Abstract
Protein molecules have evolved to adopt distinctive and well-defined functional and soluble states under physiological conditions. In some circumstances, however, proteins can self-assemble into fibrillar aggregates designated as amyloid fibrils. In vivo these processes are normally associated with severe pathological conditions but can sometimes have functional relevance. One such example is the hydrophobins, whose aggregation at air–water interfaces serves to create robust protein coats that help fungal spores to resist wetting and thus facilitate their dispersal in the air. We have performed multiscale simulations to address the molecular determinants governing the formation of functional amyloids by the class I fungal hydrophobin EAS. Extensive samplings of full-atom replica-exchange molecular dynamics and coarse-grained simulations have allowed us to identify factors that distinguish aggregation-prone from highly soluble states of EAS. As a result of unfavourable entropic terms, highly dynamical regions are shown to exert a crucial influence on the propensity of the protein to aggregate under different conditions. More generally, our findings suggest a key role that specific flexible structural elements can play to ensure the existence of soluble and functional states of proteins under physiological conditions.
Keywords: amyloid formation, protein aggregation, protein dynamics
The identification of the factors that enable proteins and macromolecules to remain functional and soluble under physiological conditions is of central importance in biology (1). The ability to avoid inappropriate protein aggregation is a distinctive property of all functional biological macromolecules and assumes a particularly crucial role in disfavoring aberrant processes such as those involving protein self-assembly into highly organized amyloid fibrils; these latter processes are associated with a range of severe pathological conditions, including neurodegenerative disorders such as Alzheimer’s and Parkinson diseases and a number of nonneuropathic conditions including type II diabetes (2–4). In vivo, amyloid formation is largely pathogenic, although in some cases it can have functional relevance, as for example in the storage of peptide hormones in mammals (5) or the response to nutrient depletion conditions in yeast (6). It has become apparent, however, that the ability to form amyloid structures is a common characteristic of polypeptide chains in vitro (7); this finding begs the question of how the majority of the peptides and proteins in a living system are able to suppress such processes under normal circumstances.
It is well established that there are multiple strategies within biological organisms to inhibit aberrant protein aggregation, including the regulation of protein expression levels (8) and the existence of quality control mechanisms to detect and degrade misfolded conformations (9–12). The principal means for controlling misfolding and aggregation are, however, intrinsic factors that proteins have optimized throughout evolution and have encoded in their amino-acid sequences (13) to define specific energy barriers that prevent their solution states from adopting aggregation-prone conformations (that we denote here as “N*”) under normal circumstances (14). The population of N* within the conformational ensemble of a protein state has a crucial impact on its rate of aggregation (15) and in this context it has been shown that negative design is an important feature of protein structures that have been specifically optimized to prevent unwanted intermolecular association (16). One of the first examples of such a phenomenon was reported by Oliveberg and coworkers, who described the effect of gatekeeper charged residues on the aggregation propensity of proteins (17). In addition, an elegant study by Richardson and Richardson revealed the nature of a variety of negative design elements that minimize intermolecular edge-to-edge β-sheet interactions (18). The specific structural elements identified in this study have been shown to be highly effective in a variety of studies but are inherently restricted to proteins that possess well-defined three-dimensional structures. Because a significant fraction of eukaryotic proteins in particular are believed to posses a partial or complete degree of intrinsic disorder (19), the question arises whether additional factors, for example sequence effects (13, 20), exist to prevent dangerous and unwanted protein aggregation.
In the present work we have used multiscale simulations to identify factors governing the aggregation process of a fungal hydrophobin from Neurospora crassa, designated as EAS (21–23). We report that flexible regions of proteins can play a major role in the avoidance of self-assembly and aggregation. In solution EAS remains monomeric but on contact with a hydrophobic-hydrophilic boundary, such as an air–water interface, EAS rapidly assembles into amyloid fibrils (24) that play a functional role in the fungal life cycle (23). In the organism, however, EAS self-assembly must be tightly controlled and restricted to specific conditions and appropriate locations. Because of its remarkable properties, EAS represents an attractive system to investigate the delicate balance governing the aggregation propensity of proteins and to understand how even an amyloidogenic protein can remain soluble within the aqueous cellular environment.
Extensive samplings of full-atom replica-exchange molecular dynamics (REMD) and Coarse-Grained (CG) simulations have allowed us to identify the energy terms that promote the significant solubility of EAS in the bulk solution yet trigger facile amyloid formation at an air–water interface. This study indicates that a flexible region, the loop between residues 19 and 45, can exert a crucial influence on the aggregation propensity of EAS by preventing unwanted aggregation in solution and suggests a general role that some dynamic elements play in the avoidance of protein aggregation.
Results
Full Atomistic Simulations of the EAS19–45 Loop in Bulk Solution and at the Air–Water Interface.
The hydrophobin EAS forms a β-barrel structure punctuated by disordered regions, the largest of which spans residues 19 to 45 (Fig. 1). The analysis of the hydrophobin EAS19–45 loop in bulk water and at an air–water interface was performed using extensive full atomic REMD simulations with explicit representation of the peptide atoms and water molecules (see Materials and Methods). All the simulations converged within the simulation time (300 ns per replica) as assessed by a variety of parameters (Fig. S1).
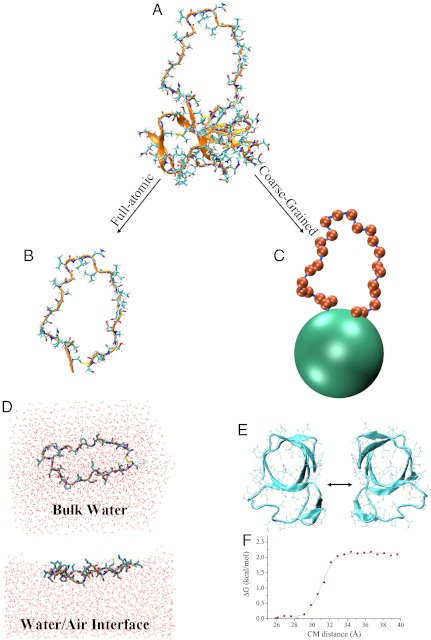
Fig. 1.
Model for multiscale simulations of EAS self-assembly. (A) The hydrophobin EAS (PDB ID code 2FMC). (B) Full-atomic model of the EAS19–45 loop. (C) Coarse-grained model of EAS. Green and silver red represent the barrel (BA) and the EAS19–45 loop (LP) moieties, respectively. (D) Full atomic model of the EAS19–45 loop at the air–water interface (top) and in the bulk solution (bottom). (E) The interaction potential between barrels has been calculated by means of PMF profiles performed in full atomic simulations with explicit waters. (F) Example of PMF along a specific interaction direction.
The sampling of the EAS19–45 loop in water resulted in a highly heterogeneous ensemble of conformations. This feature is due to the fact that in the bulk water no single conformation can shield in an effective way all of the hydrophobic side-chains from water. Instead, a variety of loop conformations are simultaneously populated with comparable Boltzmann weights (Fig. 2A). This finding is consistent with the NMR analysis of the EAS structure, which has revealed significant disorder in the fragment ranging from residues 19 to 45, with no long-range NOEs associated with this region (22). Moreover, the REMD ensemble shows a residual α-helical population at the C-terminal region of the loop (Fig. S2). In particular, the region 36–42 adopts α-helical structures with a population of 33.8%, whereas other two regions, spanning residues 29–34 and 20–23, have a population of 15.5% and 12.9% of α-helix, respectively. Remarkably, despite the possibility of intrinsic biases in the force fields, the degree of residual helical structure observed in the simulations is in good agreement with the estimate of the extent of secondary structure from the measured  secondary chemical shifts (Fig. S2). The NMR measurements were, however, carried out on the full-length protein (22), which results in the terminal regions of the loop having positive
secondary chemical shifts (Fig. S2). The NMR measurements were, however, carried out on the full-length protein (22), which results in the terminal regions of the loop having positive  secondary chemical shifts (indicative of β-sheet conformations) because in the full-length protein this loop connects two β-strands, S2 and S3.
secondary chemical shifts (indicative of β-sheet conformations) because in the full-length protein this loop connects two β-strands, S2 and S3.
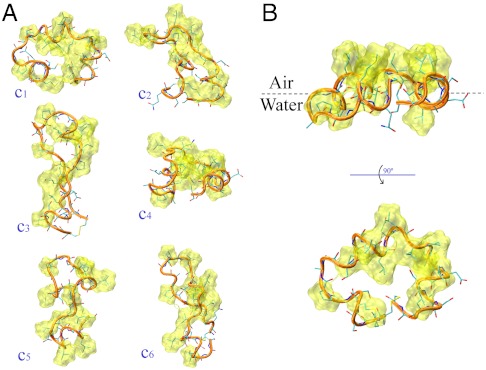
Fig. 2.
Representative conformations from the REMD simulations of the EAS19–45 loop. (A) Bulk-solution. (B) Air–water interface. The backbone trace is shown by orange ribbons, the peptide atoms by sticks. The atomic surfaces of the hydrophobic side chains are drawn in yellow. (A) The nine most populated clusters of the bulk-solution ensemble, accounting for 40% of the conformations (estimated from a cluster analysis with a cutoff of 2 Å). (B) The main conformational cluster, accounting for 74% of the conformations at the air–water interface (estimated from a cluster analysis with a cutoff of 2 Å).
We then performed a REMD conformational sampling of the EAS19–45 loop at an air–water interface. The resulting simulations reveal that the peptide is strongly localized at the interface region (see distributions in Fig. S3), indicating that the EAS19–45 loop itself has clear surfactant properties. The main characteristic of the interface ensemble is a large reduction in the conformational heterogeneity of the loop when compared to that found for the same region in bulk solution, a phenomenon arising from favourable interactions that stabilize a set of principal conformations under these conditions. In particular, at the interface a specific conformational basin exists in which the loop is able to expose efficiently the majority of the hydrophobic side chains to air and simultaneously to solvate the majority of the hydrophilic groups in water. This conformation, which also features an increased population of α-helical structure relative to that observed in the bulk solution (Fig. S4), reflects the amphipathic nature of the air-water interface. A more detailed inspection suggests that, despite the different content of residual structure, sequences showing an α-helical character at the air–water interface match the regions having a residual α-helix content in the bulk solution, as found by NMR secondary chemical shifts and REMD sampling, indicating that the air–water interface stabilizes the structured conformations of the ensemble.
The major conformational basin at the air–water interface accounts for 74% of the overall number of conformations (as estimated from a clustering analysis performed with a cutoff of 2 Å, Fig. 2B). This situation is significantly different from that of the bulk solution, in which the six most populated clusters account for only 40% of the members of the ensemble (Fig. 2A). A quantitative estimate of the reduced conformational accessibility of the ensembles representing the protein at the interface versus those characterizing the bulk solution is provided by the clustering analysis of the rmsd matrix (see Fig. 3A captions for further details). Regardless of the rmsd cutoff employed for the clustering procedure, the number of clusters sampled in solution is always very significantly higher than that sampled at the air–water interface (Fig. 3A). Similarly, root mean square fluctuation (rmsf) values are significantly larger in bulk solution than at the air–water interface, indicating enhanced fluctuations of Cα atoms when the loop is in the bulk solution (Fig. 3B).
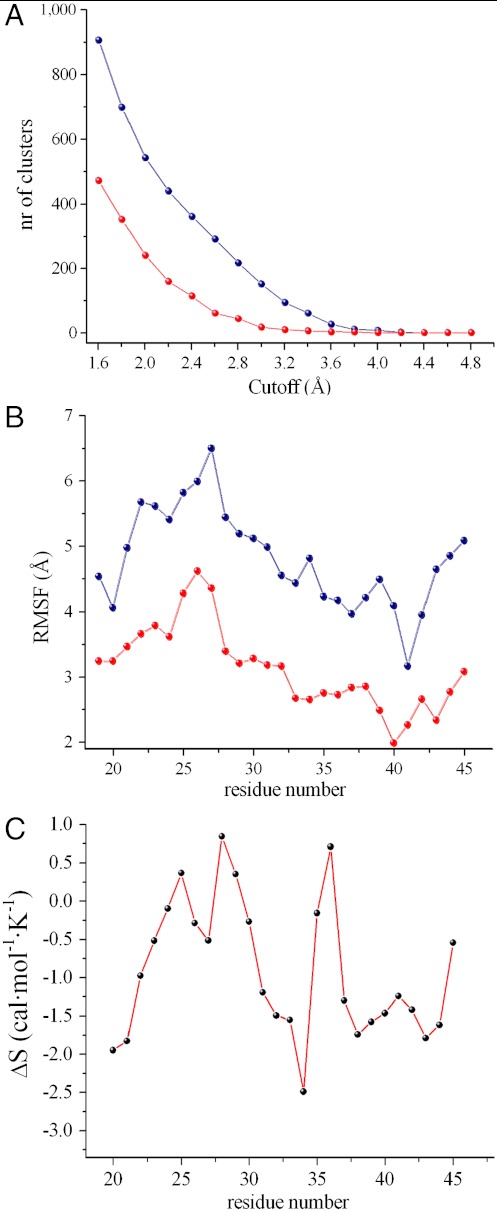
Fig. 3.
General comparison between water and air-water interface simulations. (A) Cluster analysis. This method clusters the structures of an ensemble based on the pairwise rmsd matrix, in order to ensure that pairwise rmsd values calculated among members of a cluster are lower than a given cutoff. Accordingly, the number of clusters extracted from the ensembles is dependent on the cutoff employed. This plot reports the number of clusters as a function of the cutoff values showing that the loop conformational accessibility is dramatically higher in water (blue) compared to the interface (red). The cutoff values range from 1.6 Å to 4.0 Å with a step of 0.2 Å. (B) Rmsf profiles of the loop at the interface (blue) and in solution (red). (C) Conformational entropy difference of the loop (residues 20–45). The profile is calculated as Ssolution-Sinterface. The entropic content of each residue of the loop is calculated as the sum of the mainchain and sidechain terms (Fig. S5), except for alanine and glycine residues, where the mainchain term only is considered. The overall contribution at 300 K to the conformational free energy is -TΔS = +7.11 ± 0.33 kcal/mol.
Estimation of the Entropy Difference Between Solution and Interface States.
In order to estimate the changes in the conformational entropy between the bulk solution and the air–water interface, we calculated the distribution of backbone and sidechain dihedral angles (Fig. S5 and Fig. 3C captions). The entropy difference was then estimated for residues 20 to 44, thereby excluding the disulfide bridged cysteine residues that generate the loop closure (Fig. 3C). At 300 K, the overall contribution to the free energy associated with the entropy difference between the bulk solution and the air–water interface is estimated to be 7.11 ± 0.33 kcal/mol. By assuming a total (or at least a very significant) reduction of the conformational degrees of freedom of the protein and the loop upon aggregation, this finding suggests that the process of EAS self-assembly and aggregation in the bulk solution would require an additional free energy cost of approximately 7 kcal/mol compared to that needed at the air-water interface.
Coarse-Grained Simulations of EAS Aggregation.
In order to study the collective behavior of a system containing a large number of hydrophobins, we used coarse-grained (CG) simulations. The coarse graining in this study was designed to capture the effects of flexible loops on the aggregation behavior of proteins. Therefore, although the simulations were specifically modeled on EAS, our CG implementation provides general insights for protein aggregation. Sixty-four coarse-grained EAS molecules were therefore placed in a box with dimensions of 48 × 48 × 48 nm3, and individual MD simulations were performed at temperatures ranging from 200 K to 400 K, with a spacing of 10 K. In addition to simulations of the wild-type hydrophobin EAS, we also simulated systems of truncated forms of the protein having a shortened loop (Δ15-EAS and Δ19-EAS, with 15 and 19 residues removed form the long loop, respectively) (21) and of the structured barrel without the long EAS19–45 loop (BA only).
To assess the level of aggregation of EAS molecules in the simulations, we defined an aggregation index based on a clustering algorithm. For each snapshot, the analysis identifies individual clusters of protein aggregates on the basis of the distances between the centers of mass. By calculating the average number of clusters, we quantify the aggregation propensities for the different model systems at different temperatures. The clustering index ranges from 1, corresponding to a single large aggregate, to 64, corresponding to a situation in which all the protein molecules are present as monomers; a small number of clusters therefore corresponds to a high degree of aggregation. The convergence of our coarse-grained simulations was assessed by following the time evolution of the clustering in each simulation (Fig. S6A). We performed a number of control simulations to verify that the systems that we studied reached equilibrium within 5 μs (see Fig. S6B). All simulation results that we report were obtained for well-equilibrated systems. In practice, this was achieved by not taking into account in the data analysis the first 5 μs of all simulation runs.
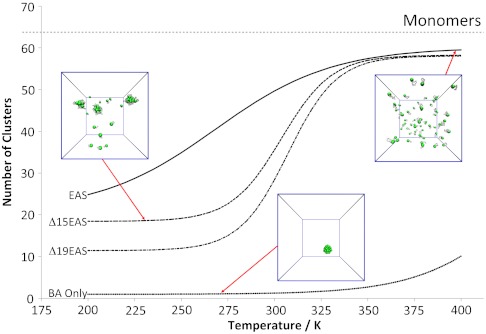
A plot of the number of clusters as a function of temperature resulted in a sigmoidal trend with a very high degree of aggregation at low temperatures and a low level of aggregation at high temperatures (Fig. 4). At 300 K the average number of clusters is 48.53, indicating that most proteins occur as monomers; closer inspection of the oligomer distribution showed that 66% of proteins are in the monomeric state, 13% occur as dimers, and 4% form trimers. Other oligomeric species are also formed, with the largest detected assemblies, 22mers, having a population of 0.0049% (Fig. S7). When the length of the EAS loop is reduced, the inflection point of the sigmoidal curve shifts to higher temperatures (Fig. 4). Hence, systems with shorter loops have a higher tendency to aggregate. This effect is maximized when we remove the entire loop, thereby effectively simulating only the β-barrel region of EAS, for which we observe extensive aggregation over a wide range of temperatures (Fig. 4, BA only).
Fig. 4.
Equilibrium aggregation profiles of EAS as function of temperature. The plot reports the average number of clusters formed at equilibrium for the different simulations of EAS aggregation. This index ranges from 1, indicating a single large aggregate, to 64, indicating that the 64 copies of EAS remain monomeric at equilibrium. The four curves refer to the full-length EAS (EAS, continuous line), EAS truncated of 15 loop residues (Δ15, dashed line), EAS truncated of 19 loop residues (Δ19, dashed/dotted line), the EAS without loop (BA only, dots).
Discussion
A detailed understanding of the factors allowing proteins and other macromolecules to remain soluble in their functional monomeric or multimeric states in the crowded environment of the cell is of central importance in biology (25, 26). The dynamic properties of proteins can play key roles in influencing their propensity for self-ssembly and aggregation (14) and have considerable significance in many aspects of their function (27–31). More generally, a range of factors discriminate between aggregation-prone and aggregation-resistant states of proteins, including charge effects, negative design elements and accessibility of aggregation-prone sequences (7, 16). The present study indicates that the presence of highly flexible regions may act to suppress very significantly the tendency of proteins to aggregate. This important conclusion results from multiscale simulations of the self-assembly process of the EAS protein, a class I hydrophobin that has high levels of solubility in an aqueous environment but which readily aggregates to form functional structures with amyloid-like characteristics at an air–water interface, where the preferential alignment of the protein may be required (23).
Because of their remarkable properties, the hydrophobins represent a powerful system in which to probe the principles that govern the balance between protein solubility and aggregation. To attain a highly soluble state in the bulk solvent under conditions where it is essential to avoid aggregation (e.g., in the cellular milieu), many hydrophobins typically form dimers or tetramers that are able to sequester hydrophobic protein surfaces from exposure to solvent (23). It has been shown, however, that the hydrophobin EAS remains largely monomeric in bulk solution (32), suggesting that a different mechanism exists to allow this protein to avoid aggregation under these conditions. The results of the present study suggest that the highly dynamical and disordered loop spanning residues 19–45 modulates the aggregation behavior of the EAS protein by inhibiting self-assembly in the bulk solution where aggregation must be avoided.
Extensive all-atom simulations show that the EAS19–45 loop spontaneously undergoes a significant conformational transition at the air–water interface resulting in a large reduction of the conformational accessibility compared to that of the protein in bulk solution. This effect can be attributed to the anisotropic environment of the interface, which stabilizes specific conformations that have surfactant-like properties by directing hydrophobic side chains into air and hydrophilic groups into water. In bulk water, no such selection is present and the conformational entropy is significantly higher than at the interface.
As a result of the significant difference in the loop dynamics and conformational entropy, the aggregation of EAS molecules in the bulk solvent would require an additional free energy cost of 7.11 kcal/mol compared to the aggregation at air-water interface. This energy barrier reduces very significantly the tendency of EAS to aggregate in the bulk solution thereby generating a highly soluble state under these conditions. It has recently been shown that driving forces for assembly in bulk water are stronger than in the vicinity of a hydrophobic surface (i.e., air–water interface), which underlines the importance of factors that can prevent aggregation when EAS is in the bulk solution (33). We suggest therefore that the major role of the loop is to disfavor aggregation in solution rather than promoting aggregation at the air–water interface. At the air–water interface, the loop loses a significant amount of its conformational entropy and can no longer act to prevent aggregation. A second shorter loop between Cys 7 and Cys 8 (22), here considered as an integral part of the folded region of the protein (barrel bead), may also contribute to this mechanism.
In conclusion, the principles emerging from this study add to our previous understanding of the strategies adopted by folded proteins to maintain soluble and functional states (16–18) by indicating the role of structural dynamics in modulating the propensity for protein aggregation. In particular, we suggest that in specific cases large flexible loops in proteins may act to inhibit aggregation as a result of their high values of conformational entropy. It is well established that the aggregation propensity of proteins depends on the properties and population of all the conformational states within the ensemble, including the N* aggregation-prone species (14, 15). The present results may provide a clear example of this concept by demonstrating that the flexibility of the ground state of EAS (the highly soluble state in bulk solution) is remarkably different from that of the aggregation-prone state (at the air–water interface). The role identified here to be played by inherent dynamics suggests that a comprehensive description of the flexibility of all the accessible conformational states must be included in the analysis of the factors influencing the solubility and self-assembly of protein molecules. Thus, the physical principles emerging from this study add to our understanding of these processes and suggest the existence of “dynamical” negative design elements to maintain protein solubility. Such elements are likely to have very general importance in biological systems in light of the fact that a large fraction of proteins in eukaryotic organisms possess substantial amount of structural disorder (19).
Material and Methods
Modeling the EAS Loop by Full Atom Simulations.
We sampled the conformations of the longest loop (residues 19 to 45) in EAS by simulating the behavior of the isolated peptide EAS19–45 with sequence CQSMSGPAGSPGLLNLIPVDLSASLGC. The initial coordinates of the loop were extracted from the NMR structure (Fig. 1A; PDB ID code 2FMC) (22). The disulfide bridge that is formed between residues Cys19 and Cys45 in full-length EAS (using the numbering of the full-length sequence) was included as an additional restraint in order to reproduce this structural feature of the loop in the native protein (Fig. 1B).
Full-atom REMD simulations (34–37) with explicit solvent were used to simulate the loop conformations in solution and at an air–water interface. In the solution simulation, the peptide was immersed in a rectangular box filled with water molecules by applying periodic boundary conditions in the three Cartesian directions (Fig. 1D), and the system was equilibrated with external temperature and pressure baths. In order to simulate the peptide at an air-water interface, the EAS19–45 loop was dissolved in a water layer with a thickness of 30 Å (Fig. 1D) that was propagated along the Cartesian x and y directions, with the z direction running perpendicular to the air–water interface. The size of the simulation box in the z direction, perpendicular to the interface, was 100 Å. These simulations were carried out by equilibrating the system with an external temperature bath. The dimensions of the box were maintained fixed during the sampling period and periodic boundary conditions were applied in the three Cartesian directions.
REMD Procedure.
The REMD methodology simulates several copies (replicas) of the same system evolving independently at different temperatures (34). Exchanges between neighboring replicas were attempted at time intervals, tswap, of 2 ps on the basis of the Metropolis criterion (38) Eq. 1:
 |
[1] |
where P(i → j) is the exchange probability, kB is Boltzmann’s constant, and Ui and Uj are the potential energies of individual configurations of the replicas at the temperatures Ti and Tj. All runs employed 24 replicas with the following temperatures (K): 297, 300, 303, 306, 309, 312.1, 315.2, 318.3, 321.5, 324.7, 327.9, 331.2, 334.5, 337.8, 341.1, 344.4, 347.8, 351.2, 354.7, 358.2, 361.8, 365.4, 369.1, and 372.9. To ensure homogeneous exchange frequencies the temperature increments, ΔTi,j, ranged from 3 K (ΔT1,2) to 3.8 K (ΔT23,24). The smaller spacings at lower temperatures were adopted to compensate for the reduced exchange frequencies, which result from the narrowing of the potential energy distributions. For each temperature bath, the initial conformation of the individual system was equilibrated for 1 ns without attempting replica exchanges, and each simulation was carried out for 300 ns per replica. The efficiency of REMD sampling depends on the rate of “diffusion” in the temperature during REMD and the choice of temperature spacings used in this study ensured that each replica explored repeatedly the entire temperature range.
Simulations were performed with the GROMACS (39) package, using the AMBER03 (40, 41) force field with an integration time step of 2 fs. Water molecules were modeled explicitly using the Tip4pEW (42) solvent model. Electrostatic interactions were evaluated using the Particle Mesh Ewald method with a grid spacing of 1.2 Å. A 14 Å cutoff distance was used for van der Waals interactions. Neutral pH was imposed by selecting the appropriate protonation states of pH sensitive side chains. The net charge of the system was neutralized by using charged Na+ and Cl- ions. All the analyses discussed in this paper refer to the conformations sampled at 300 K (i.e., the second temperature bath of the REMD samplings).
Coarse-Grained (CG) Model of the EAS Self-Assembly Process.
Coarse-grained structure of EAS.
To construct a coarse-grained model of EAS, we started from the solution structure solved by NMR spectroscopy (22). We then constructed a simple model out of two types of particles, loop and barrel beads (Fig. 1C). The latter beads represent the structured region of the protein, which in the atomistic structure assumes the topology of a β-barrel, whereas the loop region spans residues 19 to 45. Each residue of the loop was represented by one loop bead (LP beads) placed at the position of the Cα atom of the residue, with mass of the loop particles being set as the average mass of the residues in the loop. The folded β-barrel part of the protein was treated as a single bead (BA beads) located at its center of mass and having a mass equal to the total mass of this protein domain.
Interaction energies between the particles.
Using atomistic simulations, we computed protein-protein pair interaction energy profiles. To this end, we determined the reversible work needed to pull apart two copies of the folded part of EAS (Fig. 1 E and F), employing a series of different interface orientations. The final interaction energy between the folded moieties of two EAS molecules was averaged from these potential mean force (PMF) profiles thereby generating an isotropic attraction potential. The interactions of loop beads were purely repulsive to account for the excluded volume of the residues. It is worth noting that the sequence information is not included in this CG representation of the loop.
Coarse-grained simulations were performed at different temperatures ranging from 200 K to 400 K. Thermal equilibration was performed by coupling the system to an external bath and using the v-rescale method. Simulations were performed with an integration time of 5 fs, extended for 15 μs (i.e., for each temperature simulated) and performed at constant temperature and volume (NVT ensemble). The initial 5-μs segments of the simulations were used for the thermal equilibration of the systems whereas the final 10 μs were employed for the analyses.
Secondary Shifts from the NMR Resonance Assignments.
Secondary chemical shifts provide a sensitive probe for detecting secondary structures by NMR. The advantage of these measurements is that they can account for partial populations within the conformational ensembles. We calculated in this report secondary NMR chemical shifts by subtracting the random coil reference values from the measured chemical shifts (22). To estimate the chemical shift values of the reference random coil state, we used the CamCoil method (43), which is able to account for the effect of oxidized cysteine residues, as in the case of residues Cys 19 and Cys 45 of EAS, as well as the contribution of the pH.
Supplementary Material
Acknowledgments.
We are grateful to David Chandler for discussions and critical reading of the manuscript. D.F. acknowledges support from European Research Council Advanced Grant 227758, Wolfson Merit Award 2007/R3 of the Royal Society of London and Engineering and Physical Sciences Research Council (EPSRC) (Grant EP/I001352/1). A.D. acknowledges support of EPSRC (Grant EP/G049998/1). C.M.D. acknowledges support from Medical Research Council, Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118048109/-/DCSupplemental.
References
- 1.Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 4.Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 5.Maji SK, et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narayanaswamy R, et al. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci USA. 2009;106:10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobson CM. Protein misfolding, evolution and disease. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 8.Tartaglia GG, Pechmann S, Dobson CM, Vendruscolo M. A relationship between mRNA expression levels and protein solubility in E. coli. J Mol Biol. 2009;388:381–389. doi: 10.1016/j.jmb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Behrends C, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 13.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 14.De Simone A, et al. Experimental free energy surfaces reveal the mechanisms of maintenance of protein solubility. Proc Natl Acad Sci USA. 2011;108:21057–21062. doi: 10.1073/pnas.1112197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li MS, et al. Factors governing fibrillogenesis of polypeptide chains revealed by lattice models. Phys Rev Lett. 2010;105:218101. doi: 10.1103/PhysRevLett.105.218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thirumalai D, Klimov DK, Dima RI. Emerging ideas on the molecular basis of protein and peptide aggregation. Curr Opin Struct Biol. 2003;13:146–159. doi: 10.1016/s0959-440x(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 17.Otzen DE, Kristensen O, Oliveberg M. Designed protein tetramer zipped together with a hydrophobic Alzheimer homology: A structural clue to amyloid assembly. Proc Natl Acad Sci USA. 2000;97:9907–9912. doi: 10.1073/pnas.160086297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 20.Kim W, Hecht MH. Sequence determinants of enhanced amyloidogenicity of Alzheimer A{beta}42 peptide relative to A{beta}40. J Biol Chem. 2005;280:35069–35076. doi: 10.1074/jbc.M505763200. [DOI] [PubMed] [Google Scholar]
- 21.Kwan AH, et al. The Cys3-Cys4 loop of the hydrophobin EAS is not required for rodlet formation and surface activity. J Mol Biol. 2008;382:708–720. doi: 10.1016/j.jmb.2008.07.034. [DOI] [PubMed] [Google Scholar]
- 22.Kwan AH, et al. Structural basis for rodlet assembly in fungal hydrophobins. Proc Natl Acad Sci USA. 2006;103:3621–3626. doi: 10.1073/pnas.0505704103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunde M, Kwan AH, Templeton MD, Beever RE, Mackay JP. Structural analysis of hydrophobins. Micron. 2008;39:773–784. doi: 10.1016/j.micron.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Macindoe I, et al. Self-assembly of functional, amphipathic amyloid monolayers by the fungal hydrophobin EAS. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1114052109. 10.1073/pnas.1114052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tartaglia GG, Pechmann S, Dobson CM, Vendruscolo M. Life on the edge: A link between gene expression levels and aggregation rates of human proteins. Trends Biochem Sci. 2007;32:204–206. doi: 10.1016/j.tibs.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Vendruscolo M, Dobson CM. Quantitative approaches to defining normal and aberrant protein homeostasis. Faraday Discuss. 2009;143:277–291. doi: 10.1039/b905825g. discussion 359-272. [DOI] [PubMed] [Google Scholar]
- 27.Mittermaier A, Kay L. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Tzeng SR, Kalodimos CG. Dynamic activation of an allosteric regulatory protein. Nature. 2009;462:368–372. doi: 10.1038/nature08560. [DOI] [PubMed] [Google Scholar]
- 29.Gao YQ, Yang W, Karplus M. A structure-based model for the synthesis and hydrolysis of ATP by F1-ATPase. Cell. 2005;123:195–205. doi: 10.1016/j.cell.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Masterson LR, et al. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat Chem Biol. 2010;6:821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masterson LR, et al. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc Natl Acad Sci USA. 2011;108:6969–6974. doi: 10.1073/pnas.1102701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackay JP, et al. The hydrophobin EAS is largely unstructured in solution and functions by forming amyloid-like structures. Structure. 2001;9:83–91. doi: 10.1016/s0969-2126(00)00559-1. [DOI] [PubMed] [Google Scholar]
- 33.Patel AJ, et al. Extended surfaces modulate hydrophobic interactions of neighboring solutes. Proc Natl Acad Sci USA. 2011;108:17678–17683. doi: 10.1073/pnas.1110703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugita Y, Okamoto Y. Replica-exchange moleculardynamics method for protein folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 35.Camilloni C, Schaal D, Schweimer K, Schwarzinger S, De Simone A. Energy landscape of the prion protein helix 1 probed by metadynamics and NMR. Biophys J. 2012;102:158–167. doi: 10.1016/j.bpj.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Simone A, Esposito L, Pedone C, Vitagliano L. Insights into stability and toxicity of amyloid-like oligomers by replica exchange molecular dynamics analyses. Biophys J. 2008;95:1965–1973. doi: 10.1529/biophysj.108.129213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vitagliano L, Esposito L, Pedone C, De Simone A. Stability of single sheet GNNQQNY aggregates analyzed by replica exchange molecular dynamics: Antiparallel versus parallel association. Biochem Biophys Res Commun. 2008;377:1036–1041. doi: 10.1016/j.bbrc.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 38.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller EJ. Equation of state calculations by fast computing machines. J Chem Phys. 1953;21:1087–1092. [Google Scholar]
- 39.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: “A message-passing parallel molecular dynamics implementation”. Comp Phys Comm. 1995;91:43–56. [Google Scholar]
- 40.Duan Y, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 41.Sorin EJ, Pande VS. Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys J. 2005;88:2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horn HW, et al. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. J Chem Phys. 2004;120:9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- 43.De Simone A, Cavalli A, Hsu ST, Vranken W, Vendruscolo M. Accurate random coil chemical shifts from an analysis of loop regions in native states of proteins. J Am Chem Soc. 2009;131:16332–16333. doi: 10.1021/ja904937a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.