Abstract
It has been known for nearly 100 years that pressure unfolds proteins, yet the physical basis of this effect is not understood. Unfolding by pressure implies that the molar volume of the unfolded state of a protein is smaller than that of the folded state. This decrease in volume has been proposed to arise from differences between the density of bulk water and water associated with the protein, from pressure-dependent changes in the structure of bulk water, from the loss of internal cavities in the folded states of proteins, or from some combination of these three factors. Here, using 10 cavity-containing variants of staphylococcal nuclease, we demonstrate that pressure unfolds proteins primarily as a result of cavities that are present in the folded state and absent in the unfolded one. High-pressure NMR spectroscopy and simulations constrained by the NMR data were used to describe structural and energetic details of the folding landscape of staphylococcal nuclease that are usually inaccessible with existing experimental approaches using harsher denaturants. Besides solving a 100-year-old conundrum concerning the detailed structural origins of pressure unfolding of proteins, these studies illustrate the promise of pressure perturbation as a unique tool for examining the roles of packing, conformational fluctuations, and water penetration as determinants of solution properties of proteins, and for detecting folding intermediates and other structural details of protein-folding landscapes that are invisible to standard experimental approaches.
Keywords: energy landscape, fluorescence, volume change
The first observation that pressure unfolds proteins was made in 1914 by Bridgman (1). Despite numerous studies since then, the physical basis of the pressure-induced unfolding of proteins has not been explained. This difference in volume underlying pressure effects has been rationalized previously in terms of (i) increases in solvent density concomitant with solvation of exposed surfaces upon unfolding (2), (ii) modifications in the structure of bulk water leading to weakened hydrophobic interactions (3), and (iii) cavities in the folded state that are not present in the unfolded state (4–7). The goal of this study was to examine the structural origins of pressure unfolding of proteins. Twenty-five years ago Walter Kauzmann stressed the importance of understanding pressure effects (8): “Enthalpy and volume are equally fundamental properties of the (protein) unfolding process, and no model can be considered acceptable unless it accounts for the entire thermodynamic behavior.” He also noted important discrepancies between the volumetric properties of hydrophobic interactions and the pressure unfolding of proteins.
To date, no conclusive explanation for the origins of pressure unfolding of proteins has been proposed. In the present work, 10 cavity-containing variants of staphylococcal nuclease (SNase) were engineered by substitution of internal core residues to Ala. Crystal structures of the variants were obtained to verify the existence of the engineered cavities. Equilibrium unfolding studies using pressure as a denaturant were performed with the variants and with the parent protein, using tryptophan (Trp) fluorescence or the intensity of cross-peaks from 100 backbone amide detected in 1H-15N heteronuclear single quantum coherence (HSQC) spectra measured with NMR spectroscopy. The role of cavities has been studied in the past (4–6) but not as systematically nor at the level of detail afforded by NMR spectroscopy, as done here.
Previous studies have revealed a lack of correlation between protein size and volume change, demonstrating that the exposure of surface area and the accompanying changes in solvent density are not major determinants of the magnitude of pressure effects (9). In contrast, the present studies demonstrate rigorously and systematically that increasing the amount of internal void volume in the folded states of proteins leads to significant increases in the magnitude of the volume change of unfolding. Hence we conclude that internal cavities are the primary determinants of pressure unfolding of proteins. In addition, the pressure unfolding reaction monitored with NMR spectroscopy revealed significant deviation from simple two-state behavior; this deviation was different from variant to variant. Using the NMR spectroscopy data to constrain Go-model calculations allowed characterization of the structure and energetics of the folding landscape of this protein and identification of a major folding intermediate. These studies demonstrate that because pressure acts primarily to eliminate internal cavities present in the folded state, its effects are much more local and subtle than those of temperature or chemical denaturants, which act globally and in proportion to the increase in surface area exposed upon unfolding. Hence, pressure perturbation allows observation of intermediates that are normally suppressed with harsher perturbations such as high temperature or chemical denaturants. This study illustrates the potential utility of pressure perturbation coupled with NMR spectroscopy to examine with unprecedented detail many important structural aspects of folding landscapes of proteins that are inaccessible with most other experimental approaches.
Results
To increase the internal void volume of a protein (Fig. 1, Top, Left) (10, 11), 10 variants of a very stable variant of SNase referred to as Δ + PHS (11.8 kcal/mole at 298 K (10), compared to 5.4 kcal/mol for WT SNase), were engineered with substitutions of internal positions to Ala to create additional internal cavities.
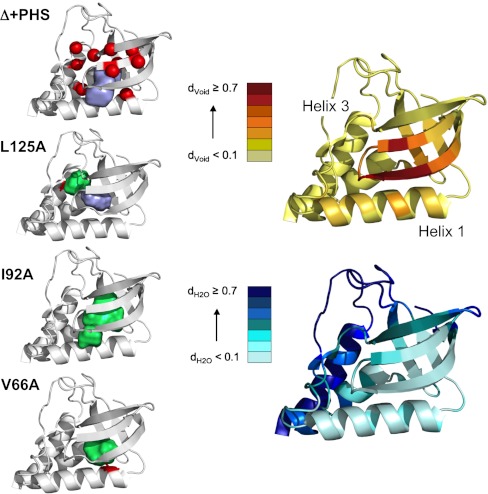
Fig. 1.
Structures of Δ + PHS SNase and cavity-containing variants. Left Panel, from Top to Bottom: Structure of Δ + PHS SNase (3BDC) with the Cα positions of the 10 cavity-containing variants indicated with red spheres and with the surface representation of the central cavity in purple. Cavity volume was calculated using a 1.1 Å sphere and McVol (11). Structures of the cavity mutants L125A (3NXW), I92A (3MEH) and V66A (3NQT) with the engineered cavities in green and the mutated residue in red. Right Panel: Estimation of the void density (Top) and hydration density (Bottom) for each Cα position of the Δ + PHS reference protein. See SI Materials and Methods in SI Appendix and (9) for the details concerning calculation of void and hydration density. Briefly, the 1,000 configurations resulting from a 10 ns all-atom MD simulation in explicit solvent were submitted to Monte Carlo point oversampling. All points that fell within the structure and not on an atom of solvent or the protein were counted for void density. Water density was calculated as the number of oxygen atoms of water molecules within 5 Å of each Cα carbon, normalized to the largest number found for a Cα carbon.
Crystal Structures.
The crystal structures of all variants, solved at 1 atm, showed that the substitutions led to the creation of internal cavities in the folded state. The structures were very similar among themselves and to that of Δ + PHS (Fig. 1 and Fig. S1 and Table S1 in SI Appendix), the main difference being either the enlargement of the naturally occurring microcavity or the introduction of a new cavity. No internal water molecules were observed in any of the cavities; not even a trace of electron density was found.
Molecular Dynamics Simulations.
Because cavities can be hydrated transiently or filled through dynamic side-chain reorganization, the persistence of the naturally occurring cavity in the Δ + PHS reference protein was investigated using molecular dynamics (MD) simulations in explicit solvent (SI Materials and Methods in SI Appendix). Void and hydration density at each Cα (Fig. 1, Right), defined as the number of Monte Carlo points (normalized to the maximum for all Cα atoms) that could be inserted within 5 Å of its position and the number of water oxygen atoms (normalized to the maximum number) within 5 Å, respectively, support the notion that even the naturally occurring microcavity observed in the structure of the wild type protein is present and dehydrated, even after dynamic reorganization and transient hydration are taken into consideration.
Equilibrium Unfolding Monitored by Trp Fluorescence.
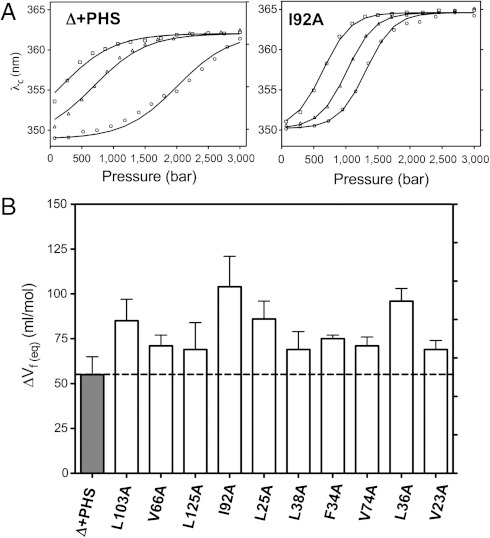
Pressure unfolding monitored by the fluorescence of Trp-140 (12) (Fig. 2A and Fig. S2 in SI Appendix) revealed that the variants with the engineered cavities all exhibited significantly larger values for the apparent volume change upon unfolding ΔVu( = -ΔVf) than did the Δ + PHS protein (Fig. 2 A and B and Table 1). The midpoints of the unfolding profiles shifted to lower pressures with increasing GuHCl, but the slopes were unaffected, indicating that the denaturant destabilized the protein without any effect on ΔVu.
Fig. 2.
Pressure unfolding monitored with Trp fluorescence. (A) High-pressure fluorescence average emission wavelength profile for (Left Panel) Δ + PHS variant at 2.0 M (circle), 2.3 M (triangle) and 2.6 M (square) GuHCl; and (Right Panel) I92A cavity mutant at 0.8 M (circle), 1.0 M (triangle) and 1.2 M (square) GuHCl. (B) Folding volume change, ΔVf( = -ΔVu) values obtained from analysis of the high-pressure fluorescence unfolding profiles for Δ + PHS and 10 cavity-containing variants assuming a two-state model. The dashed line indicates the value for the Δ + PHS protein used as reference. Measurements were performed in the presence of guanidinium chloride (GuHCl) to ensure complete unfolding in the pressure range of the instrumentation used (< 3 kbar).
Table 1.
ΔVu values obtained from high-pressure unfolding experiments
| ΔVu Fluorescence(ml/mol) | ΔVu NMR(ml/mol)* | ΔVu Trp NH(ml/mol)† | |
| Δ + PHS | −55 ± 10 | −69 ± 9 | −80 ± 8 |
| I92A | −104 ±17 | −145 ± 15 | −148 ± 21 |
| L125A | −69 ± 15 | −115 ± 5 | −108 ± 3 |
| V66A | −71 ± 6 | −106 ± 14 | −104 ± 10 |
| L103A | −85 ± 9 | −103 ± 9 | −106 ± 4 |
| V23A | −69 ± 5 | ||
| F34A | −75 ± 2 | ||
| L36A | −96 ± 7 | ||
| L38A | −69 ± 10 | ||
| L25A | −86 ± 10 | ||
| V74A | −71 ± 5 |
*For NMR data, the mean value and the standard deviation are extracted from a Gaussian fit of the ΔVu values distribution obtained from fits of the loss of HSQC peak intensity vs. pressure for the peaks that could be followed throughout the unfolding transition.
†ΔVu values for the trp NH correspond to those obtained for fits of the HSQC peak intensity of the amide NH of the tryptophan residues, for direct comparison with that obtained from the fluorescence data.
Equilibrium Unfolding Monitored by NMR Spectroscopy.
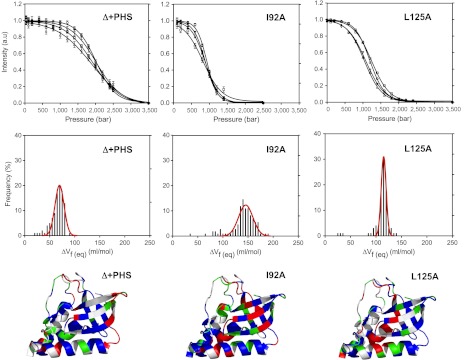
High-pressure NMR 1H-15N HSQC spectra acquired on the Δ + PHS protein and on four of the cavity variants (L125A, V66A, L103A, and I92A) yielded ΔVu values for more than100 detectable amide groups (Fig. 3 and Table 1 and Figs. S3 and S4 in SI Appendix). The average ΔVu value for each cavity-containing variant was significantly larger in magnitude than that of Δ + PHS reference protein. Note that the substitution of Leu or Ile with Ala (deletion of the same amount of van der Waals volume) resulted in effects on the magnitude of ΔVu that were different for different variants (compare L103A or L125A to I92A in Table S2 in SI Appendix). Cavities near the surface of the protein contribute less to ΔVu than cavities that are buried deeply, consistent with the results of pressure perturbation calorimetry (PPC) (Table S2 in SI Appendix) and with MD simulations showing that the central β-strands and helix-1 constitute the region of the protein least accessible to solvent (Fig. 1). We hypothesize that cavities introduced closer to the protein–water interface are likely to contribute less to ΔVu because solvent is less likely to be excluded from these regions owing to their close proximity to bulk water. Although they follow the same general trend, in all variants the average value of ΔVu obtained with NMR spectroscopy, even for the backbone and the aromatic NH resonances of the Trp itself, was larger than the value obtained from Trp fluorescence experiments (Table 1). This discrepancy can be explained by invoking pressure-induced water penetration into the region near the Trp side chain (13) in the folded state at these lower pressures, suggesting ways in which pressure can affect the properties of the folded state by promoting water penetration as suggested by previous studies with T4 lysozyme (14).
Fig. 3.
Pressure unfolding monitored with NMR spectroscopy. (Top): Examples of HSQC cross-peak intensity profiles for four representative residues of the Δ + PHS variant at 1.5 M GuHCl (Left), I92A at 0.85 M GuHCl (Middle), and L125A at 0.85 M GuHCl (Right). (Middle Panel): Distributions of the ΔVf( = -ΔVu) values obtained from high-pressure NMR spectroscopy experiments for Δ + PHS (Left), I92A (Middle), and L125A (Right). (Lower Panel): Structural mapping of the ΔVf values obtained from high-pressure NMR experiments for Δ + PHS (Left), I92A (Middle), and L125A (Right). Residues of the first quartile (the 25% lowest values) are in green; residues of the third quartile (25% highest values) are in red; and residues lying between the first and the third quartile (50% of the values) are in blue.
Structural Origins of Deviations from Two-State Unfolding.
The width and asymmetry in the distributions of ΔVu values (Fig. 3, Middle Row) revealed significant deviations from simple two-state behavior, which were unique for each variant (Fig. 3, Bottom, and Fig. S5 in SI Appendix) and independent of side-chain polarity (Fig. S6 in SI Appendix). Lower apparent values for ΔVf( = -ΔVu) in the Δ + PHS protein measured throughout the C-terminal loop, in helix-3, and in the region where this helix packs against the rest of the protein, indicate that these regions of the protein are disrupted by pressure more readily than the rest of the protein. Local destabilization by the introduction of a cavity is reflected in the very small values of ΔVf observed for residues in β-strand-2 near the site of the V66A substitution (Fig. S5 in SI Appendix). Very large values for the apparent ΔVf near the artificial cavity opposite to the site of the I92A substitution and in helix-2 between the two subdomains near the L125A substitution are consistent with the notion that residues in these deeply buried regions only sample disrupted conformations when the protein is fully unfolded.
Contact maps.
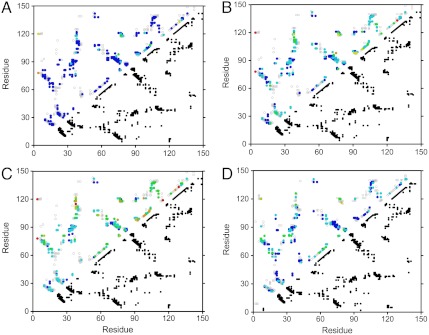
Deviations from two-state unfolding for the Δ + PHS reference protein and for the L125A variant were studied in detailed structural terms with fractional contact maps, defined as the product of the fractional intensity for the amide resonances of the two residues in contact (Fig. 4 and SI Materials and Methods in SI Appendix). The pressure dependence of the contact maps of the Δ + PHS reference protein confirmed that the regions of the protein first affected by an increase in pressure are in the C terminus and where the C-terminal helix interacts with the β-barrel and with helix-2. In contrast to the strong deviation from two-state behavior observed for the Δ + PHS variant, the contact map for the L125A variant was found to be much more homogeneous at all pressures.
Fig. 4.
Fractional contact maps. (A–C) the Δ + PHS variant at 1.5 M GuHCl and 400 bar (A), 600 bar (B), and 800 bar (C); and (D) L125A at 0.85 M GuHCl and 700 bar. The complete set of native contacts is represented by dark dots on the bottom half of the contact maps. The probabilities of contact, calculated as the product of the cross-peak fractional intensities of the implied pair of residues, are indicated by color dots on the upper half of the contact maps. The color scale is dark blue > 90% probability of contact; light blue, 80–90%; green, 70–80%; yellow, 60–70%; orange, 50–60%; and red < 50%. The heterogeneity of contact for the probabilities is most apparent for the Δ + PHS variant at 800 bar (C). It can be seen that although contacts in the core region remain dark or light blue in color (high probability of contact), those in the C terminus and the region connecting the C terminus to the core have shifted to green, yellow, red, and orange (much lower contact probability). In C and D, the fractional contacts for 1.5 M GuHCl at 800 bar for Δ + PHS and for the L125A variant at 0.85 M and 600 bar can be compared. Even if the experiments were not performed at the same concentration of GuHCl, precluding any direct correspondence of the pressures, these two conditions were found to be appropriate for comparison in terms of fractional contacts. It can be seen that compared to the contact probabilities for Δ + PHS, which are very heterogeneous, those for the L125A variant are more homogeneous, remaining mostly dark or light blue, with only a few green residues.
Go-model of the pressure unfolding landscape.
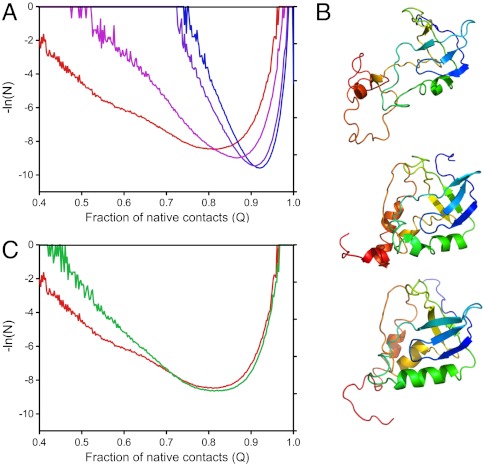
The fractional contact maps obtained experimentally were used as constraints for coarse-grained Cα Go-model simulations of the Δ + PHS reference protein and the L125A variant with the aim of obtaining structural and energetic insight into how the presence of cavities in specific regions of the folded state affected pressure sensitivity (SI Materials and Methods in SI Appendix). For the two variants studied, a total of 400,000 conformations were generated for each variant at each pressure, which were then mapped to an all-atom model (15). The free-energy profile obtained for the Δ + PHS reference protein at 800 bar (Fig. 5A and Fig. S7 in SI Appendix) revealed that at this pressure, in addition to a minimum in the folded and unfolded state basins, there is a minimum near a value of 0.55 for the fraction of native contacts (Q value). This folding intermediate retained considerable structure in the β-barrel and in helices-1 and 2; the C-terminal helix-3 and the loop preceding this helix appeared to be partially disrupted (Fig. 5B), consistent with previous studies of the unfolding of SNase (16). The simulations also suggested significant broadening and a shift of the folded state basin to lower Q value as a function of pressure, consistent with the observations based on Trp fluorescence, and supporting the notion of water penetration into the interior of the folded protein as intramolecular contacts become disrupted by pressure. In contrast to the heterogeneous behavior of the Δ + PHS protein, the L125A variant exhibited smooth free-energy profiles at all pressures (Fig. 5C), which probably arises from coupling of unfolding of the two subdomains as a result of the introduction of a second cavity at the interface between the C terminus and the rest of the protein.
Fig. 5.
Go-model calculations. (A) Pseudo free-energy profiles calculated from structure and NMR-based Go-model simulations of the Δ + PHS variant at 1 bar (blue), 400 bar (purple), 600 bar (pink), and 800 bar (red). (B) Representative conformations extracted from simulations with Δ + PHS SNase at 800 bar with (from Top to Bottom) fraction of native contacts, Q, Q = 0.55, Q = 0.83 and Q = 0.92. (C) Comparison of the pseudo free-energy profiles obtained from simulations of Δ + PHS at 800 bar (red) and L125A at 700 bar (green). Pressures shown are below the unfolding midpoint.
Discussion
Physical Origins of Pressure Effects on Protein Stability.
The most important contribution from this study is the irrefutable evidence that cavities that are present in the folded state and absent in the unfolded state provide a large contribution to the volume difference between folded and unfolded states that govern pressure-induced unfolding of proteins. Internal cavities might also determine the pressure dissociation of protein oligomers and aggregates (17).
(i). Density of hydrating solvent.
The changes in solvent density originating from hydration of exposed surfaces have been proposed as a major contributing factor to the volume change of unfolding (2). If this hypothesis were true, then like the heat capacity change, ΔCp, and the denaturant m-value (18), the magnitude of ΔVu should be correlated with the amount of exposed surface area in the unfolded state, and hence, to the size of the protein. No such correlation was found in a recent systematic study of pressure unfolding of deletion variants of a repeat protein (9) or for unfolding volume changes reported in the literature for different proteins reported at or near the same temperature (9). However, in rare cases where buried and dehydrated charged or ionizable residues become exposed to solvent upon unfolding, electrostriction of the charge can be significant. We note, as well, that, compared to real globular proteins, a small contribution from hydration density differences may be more apparent in model systems such as trp cage (19) or peptides (2), which present little or no hydrophobic core.
(ii). Structure of bulk water.
If pressure unfolding were driven by structural changes of bulk water at high pressure, ΔVu would not be an intrinsic property of the protein, but rather would result from a pressure dependence of the properties of the solvent. A number of observations argue against this notion. First, it is well understood that large changes in the structure of water occur at pressures above 10 kbar and are unlikely to contribute to unfolding as observed here with midpoints at pressures as low as 500 bar. Second, in this and in previous studies (20, 21) we find that pressure unfolding profiles shift to higher or lower pressures with changes in solution composition, but the volume changes remain constant, regardless of the pressure range over which the unfolding occurs. Moreover, measurements with PPC (22, 23), which involve pressure changes of only 5 bar, reveal volume changes upon unfolding consistent with those measured using spectroscopic approaches.
(iii). Cavities in the folded state.
In contrast to previous more anecdotal reports (5, 6), the present study shows unambiguously that the volume of cavities in the folded state, which are absent in the unfolded state, represents a major contribution to the difference in molar volume upon protein unfolding. Previous volume calculations from molecular dynamics (MD) simulations on pressure-denatured states of wild type SNase obtained by water insertion methods (24) are in reasonable agreement with this experiment (12). Although the predominant role of cavities is clear from the present work, further studies will be necessary to develop a predictive, quantitative model of volume changes based on crystal structures. This will require knowledge of the state of hydration of the cavities in the folded state and of the dependence of the cavity volume on thermal fluctuations. In addition, the value of the volume change of unfolding is a strong function of temperature, both in magnitude and even in sign (25). Positive values of ΔVu have been measured at high temperature, resulting from a large difference in the coefficient of thermal expansion of the folded and unfolded states (23, 26, 27). Improved understanding of these effects will be required before successful structure-based predictions of pressure effects on proteins are possible. These uncertainties currently preclude the prediction of the magnitude of pressure effects based on the internal void volume apparent in crystal structures of proteins.
Folding Landscape of SNase:
A second important contribution from this study concerns the demonstration of the unique utility of pressure unfolding monitored with NMR spectroscopy as an invaluable tool for quantitative examination of structural details of protein-folding landscapes. Previous studies on the effects of pressure on folded states of several proteins monitored through native state resonances in HSQC spectra (28), hydrogen exchange (29), crystallographic and computational studies (14), and MD and coarse-grained simulations (30–32) revealed pressure-induced structural reorganization and increased water penetration in the folded state. Similar observations are reported here for SNase. Beyond such observations, by focusing on the pressure unfolding reaction proper, we have characterized, with unprecedented structural and energetic resolution, the pressure unfolding landscape of SNase and its variants. Deletion mutagenesis (33), H/D exchange (34, 35), and mechanical unfolding experiments (36) all at atmospheric pressure have provided a consistent picture of the modular nature of SNase with two subdomains, an N-terminal β-barrel core domain that includes helix 1, and a C-terminal domain consisting primarily of helix 3, with helix 2 acting as an interface between the two subdomains and the C-terminal loop Leu-137 to Ser-141 locking the protein into its correct tertiary structure through long range contacts (37, 38). Besides the typical cis/trans proline isomerization observed within the unfolded states on a slow time scale, multiphasic kinetics have been detected in a stopped-flow study of a proline-free variant of SNase (39), suggesting the accumulation of on-pathway intermediate states. Both rapid mixing fluorescence experiments (40, 41) and pulsed H/D NMR experiments (42) indicated the early formation of the β-domain. The present observations are fully consistent with the known modular nature of SNase.
Conclusions
This study provides the answer to a nearly century-old fundamental question in protein chemistry concerning the origins of pressure effects on proteins. Specifically, the data presented here, taken together with our previous work showing little contribution from solvation density differences (9), demonstrate that pressure unfolds proteins because of cavities that are present in the folded state and which are eliminated in the unfolded state. Pressure-driven unfolding originates with specific, local and unique properties of the folded state. In this sense it is very different from unfolding by temperature or chemical denaturants (18, 43, 44), which act globally and depend on exposed surface area in the unfolded state. Owing to the central role of cavities and to the inherently anisotropic distribution of void volume in the folded state, pressure perturbation coupled with NMR spectroscopy and novel computational approaches can enable unprecedented exploration of structural details of protein-folding landscapes that are usually obscured in temperature or chemical denaturation.
Materials and Methods
All experimental studies were performed on the highly stable Δ + PHS form of SNase (45, 46) and on variants thereof (SI Materials and Methods in SI Appendix). Details of structural studies are reported in SI Materials and Methods in SI Appendix. High-pressure fluorescence experiments were carried out as previously described (9) and curves of average emission wavelength vs. pressure were fitted globally to a two-state unfolding equilibrium as function of pressure for values of  and
and  (SI Materials and Methods in SI Appendix). High-pressure HSQC data were acquired using a ceramic high-pressure cell system (Daedalus Innovations) (SI Materials and Methods in SI Appendix). The fitting procedure was comparable to that used for the high-pressure fluorescence experiments except for the correction for quantum yield, which was not applied. All-atom molecular dynamics simulations in explicit solvent were run as previously described (9) (SI Materials and Methods in SI Appendix). Cavity location in the folded states ensemble structures from the MD simulations, as well as the void and water density, were estimated using the McVol algorithm (11) as described in SI Materials and Methods in SI Appendix. Go-model simulations constrained by the NMR data are also described in the SI Materials and Methods in SI Appendix.
(SI Materials and Methods in SI Appendix). High-pressure HSQC data were acquired using a ceramic high-pressure cell system (Daedalus Innovations) (SI Materials and Methods in SI Appendix). The fitting procedure was comparable to that used for the high-pressure fluorescence experiments except for the correction for quantum yield, which was not applied. All-atom molecular dynamics simulations in explicit solvent were run as previously described (9) (SI Materials and Methods in SI Appendix). Cavity location in the folded states ensemble structures from the MD simulations, as well as the void and water density, were estimated using the McVol algorithm (11) as described in SI Materials and Methods in SI Appendix. Go-model simulations constrained by the NMR data are also described in the SI Materials and Methods in SI Appendix.
Supplementary Material
Acknowledgments.
This work was funded by the Agence National de la Recherche grant PiriBio number 09-455024 (C.A.R.), grants MCB-0743422 (B.G.M.E.) and MCB-0543769 and MCB-1050966 (A.E.G.) from the National Science Foundation; a fellowship from Coordination for the Training and Improvement of Higher Education Personnel (CAPES) of Brazil (D.R.N.); a fellowship from the French Ministry of Research and Higher Education (J.R.); and a Fulbright International Graduate Fellowship (J.R.). Crystallographic data were measured at beamline X25 of the National Synchrotron Light Source supported by the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health Grant P41RR012408. The authors thank Dr. Annie Heroux for assistance in data collection at Brookhaven National Laboratory.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Structure factors have been deposited in the Protein Data Bank, www.pdb.org (V23A, PDB ID code 3PMF; L25A, PDB ID code 3OSO; F34A, PDB ID code 3MVV; L36A, PDB ID code 3NP8; L38A, PDB ID code 3MHB; T62A, PDB ID codes 3MXP and 3R3O; V66A, PDB ID code 3NQT; V74A, PDB ID code 3NK9; I92A, PDB ID code 3MEH; L103A, PDB ID code 3MZ5; L125A, PDB ID code 3NXW).
See Commentary on page 6792.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200915109/-/DCSupplemental.
References
- 1.Bridgman PW. The coagulation of albumin by pressure. J Biol Chem. 1914;19:511–512. [Google Scholar]
- 2.Chalikian TV, Macgregor RB., Jr Origins of pressure-induced protein transitions. J Mol Biol. 2009;394:834–842. doi: 10.1016/j.jmb.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 3.Grigera JR, McCarthy AN. The behavior of the hydrophobic effect under pressure and protein denaturation. Biophys J. 2010;98:1626–1631. doi: 10.1016/j.bpj.2009.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ando N, et al. Structural and thermodynamic characterization of T4 lysozyme mutants and the contribution of internal cavities to pressure denaturation. Biochemistry. 2008;47:11097–11109. doi: 10.1021/bi801287m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frye KJ, Royer CA. Probing the contribution of internal cavities to the volume change of protein unfolding under pressure. Protein Sci. 1998;7:2217–2222. doi: 10.1002/pro.5560071020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lassalle MW, et al. Filling a cavity dramatically increases pressure stability of the c-Myb R2 subdomain. Proteins. 2001;45:96–101. doi: 10.1002/prot.1128. [DOI] [PubMed] [Google Scholar]
- 7.Weber G, Drickamer HG. The effect of high pressure upon proteins and other biomolecules. Q Rev Biophys. 1983;16:89–112. doi: 10.1017/s0033583500004935. [DOI] [PubMed] [Google Scholar]
- 8.Kauzmann W. Thermodynamics of unfolding. Nature. 1987;325:763–764. [Google Scholar]
- 9.Rouget JB, et al. Size and sequence and the volume change of unfolding. J Am Chem Soc. 2011;133:6020–6027. doi: 10.1021/ja200228w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Moreno B, et al. Experimental measurement of the effective dielectric in the hydrophobic core of a protein. Biophys Chem. 1997;64:211–224. doi: 10.1016/s0301-4622(96)02238-7. [DOI] [PubMed] [Google Scholar]
- 11.Till MS, Ullmann GM. McVol—a program for calculating protein volumes and identifying cavities by a Monte Carlo algorithm. J Mol Model. 2010;16:419–429. doi: 10.1007/s00894-009-0541-y. [DOI] [PubMed] [Google Scholar]
- 12.Vidugiris GJ, Markley JL, Royer CA. Evidence for a molten globule-like transition state in protein folding from determination of activation volumes. Biochemistry. 1995;34:4909–4912. doi: 10.1021/bi00015a001. [DOI] [PubMed] [Google Scholar]
- 13.Toptygin D, Woolf TB, Brand L. Picosecond protein dynamics: The origin of the time-dependent spectral shift in the fluorescence of the single Trp in the protein GB1. J Phys Chem B. 2010;114:11323–11337. doi: 10.1021/jp104425t. [DOI] [PubMed] [Google Scholar]
- 14.Collins MD, et al. Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation. Proc Natl Acad Sci USA. 2005;102:16668–16671. doi: 10.1073/pnas.0508224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rotkiewicz P, Skolnick J. Fast procedure for reconstruction of full-atom protein models from reduced representations. J Comput Chem. 2008;29:1460–1465. doi: 10.1002/jcc.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandrescu AT, Gittis AG, Abeygunawardana C, Shortle D. NMR structure of a stable “OB-fold” sub-domain isolated from staphylococcal nuclease. J Mol Biol. 1995;250:134–143. doi: 10.1006/jmbi.1995.0365. [DOI] [PubMed] [Google Scholar]
- 17.Foguel D, et al. Dissociation of amyloid fibrils of alpha-synuclein and transthyretin by pressure reveals their reversible nature and the formation of water-excluded cavities. Proc Natl Acad Sci USA. 2003;100:9831–9836. doi: 10.1073/pnas.1734009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paschek D, Hempel S, Garcia AE. Computing the stability diagram of the Trp-cage miniprotein. Proc Natl Acad Sci USA. 2008;105:17754–17759. doi: 10.1073/pnas.0804775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frye KJ, Perman CS, Royer CA. Testing the correlation between delta A and delta V of protein unfolding using m value mutants of staphylococcal nuclease. Biochemistry. 1996;35:10234–10239. doi: 10.1021/bi960693p. [DOI] [PubMed] [Google Scholar]
- 21.Frye KJ, Royer CA. The kinetic basis for the stabilization of staphylococcal nuclease by xylose. Protein Sci. 1997;6:789–793. doi: 10.1002/pro.5560060405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravindra R, Winter R. Pressure perturbation calorimetry: A new technique provides surprising results on the effects of co-solvents on protein solvation and unfolding behaviour. Chemphyschem. 2004;5:566–571. doi: 10.1002/cphc.200301080. [DOI] [PubMed] [Google Scholar]
- 23.Schweiker KL, Fitz VW, Makhatadze GI. Universal convergence of the specific volume changes of globular proteins upon unfolding. Biochemistry. 2009;48:10846–10851. doi: 10.1021/bi901220u. [DOI] [PubMed] [Google Scholar]
- 24.Sarupria S, Ghosh T, Garcia AE, Garde S. Studying pressure denaturation of a protein by molecular dynamics simulations. Proteins. 2010;78:1641–1651. doi: 10.1002/prot.22680. [DOI] [PubMed] [Google Scholar]
- 25.Brandts JF, Oliveira RJ, Westort C. Thermodynamics of protein denaturation. Effect of pressu on the denaturation of ribonuclease A. Biochemistry. 1970;9:1038–1047. doi: 10.1021/bi00806a045. [DOI] [PubMed] [Google Scholar]
- 26.Mitra L, Smolin N, Ravindra R, Royer C, Winter R. Pressure perturbation calorimetric studies of the solvation properties and the thermal unfolding of proteins in solution—experiments and theoretical interpretation. Phys Chem Chem Phys. 2006;8:1249–1265. doi: 10.1039/b516608j. [DOI] [PubMed] [Google Scholar]
- 27.Tsamaloukas AD, Pyzocha NK, Makhatadze GI. Pressure perturbation calorimetry of unfolded proteins. J Phys Chem B . 2010;114:16166–16170. doi: 10.1021/jp106294p. [DOI] [PubMed] [Google Scholar]
- 28.Lassalle MW, Akasaka K. The use of high-pressure nuclear magnetic resonance to study protein folding. Methods Mol Biol. 2007;350:21–38. doi: 10.1385/1-59745-189-4:21. [DOI] [PubMed] [Google Scholar]
- 29.Fuentes EJ, Wand AJ. Local stability and dynamics of apocytochrome b562 examined by the dependence of hydrogen exchange on hydrostatic pressure. Biochemistry. 1998;37:9877–9883. doi: 10.1021/bi980894o. [DOI] [PubMed] [Google Scholar]
- 30.Day R, Garcia AE. Water penetration in the low and high pressure native states of ubiquitin. Proteins. 2008;70:1175–1184. doi: 10.1002/prot.21562. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh T, Garcia AE, Garde S. Molecular dynamics simulations of pressure effects on hydrophobic interactions. J Am Chem Soc. 2001;123:10997–11003. doi: 10.1021/ja010446v. [DOI] [PubMed] [Google Scholar]
- 32.Hummer G, Garde S, Garcia AE, Paulaitis ME, Pratt LR. The pressure dependence of hydrophobic interactions is consistent with the observed pressure denaturation of proteins. Proc Natl Acad Sci USA. 1998;95:1552–1555. doi: 10.1073/pnas.95.4.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye K, Jing G, Wang J. Interactions between subdomains in the partially folded state of staphylococcal nuclease. Biochim Biophys Acta. 2000;1479:123–134. doi: 10.1016/s0167-4838(00)00060-1. [DOI] [PubMed] [Google Scholar]
- 34.Bedard S, Mayne LC, Peterson RW, Wand AJ, Englander SW. The foldon substructure of staphylococcal nuclease. J Mol Biol. 2008;376:1142–1154. doi: 10.1016/j.jmb.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watson E, Matousek WM, Irimies EL, Alexandrescu AT. Partially folded states of staphylococcal nuclease highlight the conserved structural hierarchy of OB-fold proteins. Biochemistry. 2007;46:9484–9494. doi: 10.1021/bi700532j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishii T, et al. Probing force-induced unfolding intermediates of a single staphylococcal nuclease molecule and the effect of ligand binding. Biochem Biophys Res Commun. 2008;375:586–591. doi: 10.1016/j.bbrc.2008.08.073. [DOI] [PubMed] [Google Scholar]
- 37.Kato S, Kamikubo H, Hirano S, Yamazaki Y, Kataoka M. Nonlocal interactions are responsible for tertiary structure formation in staphylococcal nuclease. Biophys J. 2010;98:678–686. doi: 10.1016/j.bpj.2009.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang M, Feng Y, Yao H, Wang J. Importance of the C-terminal loop L137-S141 for the folding and folding stability of staphylococcal nuclease. Biochemistry. 2010;49:4318–4326. doi: 10.1021/bi100118k. [DOI] [PubMed] [Google Scholar]
- 39.Kamagata K, Sawano Y, Tanokura M, Kuwajima K. Multiple parallel-pathway folding of proline-free Staphylococcal nuclease. J Mol Biol. 2003;332:1143–1153. doi: 10.1016/j.jmb.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Maki K, Cheng H, Dolgikh DA, Shastry MC, Roder H. Early events during folding of wild-type staphylococcal nuclease and a single-tryptophan variant studied by ultrarapid mixing. J Mol Biol. 2004;338:383–400. doi: 10.1016/j.jmb.2004.02.044. [DOI] [PubMed] [Google Scholar]
- 41.Maki K, Cheng H, Dolgikh DA, Roder H. Folding kinetics of staphylococcal nuclease studied by tryptophan engineering and rapid mixing methods. J Mol Biol. 2007;368:244–255. doi: 10.1016/j.jmb.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobs MD, Fox RO. Staphylococcal nuclease folding intermediate characterized by hydrogen exchange and NMR spectroscopy. Proc Natl Acad Sci USA. 1994;91:449–453. doi: 10.1073/pnas.91.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baldwin RL. Temperature dependence of the hydrophobic interaction in protein folding. Proc Natl Acad Sci USA. 1986;83:8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Privalov PL, Gill SJ. Stability of protein structure and hydrophobic interaction. Adv Protein Chem. 1988;39:191–234. doi: 10.1016/s0065-3233(08)60377-0. [DOI] [PubMed] [Google Scholar]
- 45.Dwyer JJ, et al. High apparent dielectric constants in the interior of a protein reflect water penetration. Biophys J. 2000;79:1610–1620. doi: 10.1016/S0006-3495(00)76411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karp DA, et al. High apparent dielectric constant inside a protein reflects structural reorganization coupled to the ionization of an internal Asp. Biophys J. 2007;92:2041–2053. doi: 10.1529/biophysj.106.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.