactate, but retained the ability to grow on pyruvate.
Conclusions/Significance
It is proposed that l-iLDH plays an indispensable function in Pseudomonas l-lactate utilization by catalyzing the conversion of l-lactate into pyruvate.
Introduction
Pseudomonas strains are Gram-negative rod-shaped bacteria commonly found in soil, water, and plant and animal tissues [1], [2]. They have very simple nutritional requirements and can grow well with a single organic molecule such as lactate as the sole carbon and energy source. In the lactate utilization processes of Pseudomonas aeruginosa, P. putida, and P. stutzeri, lactate is first converted to pyruvate, and then metabolized through the tricarboxylic acid cycle [3]–[5]. However, the enzymes responsible for the oxidation of lactate to pyruvate in these Pseudomonas strains have not been identified and further studied.
NAD-Independent l-lactate dehydrogenases (l-iLDHs), which catalyze the oxidation of l-lactate to pyruvate by an FMN-dependent mechanism, are widely distributed among bacteria, yeast, and protists [6]–[8]. These enzymes have been studied extensively in Escherichia coli and Saccharomyces cerevisiae [7], [9]–[11]. In E. coli, l-iLDH is a peripheral membrane protein, while in S. cerevisiae, it is located in the mitochondrial intermembrane space [6], [10], [11]. l-iLDHs in E. coli and S. cerevisiae have been purified and further characterized [6], [10], [11]. They catalyze the oxidation of l-lactate to pyruvate through the respiratory electron transport chain in vivo and allow these strains to grow well in medium containing l-lactate as the sole carbon source [6], [9], [12].
Previous works have also confirmed the presence of l-iLDHs in P. aeruginosa, P. putida, and P. stutzeri [3]–[5], [13], [14]. However, l-iLDHs are only induced when these Pseudomonas strains are grown aerobically with l-lactate as the carbon source [3]–[5], [13]. Based on this observation, involvement of l-iLDHs in Pseudomonas l-lactate utilization has been speculated [3]–[5], [13]. As in E. coli, l-iLDHs in Pseudomonas strains are membrane-bound proteins [13], [14] and it is difficult to purify them. Therefore, there is a lack of information on the properties and functions of l-iLDHs in Pseudomonas strains.
In this study, a membrane-bound l-iLDH from P. stutzeri strain SDM was purified, and its encoding gene, lldD, was cloned, expressed, and characterized. A mutant of strain SDM was constructed by knockout of the lldD gene. The mutant was unable to grow with l-lactate as the sole carbon source, providing evidence for an indispensable role of l-iLDH in l-lactate utilization in this Pseudomonas strain.
Results and Discussion
Purification of l-iLDH
Purification of membrane-bound l-iLDHs in Pseudomonas species has never been reported. In this study, the membrane-bound l-iLDH in P. stutzeri SDM was solubilized with Triton X-100 and purified. The results of a typical purification procedure are summarized in Figure 1 and Table 1. The specific activity at the final step was 83.0 U mg−1 of protein, which was a 364.5-fold increase over that of crude cell extract. The molecular mass of l-iLDH was found to be 42.8±0.6 kDa (Figure 1), using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 12.5% gel.
Figure 1. SDS-PAGE analysis of the purified l-iLDH from P. stutzeri SDM.

Lane M, molecular weight markers; lane 1, crude extract; lane 2, membrane fraction; lane 3, Triton X-100 extract; lane 4, active fraction after ammonium sulfate precipitation; lane 5, active fraction after DEAE Sepharose fast flow column-based extraction; lane 6, the purified l-iLDH in P. stutzeri SDM. Protein bands were visualized by Coomassie Blue staining. The arrow indicates the purified l-iLDH in P. stutzeri SDM.
Table 1. Purification of l-iLDH from P. stutzeri SDM.
| Purification step | Specific activity (U mg−1) | Purification (fold) | Yield (%) |
| Crude cell extract | 0.2 | 1 | 100 |
| Membrane fraction | 0.7 | 2.9 | 61.0 |
| Triton X-100 extract | 2.5 | 11.1 | 48.2 |
| Ammonium sulfate fractionation | 6.5 | 28.3 | 21.5 |
| DEAE-Sepharose | 12.0 | 52.7 | 8.1 |
| Source 30Q | 83.0 | 364.5 | 2.6 |
Cofactor analysis of l-iLDH
The purified l-iLDH has an intense yellow color. The absorption spectrum of l-iLDH in the visible region has peaks at around 460 nm and 380 nm, suggesting that l-iLDH is a flavoprotein (Figure 2). The flavin was released from the protein by boiling. The released flavin was confirmed to be FMN based on its identical migration with authentic FMN on high-performance liquid chromatography (HPLC) (Figure S1). Analysis of the flavin content of several preparations of homogeneous enzyme identified the ratio of l-iLDH to FMN as 1.03. Therefore, the native enzyme contains one FMN per subunit (Figure S2).
Figure 2. UV-visible absorption spectrum of l-iLDH in P. stutzeri SDM.
The UV-visible spectrum was recorded with a spectral resolution of 0.5 nm. The absorption spectrum of l-iLDH has two peaks, at around 460 nm and 380 nm, suggesting the presence of flavin in purified l-iLDH.
Sequence analysis of l-iLDH
Edman degradation analysis revealed that the N-terminal amino acid sequence of l-iLDH from P. stutzeri SDM is MIISASTDYRAAA. Two internal peptides were obtained by electrospray ionization-tandem mass spectrometry (ESI-MS/MS) of the peptides resulting from trypsin digestion (AAGVTTLVFTVDMPTPGAR and DAVTFGADGIIVSNHGGR). The three peptides exhibit high similarity to the putative FMN-dependent l-lactate dehydrogenases in P. putida KT2440 (AAN70308.1) [15], P. aeruginosa PA01 (AAG08157.1) [16], and P. entomophila L48 (CAK13685.1) [17]. The l-iLDH coding gene (denominated as lldD in this work) was cloned from the genome of P. stutzeri SDM by PCR. Its sequence also exhibited high similarity to the genes encoding the putative l-lactate dehydrogenases from P. putida KT2440 [15], P. aeruginosa PA01 [16], and P. entomophila [17]. These results implied that the putative proteins might also exhibit l-iLDH activity in these Pseudomonas strains.
Considerable sequence identity exists between l-iLDH in P. stutzeri SDM and the proteins in the family of the l-α-hydroxyacid-oxidizing flavoproteins, including flavocytochrome b2 in S. cerevisiae (29% sequence identity) [18], l-iLDH in E. coli (83% sequence identity) [10], l-lactate oxidase in Aerococcus viridans (30% sequence identity) [19], long-chain α-hydroxy acid oxidase in rat kidney (32% sequence identity) [20], l-mandelate dehydrogenase in P. putida (38% sequence identity) [21], and glycolate oxidase in Spinacia oleracea (36% sequence identity) [22]. Based on the resolved crystal structure of flavocytochrome b 2 from S. cerevisiae, six conserved amino acid residues required for flavin binding and enzymatic catalysis were identified. As shown in Figure 3, according to bioinformatic analysis with the program CLUSTAL X [23], the residues are highly conserved in all the members of the l-α-hydroxyacid-oxidizing flavoproteins family, including l-iLDH from P. stutzeri SDM.
Figure 3. Sequence alignment of l-iLDH from P. stutzeri SDM with other FMN-dependent α-hydroxy acid oxidases.
LOX, l-lactate oxidase in A. viridans [19]; LCHO, long-chain α-hydroxy acid oxidase in rat kidney [20]; MDH, l-mandelate dehydrogenase in P. putida [21]; GOX, glycolate oxidase in S. oleracea [22]; FCB2, flavocytochrome b2 (l-iLDH) in S. cerevisiae [18]; l-iLDHPs, l-iLDH in P. stutzeri SDM; l-iLDHEc, l-iLDH in E. coli [9]. Red arrows indicate the highly conserved residues important for FMN binding and enzymatic catalysis across this class of enzymes. The boxed segments in l-mandelate dehydrogenase represent the internal sequences that were implicated in membrane association. The lined segments in l-iLDH in P. stutzeri SDM represent the peptides identified through Edman degradation analysis and ESI-MS/MS. The sequences were aligned with the program CLUSTAL X [23]. Parameters and color scheme were set as default.
Among the proteins compared in Figure 3, l-mandelate dehydrogenase in P. putida and l-iLDHs in P. stutzeri SDM and E. coli are all membrane-bound proteins [10], [13], [21]. In an earlier study, it was shown that l-mandelate dehydrogenase has an internal insertion which is responsible for membrane association [24], [25]. l-iLDHs from P. stutzeri SDM and E. coli also have an internal insertion very similar to that found in l-mandelate dehydrogenase, in contrast to the soluble l-α-hydroxyacid-oxidizing flavoproteins (Figure 3). Therefore, like l-mandelate dehydrogenase, l-iLDH from P. stutzeri SDM might also be anchored in the membrane through an internal membrane-anchoring segment (Figure 3).
Properties of l-iLDH
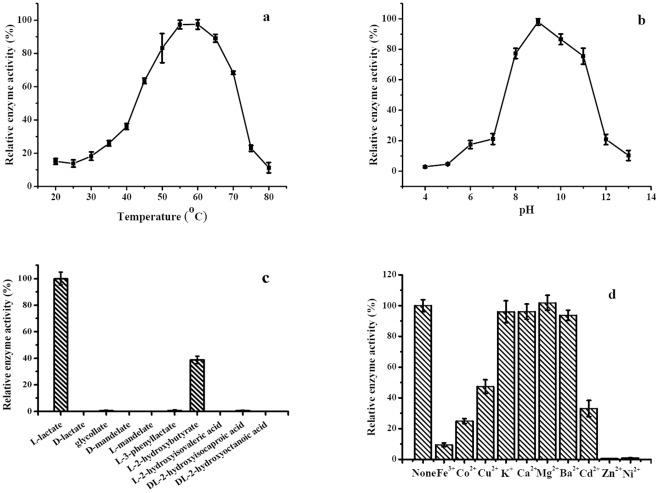
l-iLDH activities of various Pseudomonas strains have been characterized using the crude cell extract [3]–[5], [13], [14]. l-iLDH was expressed in E. coli C43 (DE3) and purified to homogeneity, as described in the “Materials and Methods” (Figure S3). The purified recombinant l-iLDH showed a UV-visible absorption spectrum and cofactor ratio similar to that of the native protein. Effect of temperature on activity of the recombinant l-iLDH in P. stutzeri SDM was investigated over a range of 20–80°C; maximum activity was observed at 55°C (Figure 4a). The pH dependence of the recombinant l-iLDH was also investigated over a range of 4.0–13.0. The results showed that l-iLDH exhibited maximum activity at pH 9.0 (Figure 4b).
Figure 4. Basic enzymatic properties of l-iLDH from P. stutzeri SDM.
The reaction mixture contained 20 mM l-lactate, 0.2 mM MTT, 50 mM Tris-HCl (pH 7.5), and 0.1 µg purified l-iLDH. (a) Effect of temperature on l-iLDH activity. Enzyme reactions were carried out at pH 7.5 at different temperatures (20–80°C). (b) Effect of pH on l-iLDH activity. The optimum pH was assessed in a buffer containing citric acid, KH2PO4, boric acid, and barbital (CKBB buffer) at various pHs (3.0–13.0) at 30°C. (c) Substrate specificity of l-iLDH from P. stutzeri SDM. The sodium salts of different α-hydroxy acids were used at a concentration of 20 mM. (d) Effect of different metal ions on l-iLDH activity. The concentration of metal ions was 5 mM in 50 mM Tris-HCl (pH 7.5). Values are the average ± SD of three separate determinations.
Substrate specificity of the recombinant l-iLDH from P. stutzeri SDM was examined with 20 mM α-hydroxy acids (l-lactate, d-lactate, glycollate, d-mandelate, l-mandelate, l-3-phenyllactate, l-2-hydroxybutyrate, l-2-hydroxyisovaleric acid, dl-2-hydroxyisocaproic acid, dl-2-hydroxyoctanoic acid) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) as the electron acceptor (Figure 4c). The P. stutzeri SDM l-iLDH seems to have narrow substrate specificity. Only l-lactate and l-2-hydrobutyrate were clearly oxidized by the enzyme. The same results were observed in E. coli, implying similar functions of l-iLDHs in l-lactate metabolism of E. coli and P. stutzeri SDM [9], [10]. In contrast, l-iLDH in Neisseria gonorrhoeae exhibited broader substrate specificity, which allowed the strain to catalyze the conversion of l-phenyllactate to phenylpyruvate [26].
The rate of dehydrogenation of l-lactate and l-2-hydrobutyrate catalyzed by the recombinant l-iLDH from P. stutzeri SDM followed Michaelis-Menten kinetics. Double-reciprocal plots of the initial rates plotted against the concentrations of l-lactate and l-2-hydroxybutylate were linear at a fixed concentration of MTT (0.2 mM), and yielded Km values of 29±0.65 μM and 99±3.9 μM, respectively, at 30°C. V max was estimated to be 332.3±5.4 μmol·min−1·mg−1 for l-lactate and 305.4±7.9 μmol·min−1·mg−1 for l-2-hydroxybutylate with MTT as the electron acceptor (Figure S4). The Km for l-lactate of l-iLDH in P. stutzeri SDM was lower than that of other homologous enzymes like l-iLDH in E. coli [10], flavocytochrome b 2 in S. cerevisiae [18], and l-lactate oxidase in A. viridans [19], in spite of some differences in the experimental conditions (Table S2). Inhibition of l-iLDH by oxalate and oxamate, which are canonical inhibitors of iLDHs [8], was also studied. Both oxalate and oxamate competitively inhibited l-iLDH activity (Figure S5), and their K i values were 1.9±0.1 mM and 29±1.7 mM, respectively.
The role of metal ions on activity of the recombinant P. stutzeri SDM l-iLDH was tested by adding metal salts (final concentration, 5 mM) to the assay buffer. As shown in Figure 4d, Ca2+, K+, Mg2+, and Ba2+ did not affect l-iLDH activity. However, Fe3+, Co2+, Cu2+, and Cd2+ partially decreased l-iLDH activity, while Zn2+ and Ni2+ completely inhibited l-iLDH activity. Experiments carried out with native protein gave similar results (data not shown).
Kinetic analysis of l-iLDH
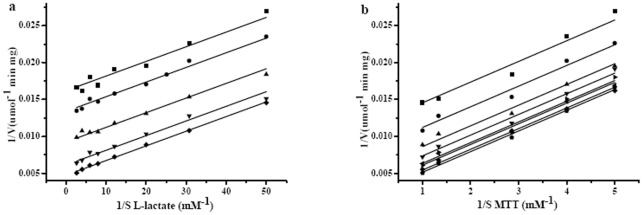
A set of parallel straight lines was obtained in the double-reciprocal plot of different concentrations of MTT plotted against the activity at fixed concentrations of l-lactate or different concentrations of l-lactate plotted against the activity at fixed concentrations of MTT (Figure 5a, 5b). This kinetic behavior is typical of a double-displacement kinetic (ping-pong) mechanism. A ping-pong kinetic mechanism for l-iLDH in P. stutzeri SDM is in agreement with previous reports describing the same mechanism for other l-α-hydroxyacid-oxidizing flavoproteins [19].
Figure 5. Kinetic mechanism of l-iLDH from P. stutzeri SDM.
Purified l-iLDH (0.1 µg) was incubated in a reaction mixture containing 50 mM Tris-HCl (pH 7.5) at 30°C. (a) The reaction was started with different l-lactate concentrations at variable MTT concentrations. ▪, 0.2 mM MTT; •, 0.25 mM MTT, ▴, 0.35 mM MTT; ▾, 0.75 mM MTT; ⧫, 0.1 mM MTT. (b) The reaction was started with different MTT concentrations at variable l-lactate concentrations. ▪, 0.02 mM l-lactate; •, 0.0325 mM l-lactate; ▴, 0.05 mM l-lactate; ▾, 0.0825 mM l-lactate; ◂, 0.125 mM l-lactate; ▸, 0.1675 mM l-lactate; ⧫, 0.25 mM l-lactate; <$>\scale 80%\raster="rg1"<$>, 0.375 mM l-lactate.
Function of l-iLDH in vivo
Many microorganisms can use l-lactate as their sole carbon source for growth [6]–[9], [27]. In these l-lactate utilization processes, l-lactate is first converted to pyruvate, which subsequently enters the tricarboxylic acid cycle [6]–[9], [12], [27]. Thus, the enzymes catalyzing l-lactate into pyruvate are very important for the survival of these microorganisms in habitats containing l-lactate [6]–[9], [12], [27]. In E. coli and Corynebacterium glutamicum, the l-lactate induced l-iLDHs oxidized l-lactate in vivo. Disruption of l-iLDH resulted in the loss of ability to utilize l-lactate as the sole carbon source [9], [28]. Constitutively expressed NAD-dependent l-lactate dehydrogenase would enable the l-iLDH inactivation mutant of C. glutamicum to grow on l-lactate [29]. In the present study, l-iLDH from P. stutzeri SDM was purified and characterized. The enzyme was able to catalyze the conversion of l-lactate into pyruvate in vitro (Figure S6). To identify the function of l-iLDH in vivo, insertional inactivation of the l-iLDH encoding gene, lldD, in P. stutzeri SDM was conducted using the pK18mob system.
We investigated whether the mutant is impaired in growth on solid minimal salt medium (MSM) with l-lactate as the sole carbon source. The mutant P. stutzeri SDM-dlldD exhibited little growth compared to the wild type (Figure S7a). As a control, both the wild-type and P. stutzeri SDM-dlldD strains grew equally well on solid MSM with 0.5% pyruvate as the sole carbon source (Figure S7b).
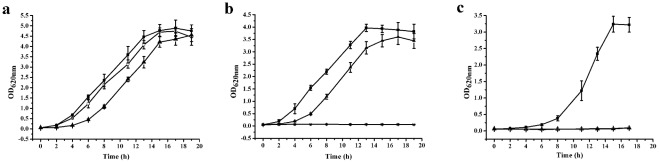
As with growth on solid MSM, the wild-type and P. stutzeri SDM-dlldD strains grew well in liquid MSM with 0.5% pyruvate as the sole carbon source (Figure 6a). However, compared with the wild type, P. stutzeri SDM-dlldD exhibited no growth in liquid MSM with 0.5% l-lactate as the sole carbon source (Figure 6b). Growth in MSM with l-lactate as the sole carbon source could be restored when P. stutzeri SDM-dlldD were complemented with a broad-host-range plasmid pBSP II SK(–) harboring the lldD gene. Altogether, these results suggest that the lldD gene encodes l-iLDH, which catalyzes the indispensable transformation of l-lactate to pyruvate in the P. stutzeri SDM l-lactate utilization process.
Figure 6. Time-course study of P. stutzeri SDM growth on media with.
(a) pyruvate, (b) l-lactate, and (c) d-lactate. (▪) Wild-type P. stutzeri SDM; (□) P. stutzeri SDM-dlldD; (▴) P. stutzeri SDM-dlldD harboring the plasmids pBSP II SKlldD. Data are presented as the average ± standard deviation values of 3 parallel replicates.
Interestingly, the l-iLDH inactivation mutant of P. stutzeri SDM also lost its ability to grow in liquid MSM with 0.5% d-lactate as the sole carbon source. Additionally, the expression of gene lldD was not able to complement the phenotype of d-lactate utilization (Figure 6c). In E. coli and C. glutamicum, the NAD-independent d-lactate dehydrogenase (d-iLDH) that catalyzes d-lactate oxidation in vivo was constitutively expressed, while l-iLDH was in the l-lactate utilization operon and controlled by the regulator LldR [9], [28], [30], [31]. In a previous study, d-iLDH activity in P. stutzeri SDM was studied. Both l-iLDH and d-iLDH were induced by either enantiomer of lactate in P. stutzeri SDM [13]. Because the insertion of pK18mob into the genome of P. stutzeri SDM influenced the transcription of the l-iLDH encoding gene and resulted in the inability of P. stutzeri SDM to use d-lactate as the sole carbon source, it was hypothesized that the l-iLDH and d-iLDH encoding genes might be in the same operon. Indeed, four adjacent genes (lldR, lldP, lldD, and dld-II) encoding a lactate-responsive regulator LldR, an l-lactate permease, l-iLDH, and a predicted d-iLDH, were annotated in the draft genome sequence of P. stutzeri SDM [32].
Many Pseudomonas strains have been isolated as opportunistic human pathogens. Pseudomonas infections often elevate human fluid NAD-dependent l-lactate dehydrogenase (l-nLDH) levels [1], [2], [33], [34]. The concentration of l-lactate, the product of l-nLDH-catalyzed pyruvate reduction, might also increase in human fluid during infection. Studies on some microorganisms such as Neisseria meningitidis and N. gonorrhoeae showed that, in addition to the general stimulation of metabolism, utilization of l-lactate in human hosts also promotes the production of some determinants of pathogenicity [35]–[38]. Thus, the utilization of l-lactate in human hosts has been found to increase the pathogenicity of these organisms [35]. As for the situation in different Pseudomonas strains, it has not been determined whether the metabolism of l-lactate would affect their pathogenicity. If a mutant pathogenic Pseudomonas strain incapable of utilizing l-lactate is isolated, a comparison of the pathogenicity between the wild-type and mutant strains might provide useful information related to the pathogenic mechanisms of Pseudomonas strains.
In summary, l-iLDH in l-lactate utilization strain P. stutzeri SDM was purified and further characterized. This key enzyme was confirmed to be required for the l-lactate utilization of strain SDM. Considering the high sequence identity among the lldD genes in various Pseudomonas strains, l-iLDHs might also play an indispensable function in the l-lactate utilization of other Pseudomonas strains.
Materials and Methods
Chemicals
l-Lactate, glycollate, d-mandelate, l-mandelate, l-3-phenyllactate, l-2-hydroxybutyrate, l-2-hydroxyisovaleric acid, dl-2-hydroxyisocaproic acid, dl-2-hydroxyoctanoic acid MTT, FMN, bovine serum albumin (BSA), isopropyl-β-d-1-thiogalactopyranoside (IPTG), phenylmethanesulfonyl fluoride (PMSF), and dithiothreitol (DTT) were purchased from Sigma. d-Lactate was purchased from Fluka. All other chemicals were of the reagent grade.
Bacteria and culture conditions
Bacterial strains used in this study are listed in Table 2. The P. stutzeri SDM (CCTCC no: M206010), isolated from soil, was cultured in MSM supplemented with different sole carbon source (10 g·L−1) at 30°C [13]. E. coli strains were grown in lysogenic broth (LB) medium at 37°C. Antibiotics were used, when appropriate, at the following concentrations: ampicillin (100 μg·mL−1), chloramphenicol (50 μg·mL−1), and kanamycin (50 μg·mL−1).
Table 2. The strains used in this work.
| Strain | Characteristics a | Source or reference |
| P. stutzeri SDM | Undomesticated wild strain capable of l-Lactate utilizing | [13] |
| P. stutzeri SDM-dlldD | Kmr; P. stutzeri SDM mutant obtained by disruption of the lldD gene | This study |
| P. stutzeri SDM-dlldD(pBSP II SKlldD) | Kmr; Apr; P. stutzeri SDM-dlldD harboring the plasmids pBSP II SKlldD | This study |
| E. coli DH5α | F− SupE44, Δ(argF lacZya)U169 ϕ80lacZΔM15 hsdR17(rk −,mk +) recA1 endA1 gyrA96 thi-1 relA1 λ− | Invitrogen |
| E. coli DH10B | F− endA1 recA1 galE15 galK16 nupG rpsL ΔlacX74 Φ80lacZΔM15 araD139 Δ(ara,leu)7697 mcrA Δ(mrr-hsdRMS-mcrBC) λ− | Invitrogen |
| E. coli HB101 | F− mcrB mrr hsdS20(rB − mB −) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20(SmR) glnV44 λ− | Invitrogen |
| E. coli C43 (DE3) | Mutant of E. coli BL21 (DE3) for membrane-bound protein expression | [39] |
Kmr and Apr indicate resistance to kanamycin and ampicillin, respectively.
Purification of l-iLDH
The membrane fraction of P. stutzeri SDM was prepared as described by Ma et al. [13]. Triton X-100 (10%, w/v) was added to the membrane fraction to a final concentration of 1 mg·mg−1 protein. The suspensions were stirred gently for 30 min, and then centrifuged at 140,000× g for 180 min at 4°C. The supernatants (detergent extracts) were gently transferred into other containers. Solid (NH4)2SO4 was slowly added to the detergent extracts to yield 30% saturation. When (NH4)2SO4 was completely dissolved, the solution was stirred for another 30 min before being centrifuged at 140,000 × g for 30 min at 4°C. More (NH4)2SO4 was added to the supernatant to reach 40% saturation, and the solution was stirred and centrifuged as before. The pellet was dissolved in buffer A: 50 mM Tris-HCl (pH 8.6) containing 1 mM DTT, 5 mM MgSO4, 0.1% Triton X-100, and 1 mM EDTA, and applied to a column of DEAE Sepharose Fast Flow equilibrated with buffer A. The column was washed with buffer B: 50 mM Tris-HCl (pH 8.6) containing 1 mM DTT, 200 mM KCl, 5 mM MgSO4, 0.1% Triton X-100, and 1 mM EDTA, at a flow rate of 5 mL·min−1. The fractions containing l-iLDH were concentrated by ultrafiltration and desalted with gel Sephadex G-25. The l-iLDH pool was then applied to a column of SOURCE 30Q pre-equilibrated with buffer A. The column was washed with a linear gradient of 0%–100% buffer B at a flow rate of 5 mL·min−1. The fractions containing l-iLDH were concentrated by ultrafiltration and analyzed by SDS-PAGE.
Analysis of N-terminal amino acid and microsequence
The purified l-iLDH was subjected to 12.5% SDS-PAGE for analysis. After electrophoresis, proteins in the gel were electrophoretically transferred to an Immobilon-PSQ PVDF membrane (Millipore) by using 25 mM Tris, 192 mM glycine, and 0.1% SDS as the cathode buffer, and 25 mM Tris, 192 mM glycine, and 20% methanol as the anode buffer. The bands on the PVDF membrane were excised, and sent to the Shanghai GeneCore BioTechnologies Co., Ltd., for N-terminal sequencing by automated Edman degradation.
For microsequencing of the internal peptides, the protein bands were cut from SDS-polyacrylamide gels and washed 3 times with water before being digested with trypsin. The resulting peptides were sent to the Shandong Academy of Medical Sciences for ESI-MS/MS.
Cloning of the l-iLDH encoding gene
The plasmids and primers used in this study are listed in Table 3. Degenerate primers P1, P2, and P3 were constructed according to the N-terminal and internal peptides resulting from the sequencing of P. stutzeri SDM l-iLDH. Primer P4 was constructed according to the conserved region of NAD-independent l-lactate dehydrogenase close to l-iLDH in Pseudomonas strains. The total genomic DNA of P. stutzeri SDM, extracted with the Wizard genomic DNA purification kit (Promega, Madison, WI), was used as the template for the first round of PCR with the primers P1 and P3. One microliter of the PCR products from the reaction mixture was used as the template for the second round of PCR with the primers P1 and P2. Using the genomic DNA of P. stutzeri SDM as the template, the third PCR was conducted with the primers P4 and P5. P5 was constructed according to the product of the second round of PCR. After sequencing of the three PCR products, the gene sequence of l-iLDH in P. stutzeri SDM was identified (Figure S8).
Table 3. The primers and plasmids used in this work.
| Plasmid or primer | Characteristics | Source or reference |
| Plasmid | ||
| pK18mob | Kmr; oriColE1 Mob+, lacZα; used for directed insertional disruption | [40] |
| pK18moblldD | Kmr; a pK18mob derivative containing an BamHI/HindIII 322bp internal fragment of the lldD gene from P. stutzeri SDM | This study |
| pETDuet-1 | Apr; vector for protein expression | Novagen |
| pET-LDH | pETDuet-1 with lldD gene of P. stutzeri SDM | This study |
| pBSP II SK(–) | Apr; broad-host-range expression vector | [41] |
| pBSP II SKlldD | Apr; pBSP II SK(–) with lldD gene of P. stutzeri SDM | This study |
| pRK600 | Cmr; oriColE1, RK2-Mob+, RK2-Tra+; helper plasmid for triparental matings | [42] |
| Primer | ||
| P1 | 5′- ATGATCATTWSNGCNGC -3′ b | This study |
| P2 | 5′- CGGCATRTCSACGGTGAASAC -3′ b | This study |
| P3 | 5′- GATGCCRTCSGCGCCGAAGGT -3′ b | This study |
| P4 | 5′- GCGCGCCGTCGGCGCGGATTTC -3′ | This study |
| P5 | 5′- CCAGCATCGCCCTGCGCCAG -3′ | This study |
| Pc1 | 5′- GGCAAGCTTATGATCATTTCCGCCTCTACC -3′ c | This study |
| Pc2 | 5′- ATTCTCGAGTCAGACGTCAGCAGACGTTGT -3′ c | This study |
| MF | 5′- GCGAAGCTTTAACATGTCGGAACTCAGCC -3′ c | This study |
| MR | 5′- ATAGGATCCGGTGGGCATGTCGACGGTGA -3′ c | This study |
| CF | 5′- CCGGTACCATGATCATTTCTGCTTCCACG -3′ c | This study |
| CR | 5′- CTGAGCTCTCAGACGTCAGCAGACGTTGT -3′ c | This study |
Kmr, Apr, and Cmr indicate resistance to kanamycin, ampicillin, and chloramphenicol, respectively.
W stands for one of A or T, S stands for one of C or G, R stands for one of A or G, and N stands for A, C, G, or T.
HindIII, XhoI, BamHI, KpnI and SacI restriction sites introduced in the primers are underlined.
Purification of recombinant l-iLDH
The l-iLDH encoding gene lldD, was amplified by PCR from the genome of P. stutzeri SDM by using the primers Pc1 and Pc2. The PCR product was digested by XhoI and HindIII, and cloned into the HindIII/XhoI sites of pETDuet-1 to construct the plasmid pET-LDH. E. coli C43 (DE3) carrying the pET-LDH plasmid was grown in LB medium (100 μg·mL−1 ampicillin) at 37°C to an optical density of 0.6 at 600 nm. Then, 1 mM IPTG was added to induce the expression of l-iLDH. Cells were harvested by centrifugation at 14,000× g for 5 min at 4°C and washed with 0.85% (w/v) sodium chloride (NaCl) solution. The cell pellets were subsequently suspended in the binding buffer (pH 7.4, 20 mM sodium phosphate, 20 mM imidazole, 500 mM NaCl, 1 mM PMSF, 0.1% Triton X-100, and 10% glycerol). Approximately 2–2.5 g of wet cells (suspended in 50 mL of binding buffer) were sonicated for 150 cycles (with 5 s at 40% of maximal output and 5 s rest in each cycle) with a Sonics sonicator (500 W/20 KHz, USA) in an ice bath. The sonicate was then centrifuged at 14,000× g for 20 min at 4°C. The supernatant was loaded onto a HisTrap HP column (5 mL), and eluted with 80% binding buffer and 20% elution buffer (pH 7.4, 20 mM sodium phosphate, 500 mM imidazole, 500 mM NaCl, 1 mM PMSF, 0.1% Triton X-100, and 10% glycerol) at a flow rate of 5 mL·min−1. The fractions containing l-iLDH were concentrated by ultrafiltration, desalted with Sephadex G-25, and then stored in 100 mM sodium phosphate buffer (pH 9.0, containing 50% glycerol and 0.1% Triton X-100) at −20°C. Under these conditions, the enzyme was stable for months with no apparent loss of activity.
Cofactor analysis of l-iLDH
UV-visible spectra of l-iLDH were recorded at 320–550 nm with an Ultrospec™ 2100 pro UV-visible Spectrophotometer (GE Healthcare Life Sciences). l-iLDH was heated to 100°C for 3 min, and then centrifuged at 10,000 × g for 10 min to remove denatured protein. The cofactor released from the purified protein was analyzed by HPLC (Agilent 1100 series; Hewlett-Packard, USA) using an ODS C18 column (4.6×150 mm, particle size: 5 μm) [43]. The eluent was 100 mM ammonium bicarbonate in 82%–18% methanol [43]. Standard FMN solutions of 0.05, 0.1, 0.15, 0.2, 0.25, and 0.3 mM were used for quantitative analysis and detected at 450 nm.
Knockout of gene lldD
To construct the P. stutzeri SDM-dlldD mutant strains, an internal fragment (322 bp) of the target lldD gene was PCR amplified by using the primers MF and MR, which contain HindIII and BamHI restriction enzyme sites, respectively. The PCR product was cloned into HindIII/BamHI-cut pK18mob, a mobilizable plasmid that does not replicate in Pseudomonas, to form pK18moblldD [44]. To transfer plasmid pK18moblldD into P. stutzeri SDM, a triparental filter mating was performed as previously described by using E. coli DH10B (pK18moblldD) as the donor strain, E. coli HB101 (pRK600) as the helper strain, and P. stutzeri SDM as the recipient strain [44]. P. stutzeri SDM exconjugants harboring the disrupted lldD gene (P. stutzeri SDM-dlldD) were isolated on minimal medium plates containing citrate that was selected for the Pseudomonas recipient cells and kanamycin that was selected for the insertion of the suicide vector after incubation at 30°C for 16 h [44]. The mutant strain was analyzed by PCR to confirm disruption of the target gene (Table S1 and Figure S9).
Complementation of P. stutzeri SDM-dlldD
To achieve complementation of the P. stutzeri SDM-dlldD mutant, the lldD gene of P. stutzeri SDM was amplified by PCR using the primers CF and CR, which contain KpnI and SacI restriction enzyme sites, respectively. The PCR products were cloned into the KpnI/SacI restriction sites (indicated in the primers) of the vector pBSP II SK(–) [41], which contains an IPTG inducible promoter. The recombinant plasmid pBSP II SKlldD was then introduced into the P. stutzeri SDM-dlldD mutant by triparental filter mating [44]. The P. stutzeri SDM-dlldD harboring the plasmid pBSP II SKlldD was isolated on minimal medium plates containing citrate, ampicillin, and kanamycin.
Growth in MSM
To test bacterial growth on solid media, wild-type and mutant strains were first streaked on LB agar medium and grown at 30°C. The next day, cells from single colonies on LB agar medium were picked and streaked onto solid MSM. Solid MSM was supplemented with l-lactate, d-lactate, or pyruvate as the sole carbon source.
To test bacterial growth in liquid medium, cells were first grown in LB medium until log phase, and then diluted with 20 mL of minimal medium containing l-lactate, d-lactate, or pyruvate as the sole carbon source. Cells were then grown at 30°C with shaking. IPTG was added at a final concentration of 1 mM to the minimal media. At various time points, the cell densities (OD620nm) of the cultures were recorded with an Ultrospec™ 2100 pro UV/visible spectrophotometer.
Biochemical assays
The activity of l-iLDH was determined at 30°C in 1 mL of 50 mM Tris-HCl (pH 7.5) and 0.2 mM MTT. The reaction was started by addition of l-lactate, and the rate of MTT reduction was determined by measuring the changes in absorbance at 578 nm [45]. One unit of l-iLDH activity was defined as the amount reducing 1.0 μmol of electron acceptor per minute under the test conditions. Protein concentrations were determined by the Lowry method, with BSA as the standard [46].
Polyacrylamide gel electrophoresis
SDS-PAGE was performed using a 12.5% polyacrylamide resolving gel and a 4% polyacrylamide stacking gel. Electrophoresis was run at a constant voltage of 80 V when the samples were in the stacking gel. When the dye front reached the resolving gel, the voltage was increased to 120 V. The run was stopped when the dye front was 2–3 mm away from the bottom edge of the gel.
Nucleotide sequence accession number
The nucleotide sequence of the lldD gene has been deposited in the GenBank nucleotide sequence databases under the accession no. GU373722.
Supporting Information
HPLC analysis of (a) authentic FMN, and (b) the cofactor released from the purified l-iLDH in P. stutzeri SDM. For the identification of the cofactor of l-iLDH, the purified l-iLDH was heated to 100°C for 3 min and then centrifuged at 10,000× g for 10 min to remove denatured protein. Cofactor released from purified protein was analyzed by HPLC (Agilent 1100 series, Hewlett-Packard, USA) using an ODS C18 column (4.6×150 mm, particle size: 5 μm). The eluent was 100 mM ammonium bicarbonate 82–18% methanol. As shown in Figure S1, a compound identical to authentic FMN was produced. Therefore, l-iLDH in P. stutzeri SDM used FMN as the cofactor.
(PDF)
Standard curve of FMN concentration related to peak area obtained by HPLC analysis. For the determination of the ratio between l-iLDH and FMN, 0.1171 mM l-iLDH was heated to 100°C for 3 min and then centrifuged at 10,000× g for 10 min to remove denatured protein. Cofactor released from purified protein was analyzed by HPLC (Agilent 1100 series, Hewlett-Packard, USA) using an ODS C18 column (4.6×150 mm, particle size: 5 μm). The eluent was 100 mM ammonium bicarbonate 82–18% methanol. Standard FMN solutions of 0.05, 0.1, 0.15, 0.2, 0.25, and 0.3 mM were used for quantitative analysis and detected at 450 nm. The concentration of the cofactor released from purified protein was determined to be 0.1136 mM. Thus, the ratio between l-iLDH and FMN was 0.1171/0.1136 = 1.03. Therefore, the native enzyme contains one FMN per subunit.
(PDF)
SDS-PAGE analysis of over-expression and purification of l-iLDH. Lane M, molecular weight markers; lane 1, whole cell proteins of E. coli C43(DE3) (pET-LDH); lane 2, crude extract of E. coli C43(DE3) (pET-LDH); 3, the purified recombinant L-iLDH.
(PDF)
Apparent Km of l-lactate and l-2-hydroxybutylate for l-iLDH in P. stutzeri SDM. The reaction mixture contained 0.2 mM MTT, 50 mM Tris–HCl (pH 7.5), and 0.1 μg purified l-iLDH. The reaction was started with variable l-lactate and l-2-hydroxybutylate concentrations. Double-reciprocal plots of the initial rates versus the concentration of l-lactate (a) and l-2-hydroxybutylate (b) were linear and yielded = the K m values of 29±0.65 μM and 99±3.9 μM, respectively, at 30°C. The V max values were estimated to be 332.3±5.4 μmol min−1 mg−1 for l-lactate, and 305.4±7.9 μmol min−1 mg−1 for l-2-hydroxybutylate, with MTT as the electron acceptor. The dispersion values indicate the standard errors of the mean (SEM) of the linear regression analysis of one experiment.
(PDF)
Inhibition of l-iLDH by oxalate and oxamate. Purified l-iLDH (0.1 μg) was incubated in the reaction mixture contained 0.2 mM MTT and 50 mM Tris–HCl (pH 7.5) at 30°C. (a) The reaction was started with different l-lactate concentrations at variable oxalate concentrations. ▪, no oxalate; •, 1.25 mM oxalate; ▴, 2.5 mM oxalate; ▾, 3.75 mM oxalate; ◂, 5 mM oxalate. (b) The reaction was started with different l-lactate concentrations at variable oxamate concentrations. ▪, no oxamate; •, 2.5 mM oxamate; ▴, 5 mM oxamate; ▾, 20 mM oxamate; ◂, 31.25 mM oxamate. The patterns of double-reciprocal plots indicate a competitive inhibition for both chemicals. The K i values of oxalate and oxamate were estimated to be 1.9±0.1 mM and 29±1.7 mM, respectively. The dispersion values indicate the SEM of the linear regression analysis of one experiment.
(PDF)
HPLC analysis of the product of the reaction catalyzed by l-iLDH. (a), authentic l-lactate; (b), authentic pyruvate; (c) the reaction mixture after 4 h. To determine the l-lactate oxidation product of the l-iLDH in P. stutzeri SDM, biotransformation was carried out using l-lactate (10 mM) and MTT (5 mM) as the substrate and purified l-iLDH as the biocatalyst in 50 mM Tris–HCl (pH 7.5) at 30°C. After 4 h of reaction, the mixture was centrifuged at 20,000× g for 15 min. The supernatant was analyzed by a HPLC system (Agilent 1100 series, Hewlett-Packard, USA) equipped with an Aminex HPX-87H column (Bio-Rad). The mobile phase (10 mM H2SO4) was pumped at 0.4 ml min−1 (55°C). As shown in Figure S6c, a compound identical to authentic pyruvate was produced. Similar to other reported l-iLDH, the l-iLDH in P. stutzeri SDM also catalyzes the conversion of l-lactate into pyruvate.
(PDF)
The lldD gene is required for growth on l-lactate. (a), growth of wild-type and mutant strains of P. stutzeri SDM on solid minimal media containing 0.5% pyruvate as the sole carbon source. (b), growth of the same set of strains (shown in panels a) on solid minimal media containing 0.5% l-lactate as the sole carbon source. We constructed the P. stutzeri SDM mutants lacking the lldD. Whether the mutants were impaired in growth on solid minimal medium with 0.5% l-lactate as the sole carbon source was tested. As shown in Figure S7b, the mutant exhibited little growth compared to the wild type. As a control, both the wild type and the mutant grew equally well on solid minimal medium with 0.5% pyruvate as the sole carbon source (Figure S7a).
(PDF)
Scheme for the lldD gene cloning procedure.
(PDF)
PCR analysis for verification of the insertional inactivation of the gene encoding l-iLDH. the l-iLDH encoding gene lldD . (A) Structure of pK18moblldD (left), the l-iLDH encoding gene lldD in P. stutzeri SDM genome (right), the insertional inactivated l-iLDH encoding gene lldD in P. stutzeri SDM genome (bottom). (B) PCR verification of the mutant strains (Figure S7) with the insertion of pK18mob in P. stutzeri SDM genome. In all cases, the primer pair VF1/VR1 (arrows; for the sequence, see Table S1) was used. (C) PCR verification of the mutant strains (Figure S7) with the homologus recombination between pK18moblldD and P. stutzeri SDM genome. In all cases, the primer pair VF2/VR2 (arrows; for the sequence, see Table S1) was used.
(PDF)
Primers used in the verification of the insertional inactivation of the l-iLDH encoding gene.
(DOC)
Comparison of K m values estimated for different enzymes.
(DOC)
Acknowledgments
The authors thank Professor Jan-Willem de Gier, for providing E. coli C43 (DE3) for protein expression.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The work was supported by the National Natural Science Foundation of China (31000014 and 31070062), and by the grants from the Ministry of Science and Technology of China, National Basic Research Program of China (2007CB707803), and the Research Fund for the Doctoral Program of Higher Education of China (Grant no. 20090131110036). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lalucat J, Bennasar A, Bosch R, García-Valdés E, Palleroni NJ, et al. Biology of Pseudomonas stutzeri. Microbiol Mol Biol Rev. 2006;70:510–547. doi: 10.1128/MMBR.00047-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev. 2011;35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- 3.Brown PR, Tata R. Glycollate inhibition of growth of Pseudomonas aeruginosa on lactate medium. J Gen Microbiol. 1987;133:1521–1526. doi: 10.1099/00221287-133-6-1521. [DOI] [PubMed] [Google Scholar]
- 4.Hao J, Ma C, Gao C, Qiu J, Wang M, et al. Pseudomonas stutzeri as a novel biocatalyst for pyruvate production from dl-lactate. Biotechnol Lett. 2007;29:105–110. doi: 10.1007/s10529-006-9204-6. [DOI] [PubMed] [Google Scholar]
- 5.O'Brien RW. Metabolism of d- and l-lactate by Pseudomonas putida. Aust J Biol Sci. 1977;30:553–558. doi: 10.1071/bi9770553. [DOI] [PubMed] [Google Scholar]
- 6.Chapman SK, Reid GA, Bell C, Short D, Daff S. Flavocytochrome b2: an ideal model for studying protein-mediated electron transfer. Biochem Soc Trans. 1996;24:73–77. doi: 10.1042/bst0240073. [DOI] [PubMed] [Google Scholar]
- 7.Garvie EI. Bacterial lactate dehydrogenases. Microbiol Rev. 1980;44:106–139. doi: 10.1128/mr.44.1.106-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jasso-Chávez R, Torres-Márquez ME, Moreno-Sánchez R. The membrane-bound l- and d-lactate dehydrogenase activities in mitochondria from Euglena gracilis. Arch Biochem Biophys. 2001;390:295–303. doi: 10.1006/abbi.2001.2353. [DOI] [PubMed] [Google Scholar]
- 9.Dong JM, Taylor JS, Latour DJ, Iuchi S, Lin ECC. Three overlapping lct genes involved in l-lactate utilization by Escherichia coli. J Bacteriol. 1993;175:6671–6678. doi: 10.1128/jb.175.20.6671-6678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Futai M, Kimura H. Inducible membrane-bound l-lactate dehydrogenase from Escherichia coli: purification and properties. J Biol Chem. 1977;252:5820–5827. [PubMed] [Google Scholar]
- 11.Xia ZX, Mathews FS. Molecular structure of flavocytochrome b2 at 2.4 Å resolution. J Mol Biol. 1990;212:837–863. doi: 10.1016/0022-2836(90)90240-M. [DOI] [PubMed] [Google Scholar]
- 12.Philippe G, Frédérique L, Michiel K, Pascal H. Major role of NAD-dependent lactate dehydrogenases in aerobic lactate utilization in Lactobacillus plantarum during early stationary phase. J Bacteriol. 2004;186:6661–6666. doi: 10.1128/JB.186.19.6661-6666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma C, Gao C, Qiu J, Hao J, Liu W, et al. Membrane-bound l- and d-lactate dehydrogenase activities of a newly isolated Pseudomonas stutzeri strain. Appl Microbiol Biotechnol. 2007;77:91–98. doi: 10.1007/s00253-007-1132-4. [DOI] [PubMed] [Google Scholar]
- 14.Kemp MB. d- and l-lactate dehydrogenases of Pseudomonas aeruginosa. Biochem J. 1972;130:307–309. doi: 10.1042/bj1300307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;12:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 16.Erdos G, Sayeed S, Hu FZ, Antalis PT, Shen K, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 17.Vodovar N, Vallenet D, Cruveiller S, Rouy Z, Barbe V, et al. Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat Biotechnol. 2006;24:673–679. doi: 10.1038/nbt1212. [DOI] [PubMed] [Google Scholar]
- 18.Black MT, White SA, Reid GA, Chapman SK. High-level expression of fully active yeast flavocytochrome b2 in Escherichia coli. Biochem J. 1989;258:255–259. doi: 10.1042/bj2580255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yorita K, Aki K, Sagai H, Misaki H, Massey V. l-Lactate oxidase and l-lactate monooxygenase: mechanistic variations on a common structural theme. Biochimie. 1995;77:631–642. doi: 10.1016/0300-9084(96)88178-8. [DOI] [PubMed] [Google Scholar]
- 20.Diep LeKH, Lederer F. Amino acid sequence of long chain alpha-hydroxy acid oxidase from rat kidney, a member of the family of FMN-dependent alpha-hydroxy acid-oxidizing enzymes. J Biol Chem. 1991;266:20877–20881. [PubMed] [Google Scholar]
- 21.Tsou AY, Ransom SC, Gerlt JA, Buechter DD, Babbitt PC, et al. Mandelate pathway of Pseudomonas putida: sequence relationships involving mandelate racemase, (S)-mandelate dehydrogenase, and benzoylformate decarboxylase and expression of benzoylformate decarboxylase in Escherichia coli. Biochemistry. 1990;29:9856–9862. doi: 10.1021/bi00494a015. [DOI] [PubMed] [Google Scholar]
- 22.Volokita M, Somerville CR. The primary structure of spinach glycolate oxidase deduced from the DNA sequence of a cDNA clone. J Biol Chem. 1987;262:15825–15828. [PubMed] [Google Scholar]
- 23.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sukumar N, Xu Y, Gatti DL, Mitra B, Mathews FS. Structure of an active soluble mutant of the membrane-associated (S)-mandelate dehydrogenase. Biochemistry. 2001;40:9870–9878. doi: 10.1021/bi010938k. [DOI] [PubMed] [Google Scholar]
- 25.Mitra B, Gerlt JA, Babbitt PC, Koo CW, Kenyon GL, et al. A novel structural basis for membrane association of a protein: construction of a chimeric soluble mutant of (S)-mandelate dehydrogenase from Pseudomonas putida. Biochemistry. 1993;32:12959–12967. doi: 10.1021/bi00211a003. [DOI] [PubMed] [Google Scholar]
- 26.Bhatnagar RK, HendryAT, Shanmugam KT, Jensen RA. The broad-specificity, membrane-bound lactate dehydrogenase of Neisseria gonorrhoeae: ties to aromatic metabolism. J Gen Microbiol. 1989;135:353–360. doi: 10.1099/00221287-135-2-353. [DOI] [PubMed] [Google Scholar]
- 27.Seki M, Iida K-I, Saito M, Nakayama H, Yoshida S-I. Hydrogen peroxide production in Streptococcus pyogenes: involvement of lactate oxidase and coupling with aerobic utilization of lactate. J Bacteriol. 2004;186:2046–2051. doi: 10.1128/JB.186.7.2046-2051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stansen C, Uy D, Delaunay S, Eggeling L, Goergen JL, et al. Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl Environ Microbiol. 2005;71:5920–5928. doi: 10.1128/AEM.71.10.5920-5928.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharkey MA, Maher MA, Guyonvarch A, Engel PC. Kinetic characterisation of recombinant Corynebacterium glutamicum NAD+-dependent LDH over-expressed in E. coli and its rescue of an lldD-phenotype in C. glutamicum: the issue of reversibility re-examined. Arch Microbiol. 2011;193:731–740. doi: 10.1007/s00203-011-0711-z. [DOI] [PubMed] [Google Scholar]
- 30.Kato O, Youn JW, Stansen KC, Matsui D, Oikawa T, et al. Quinone-dependent d-lactate dehydrogenase Dld (Cg1027) is essential for growth of Corynebacterium glutamicum on d-lactate. BMC Microbiol. 2010;10:321. doi: 10.1186/1471-2180-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neuner A, Heinzle E. Mixed glucose and lactate uptake by Corynebacterium glutamicum through metabolic engineering. Biotechnol J. 2011;6:318–329. doi: 10.1002/biot.201000307. [DOI] [PubMed] [Google Scholar]
- 32.Jiang TY, Gao C, Su F, Zhang W, Hu CH, et al. J Bacteriol; 2011. Genome sequence of Pseudomonas stutzeri SDM-LAC, a typical strain for studying the molecular mechanism of lactate utilization. doi: 10.1128/JB.06478-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Köse M, Oztürk M, Kuyucu T, Güneş T, Akçakuş M, et al. Community-acquired pneumonia and empyema caused by Pseudomonas stutzeri: a case report. Turk J Pediatr. 2004;46:177–178. [PubMed] [Google Scholar]
- 34.Rejman J, Di Gioia S, Bragonzi A, Conese M. Pseudomonas aeruginosa infection destroys the barrier function of lung epithelium and enhances polyplex-mediated transfection. Hum Gene Ther. 2007;18:642–652. doi: 10.1089/hum.2006.192. [DOI] [PubMed] [Google Scholar]
- 35.Smith H, Yates EA, Cole JA, Parsons NJ. Lactate stimulation of gonococcal metabolism in media containing glucose: mechanism, impact on pathogenicity, and wider implications for other pathogens. Infect Immun. 2001;69:6565–6572. doi: 10.1128/IAI.69.11.6565-6572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu K, Hassett DJ, Cohen MS. Oxidant stress in Neisseria gonorrhoeae: adaptation and effects on l-(-)-lactate dehydrogenase activity. Infect Immun. 1989;57:2173–2178. doi: 10.1128/iai.57.7.2173-2178.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yates E, Gao L, Woodcock N, Parsons NJ, Cole J, et al. In a medium containing glucose, lactate carbon is incorporated by gonococci predominantly into fatty acids and glucose carbon incorporation is increased; implications regarding lactate stimulation of metabolism. Int J Med Microbiol. 2000;290:627–639. doi: 10.1016/S1438-4221(00)80012-0. [DOI] [PubMed] [Google Scholar]
- 38.Parsons NJ, Emond JP, Goldner M, Bramley J, Crooke H, et al. Lactate enhancement of sialylation of gonococcal lipopolysaccharide and induction of serum resistance by CMP-NANA is not due to direct activation of the sialyltransferase: metabolic events are involved. Microb Pathog. 1996;21:193–204. doi: 10.1006/mpat.1996.0054. [DOI] [PubMed] [Google Scholar]
- 39.Dumon-Seignovert L, Cariot G, Vuillard L. The toxicity of recombinant proteins in Escherichia coli: a comparison of overexpression in BL21(DE3), C41(DE3), and C43(DE3). Protein Expr Purif. 2004;37:203–206. doi: 10.1016/j.pep.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 40.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 41.Schweizer HP. Vectors to express foreign genes and techniques to monitor gene expression in Pseudomonads. Curr Opin Biotechnol. 2001;12:439–445. doi: 10.1016/s0958-1669(00)00242-1. [DOI] [PubMed] [Google Scholar]
- 42.de Lorenzo V, Eltis L, Kessler B, Timmis KN. Analysis of Pseudomonas gene products using lacI q/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 43.Eppink MHM, Boeren SA, Vervoort J, Berkel WJHV. Purification and properties of 4-hydroxybenzoate 1-hydroxylase (Decarboxylating), a novel flavin adenine dinucleotide dependent monooxygenase from Candida parapsilosis CBS604. J Bacteriol. 1997;179:6680–6687. doi: 10.1128/jb.179.21.6680-6687.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiménez JI, Canales A, Jiménez-Barbero J, Ginalski K, Rychlewski L, et al. Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc Natl Acad Sci USA. 2008;105:11329–11334. doi: 10.1073/pnas.0802273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagel M, Andreesen JR. Purification and characterization of the molybdoenzymes nicotinate dehydrogenase and 6-hydroxynicotinate dehydrogenase from Bacillus niacini. Arch Microbiol. 1990;154:605–613. [Google Scholar]
- 46.Markwell MAK, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HPLC analysis of (a) authentic FMN, and (b) the cofactor released from the purified l-iLDH in P. stutzeri SDM. For the identification of the cofactor of l-iLDH, the purified l-iLDH was heated to 100°C for 3 min and then centrifuged at 10,000× g for 10 min to remove denatured protein. Cofactor released from purified protein was analyzed by HPLC (Agilent 1100 series, Hewlett-Packard, USA) using an ODS C18 column (4.6×150 mm, particle size: 5 μm). The eluent was 100 mM ammonium bicarbonate 82–18% methanol. As shown in Figure S1, a compound identical to authentic FMN was produced. Therefore, l-iLDH in P. stutzeri SDM used FMN as the cofactor.
(PDF)
Standard curve of FMN concentration related to peak area obtained by HPLC analysis. For the determination of the ratio between l-iLDH and FMN, 0.1171 mM l-iLDH was heated to 100°C for 3 min and then centrifuged at 10,000× g for 10 min to remove denatured protein. Cofactor released from purified protein was analyzed by HPLC (Agilent 1100 series, Hewlett-Packard, USA) using an ODS C18 column (4.6×150 mm, particle size: 5 μm). The eluent was 100 mM ammonium bicarbonate 82–18% methanol. Standard FMN solutions of 0.05, 0.1, 0.15, 0.2, 0.25, and 0.3 mM were used for quantitative analysis and detected at 450 nm. The concentration of the cofactor released from purified protein was determined to be 0.1136 mM. Thus, the ratio between l-iLDH and FMN was 0.1171/0.1136 = 1.03. Therefore, the native enzyme contains one FMN per subunit.
(PDF)
SDS-PAGE analysis of over-expression and purification of l-iLDH. Lane M, molecular weight markers; lane 1, whole cell proteins of E. coli C43(DE3) (pET-LDH); lane 2, crude extract of E. coli C43(DE3) (pET-LDH); 3, the purified recombinant L-iLDH.
(PDF)
Apparent Km of l-lactate and l-2-hydroxybutylate for l-iLDH in P. stutzeri SDM. The reaction mixture contained 0.2 mM MTT, 50 mM Tris–HCl (pH 7.5), and 0.1 μg purified l-iLDH. The reaction was started with variable l-lactate and l-2-hydroxybutylate concentrations. Double-reciprocal plots of the initial rates versus the concentration of l-lactate (a) and l-2-hydroxybutylate (b) were linear and yielded = the K m values of 29±0.65 μM and 99±3.9 μM, respectively, at 30°C. The V max values were estimated to be 332.3±5.4 μmol min−1 mg−1 for l-lactate, and 305.4±7.9 μmol min−1 mg−1 for l-2-hydroxybutylate, with MTT as the electron acceptor. The dispersion values indicate the standard errors of the mean (SEM) of the linear regression analysis of one experiment.
(PDF)
Inhibition of l-iLDH by oxalate and oxamate. Purified l-iLDH (0.1 μg) was incubated in the reaction mixture contained 0.2 mM MTT and 50 mM Tris–HCl (pH 7.5) at 30°C. (a) The reaction was started with different l-lactate concentrations at variable oxalate concentrations. ▪, no oxalate; •, 1.25 mM oxalate; ▴, 2.5 mM oxalate; ▾, 3.75 mM oxalate; ◂, 5 mM oxalate. (b) The reaction was started with different l-lactate concentrations at variable oxamate concentrations. ▪, no oxamate; •, 2.5 mM oxamate; ▴, 5 mM oxamate; ▾, 20 mM oxamate; ◂, 31.25 mM oxamate. The patterns of double-reciprocal plots indicate a competitive inhibition for both chemicals. The K i values of oxalate and oxamate were estimated to be 1.9±0.1 mM and 29±1.7 mM, respectively. The dispersion values indicate the SEM of the linear regression analysis of one experiment.
(PDF)
HPLC analysis of the product of the reaction catalyzed by l-iLDH. (a), authentic l-lactate; (b), authentic pyruvate; (c) the reaction mixture after 4 h. To determine the l-lactate oxidation product of the l-iLDH in P. stutzeri SDM, biotransformation was carried out using l-lactate (10 mM) and MTT (5 mM) as the substrate and purified l-iLDH as the biocatalyst in 50 mM Tris–HCl (pH 7.5) at 30°C. After 4 h of reaction, the mixture was centrifuged at 20,000× g for 15 min. The supernatant was analyzed by a HPLC system (Agilent 1100 series, Hewlett-Packard, USA) equipped with an Aminex HPX-87H column (Bio-Rad). The mobile phase (10 mM H2SO4) was pumped at 0.4 ml min−1 (55°C). As shown in Figure S6c, a compound identical to authentic pyruvate was produced. Similar to other reported l-iLDH, the l-iLDH in P. stutzeri SDM also catalyzes the conversion of l-lactate into pyruvate.
(PDF)
The lldD gene is required for growth on l-lactate. (a), growth of wild-type and mutant strains of P. stutzeri SDM on solid minimal media containing 0.5% pyruvate as the sole carbon source. (b), growth of the same set of strains (shown in panels a) on solid minimal media containing 0.5% l-lactate as the sole carbon source. We constructed the P. stutzeri SDM mutants lacking the lldD. Whether the mutants were impaired in growth on solid minimal medium with 0.5% l-lactate as the sole carbon source was tested. As shown in Figure S7b, the mutant exhibited little growth compared to the wild type. As a control, both the wild type and the mutant grew equally well on solid minimal medium with 0.5% pyruvate as the sole carbon source (Figure S7a).
(PDF)
Scheme for the lldD gene cloning procedure.
(PDF)
PCR analysis for verification of the insertional inactivation of the gene encoding l-iLDH. the l-iLDH encoding gene lldD . (A) Structure of pK18moblldD (left), the l-iLDH encoding gene lldD in P. stutzeri SDM genome (right), the insertional inactivated l-iLDH encoding gene lldD in P. stutzeri SDM genome (bottom). (B) PCR verification of the mutant strains (Figure S7) with the insertion of pK18mob in P. stutzeri SDM genome. In all cases, the primer pair VF1/VR1 (arrows; for the sequence, see Table S1) was used. (C) PCR verification of the mutant strains (Figure S7) with the homologus recombination between pK18moblldD and P. stutzeri SDM genome. In all cases, the primer pair VF2/VR2 (arrows; for the sequence, see Table S1) was used.
(PDF)
Primers used in the verification of the insertional inactivation of the l-iLDH encoding gene.
(DOC)
Comparison of K m values estimated for different enzymes.
(DOC)