Abstract
The P-cluster of nitrogenase is largely known for its function to mediate electron transfer to the active cofactor site during catalysis. Here, we show that a P-cluster variant (designated P*-cluster), which consists of paired [Fe4S4]-like clusters, can catalyze ATP-independent substrate reduction in the presence of a strong reductant, europium(II) diethylenetriaminepentaacetate [Eu(II)-DTPA]. The observation of a decrease of activity in the rank ΔnifH, ΔnifBΔnifZ, and ΔnifB MoFe protein, which corresponds to a decrease of the amount of P*-clusters in these cofactor-deficient proteins, firmly establishes P*-cluster as a catalytically active metal center in Eu(II)-DTPA–driven reactions. More excitingly, the fact that P*-cluster is not only capable of catalyzing the two-electron reduction of proton, acetylene, ethylene, and hydrazine, but also capable of reducing cyanide, carbon monoxide, and carbon dioxide to alkanes and alkenes, points to a possibility of developing biomimetic catalysts for hydrocarbon production under ambient conditions.
Keywords: Fischer–Tropsch, CN−, CO, CO2
Nitrogenases are a family of metalloenzymes that catalyze the nucleotide-dependent reduction of dinitrogen to ammonia under ambient conditions. The best-characterized member of this enzyme family is the molybdenum (Mo)-nitrogenase of Azotobacter vinelandii, which consists of two redox-active component proteins (1). One of the proteins, termed iron (Fe) protein (encoded by nifH), is a γ2-dimer that contains a [Fe4S4] cluster between the two subunits and a MgATP binding site within each subunit; the other, termed molybdenum-iron (MoFe) protein (encoded by nifDK), is an α2β2-heterotetramer that houses two complex metal clusters per αβ-dimer: the P-cluster ([Fe8S7]), which is located at each α/β-subunit interface; and the iron-molybdenum (FeMo) cofactor, or FeMoco ([MoFe7S9C-homocitrate]), which is buried within each α-subunit (2–4). Catalysis by nitrogenase presumably involves repeated association and dissociation between the two component proteins and ATP-dependent electron transfer from the [Fe4S4] cluster of the Fe protein, through the P-cluster, to the FeMoco of MoFe protein, where substrate reduction occurs (5).
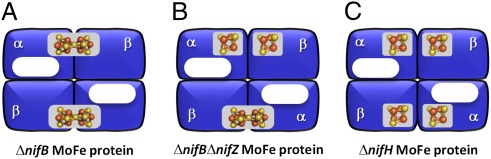
The P-cluster has long been regarded as a “capacitor” that mediates the electron transfer between the [Fe4S4] cluster of Fe protein and the cofactor site of MoFe protein during catalysis (1). Although the question of whether the P-cluster can directly reduce substrates was raised previously, it has remained a topic of debate because of the difficulty of distinguishing the contribution of the P-cluster from that of the cofactor to catalysis. As such, MoFe protein variants carrying the P-cluster species alone need to be generated to address the catalytic capacity of P-cluster. Recent studies of the biosynthesis of nitrogenase have led to the identification of three cofactor-deficient forms of MoFe protein (Fig. 1). One form, designated ΔnifB MoFe protein, contains two intact, [Fe8S7] P-clusters (6); another, designated ΔnifH MoFe protein, contains two P-cluster variants—each comprising a pair of [Fe4S4]-like clusters—in place of the two P-clusters (7–9); and the third, designated ΔnifBΔnifZ MoFe protein, contains one P-cluster and one pair of [Fe4S4]-like clusters (10, 11). Combined biochemical, electron paramagnetic resonance (EPR), X-ray absorption (XAS)/extended X-ray absorption fine structure (EXAFS), and magnetic circular dichroism (MCD) analyses (7–9) show that the [Fe4S4]-like cluster pair (designated P*-cluster) is in fact a precursor to the P-cluster, which can be reductively coupled into a [Fe8S7] structure upon incubation with dithionite, MgATP, and either Fe protein alone (in the case of ΔnifH MoFe protein) (12) or Fe protein plus NifZ (in the case of ΔnifBΔnifZ MoFe protein) (13).
Fig. 1.
Schematic presentations of cofactor-deficient MoFe proteins. The ΔnifB MoFe protein (A) contains two P-clusters (i.e., [Fe8S7] clusters); the ΔnifBΔnifZ MoFe protein (B) contains one P-cluster and one P*-cluster (i.e., paired [Fe4S4]-like clusters); and ΔnifH MoFe protein (C) contains two P*-clusters. The [Fe8S7] and [Fe4S4] clusters are represented by gray ovals and cubes, respectively; whereas the vacant cofactor sites are represented by white ovals.
The capture of these cofactor-deficient forms of MoFe protein affords an unprecedented opportunity to examine the catalytic competence of P-cluster species without the interference of the cofactor. Moreover, the distinct P-cluster contents of these MoFe proteins permit a comparison between the catalytic capacities of P- and P*-clusters, as well as a correlation between the structure and function of the P-cluster species. Here, we present a study of the substrate-reducing activities of ΔnifH, ΔnifBΔnifZ, and ΔnifB MoFe proteins in Fe protein/ATP-free, europium (II) diethylenetriaminepentaacetate [Eu(II)-DTPA]-driven reactions. The observation of a decrease of activity in the rank ΔnifH, ΔnifBΔnifZ, and ΔnifB MoFe protein, which corresponds to a decrease of P*-cluster content in these proteins, firmly establishes P*-cluster as the catalytically active metallocenter in these reactions. More excitingly, the fact that P*-cluster is not only capable of catalyzing the two-electron reduction of proton, acetylene, ethylene, and hydrazine, but also capable of reducing cyanide, carbon monoxide, and carbon dioxide to alkanes and alkenes, points to a possibility of developing novel biomimetic catalysts for hydrocarbon production under ambient conditions.
Results
Because the conversion of the P*-cluster to P-cluster (12) requires the same factors as those for the substrate turnover (i.e., Fe protein, MgATP, and dithionite), the activity of P*-cluster cannot be assessed by a conventional nitrogenase assay. To circumvent this problem, a strong reductant, Eu(II)-DTPA (14), was used to drive the reactions in the absence of Fe protein, ATP, and dithionite. Eu(II)-DTPA has been used previously to drive substrate reduction by both a MoFe protein variant (15) and the isolated cofactors of nitrogenase (16) and, therefore, is an appropriate choice of reducing agent in this case. Indeed, the EPR spectra of ΔnifH, ΔnifBΔnifZ, and ΔnifB MoFe proteins show little or no change following the treatment with Eu(II)-DTPA (Fig. S1), suggesting that (i) these proteins and their associated clusters remain intact in the presence of Eu(II)-DTPA; and (ii) the P*-clusters in ΔnifH and ΔnifBΔnifZ MoFe proteins are not converted to the P-clusters by Eu(II)-DTPA.
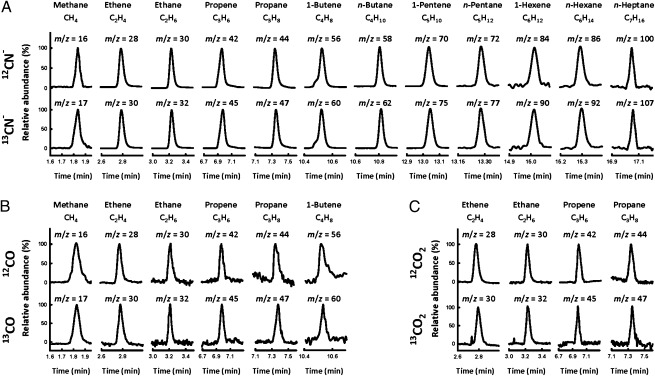
Driven by Eu(II)-DPTA (E0′ = −1.14 V at pH 8), the ΔnifH MoFe protein can reduce cyanide (CN−) to methane (CH4), ethene (C2H4), ethane (C2H6), propene (C3H6), propane (C3H8), 1-butene (C4H8), n-butane (C4H10), 1-pentene (C5H10), n-pentane (C5H12), 1-hexene (C6H12), n-hexane (C6H14), n-heptane (C7H16), and ammonia (NH3) under ambient conditions (Fig. 2A and Table 1). Thus, contrary to the wild-type MoFe protein, which generates CH4 and NH3 as the sole products of CN−-reduction in a Fe protein/ATP-driven reaction (1), the ΔnifH MoFe protein generates hydrocarbons of up to seven carbon-length as additional products in the same reaction driven by Eu(II)-DTPA. Consistent with this observation, the CH4:NH3 ratio of the reaction catalyzed by ΔnifH MoFe protein (1:8) is considerably lower than that by the wild-type MoFe protein (1:1), suggesting a redistribution of electrons available for CH4 formation toward C-C coupling in the case of the former.
Fig. 2.
Hydrocarbon formation by ΔnifH MoFe protein. GC-MS analysis of hydrocarbons generated from the reduction of CN− (A), CO (B), and CO2 (C). Products were generated from the 12C (Upper)- or 13C (Lower)-labeled CN− (A), CO (B), and CO2 (C). The mass-to-charge (m/z) ratios at which the products were traced are indicated in the figure.
Table 1.
Activities of hydrocarbon formation by cofactor-deficient MoFe proteins in Eu(II)-DTPA–driven reactions
| Activities |
|||
| Products | ΔnifB MoFe protein | ΔnifBΔnifZ MoFe protein | ΔnifH MoFe protein |
| From CN− | |||
| CH4 | 91 ± 14 | 468 ± 49 | 1,280 ± 204 |
| C2H4 | 85 ± 9 | 515 ± 51 | 1,230 ± 114 |
| C2H6 | 18 ± 2 | 117 ± 10 | 250 ± 20 |
| C3H6 | 20 ± 3 | 182 ± 13 | 475 ± 51 |
| C3H8 | 2.7 ± 0.3 | 36 ± 3 | 72 ± 9 |
| C4H8 | ND | 72 ± 8 | 160 ± 19 |
| C4H10 | ND | 7 ± 1 | 24 ± 2 |
| C5H10 | ND | 2.1 ± 0.3 | 67 ± 9 |
| C5H12 | ND | ND | 13 ± 1 |
| NH3 | 1,193 ± 114 | 4,648 ± 410 | 10,180 ± 1131 |
| From CO | |||
| CH4 | 6 ± 1 | 25 ± 3 | 83 ± 11 |
| C2H4 | 2.7 ± 0.3 | 11 ± 1 | 29 ± 4 |
| C2H6 | 0.9 ± 0.1 | 4 ± 4 | 14 ± 2 |
| C3H6 | ND | 1.4 ± 0.2 | 5 ± 1 |
| C3H8 | ND | 0.5 ± 0.1 | 2.7 ± 0.3 |
Activities are expressed in nanomole product/micromole protein per hour. See Materials and Methods for assay conditions. ND, not detectable.
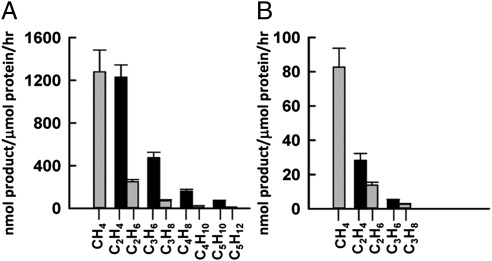
When CN− is replaced by CO, an isoelectronic carbonaceous molecule, a similar yet narrower set of hydrocarbons is generated by ΔnifH MoFe protein in the Eu(II)-DTPA–driven reaction. Only CH4, C2H4, C2H6, C3H6, C3H8, and 1-C4H8 are detected as products in this case, and the rates of product formation from CO-reduction are considerably lower than those from CN−-reduction by the ΔnifH MoFe protein (Fig. 2B and Table 1). The same discrepancy in reaction efficiency and product range is observed when CO and CN− are reduced by the isolated nitrogenase cofactors in Eu(II)-DTPA–based reactions (16) and, given the topological analogy between the P-cluster and cofactor (17, 18), it could be explained by the previous observation of a stabilizing effect of CN−-binding on certain redox states of these clusters (19), such that the clusters are more effective in substrate reduction. Nevertheless, the two reactions share a similar product profile, showing a decrease in the efficiency of product formation with an increase in product length, as well as a preference for the formation of unsaturated 1-alkenes over saturated n-alkanes (Fig. 3), both of which are consistent with the earlier proposal of a common reaction pathway for CN−- and CO-reduction (20). GC-MS analysis further confirms that CO and CN− are the carbon sources of hydrocarbons formed in these reactions, because all products display the expected mass shifts when 12CO and 12CN− are replaced by 13CO and 13CN−, respectively (Fig. 2 A and B).
Fig. 3.
Product distributions of reactions catalyzed by ΔnifH MoFe protein. Specific activities of hydrocarbon formation from the reduction of CN− (A) and CO (B), which were calculated based on the rates of product formation in the linear phases (first 15 min) of these reactions (Fig. S2). Alkane and alkene products are presented in gray and black bars, respectively, in the figure. The alkene/alkane ratios are 4.9 (C2H4/C2H6), 6.6 (C3H6/C3H8), 6.7 (C4H8/C4H10), and 5.2 (C5H10/C5H12) in the reaction of CN−-reduction (A) and 2.1 (C2H4/C2H6) and 1.9 (C3H6/C3H8) in the reaction of CO-reduction (B).
The observation that ΔnifH MoFe protein is capable of reducing CO to hydrocarbons raises the question of whether this protein can generate hydrocarbons from CO2, particularly given the previous observation that the wild-type MoFe protein can reduce CO2 to CO (21). Indeed, when a mixture of carbon dioxide/bicarbonate (CO2/HCO3−) is used as a substrate, C2H4, C2H6, C3H6, and C3H8 are detected as products in the Eu(II)-DTPA–driven, ΔnifH MoFe protein-based reaction (Fig. 2C). The same set of products can be identified with the expected mass shifts when 12CO2/H12CO3− is replaced by 13CO2/H13CO3− (Fig. 2C). However, the rate of CO2-reduction by ΔnifH MoFe protein is too low to be accurately determined by GC analysis. Additionally, CH4, the major product in ΔnifH MoFe protein-catalyzed reduction of CN− and CO, cannot be clearly identified in this case (Fig. 2C).
Apart from CN−, CO, and CO2, other nitrogenase substrates can also be reduced by the ΔnifH MoFe protein in the presence of Eu(II)-DTPA. Like the wild-type MoFe protein, the ΔnifH MoFe protein can reduce proton (H+) to hydrogen (H2), acetylene (C2H2) to C2H4, C2H4 to C2H6, azide (N3−) to NH3, and hydrazine (N2H4) to NH3 (Table S1). However, contrary to the wild-type MoFe protein, the ΔnifH MoFe protein: (i) does not reduce N2 to NH3; (ii) generates C2H6 in addition to C2H4 from C2H2; and (iii) reduces C2H4 to C2H6 in the Eu(II)-DTPA–driven reaction (Table S1). Moreover, the ΔnifH MoFe protein/Eu(II)-DTPA system is far less efficient than the wild-type MoFe protein/Fe protein/ATP system in reducing H+, N2H4, N3− and C2H2 (Table S1), as the rates of product formation from these substrates by the Eu(II)-DTPA–based system range between 0.5% and 3.5% of those by the Fe protein/ATP-based system.† Interestingly, although a similar discrepancy in efficiency is also observed between the ΔnifH and wild-type MoFe protein-catalyzed reactions of CO-reduction, the rate of product formation by the former is still 20% of that by the latter (see comment regarding calculations, above), suggesting a preference of CO over other substrates by the ΔnifH MoFe protein/Eu(II)-DTPA system.
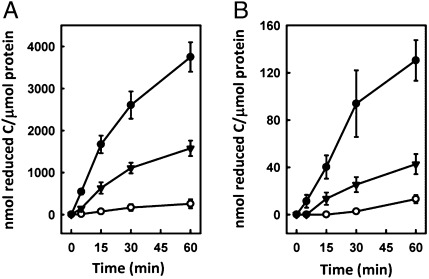
The distinct reactivity of ΔnifH MoFe protein is clearly associated with its unique P*-cluster species. The rates of product formation by the three cofactor-deficient forms of MoFe protein are ranked in the order of ΔnifB MoFe protein (containing two P-clusters) < ΔnifBΔnifZ MoFe protein (containing one P-cluster and one P*-cluster) < ΔnifH MoFe protein (containing two P*-clusters), illustrating that the [Fe4S4]-like cluster pair (i.e., P*-cluster) is a much better catalyst than the [Fe8S7] cluster (i.e., P-cluster) in the Eu(II)-DTPA–driven reactions (Table 1 and Table S1). In particular, the overall efficiencies of CN−- and CO-reduction by ΔnifBΔnifZ MoFe protein are 39% and 31%, respectively, of those by ΔnifH MoFe protein (Fig. 4), both of which are close to—albeit lower than—the theoretical number of 50% that is derived from the half P*-cluster content in the ΔnifBΔnifZ MoFe protein (Fig. 1). In contrast, the ΔnifB MoFe protein is much less effective in substrate reduction than either ΔnifBΔnifZ or ΔnifH MoFe protein, suggesting that the P-cluster is minimally active in the reactions driven by Eu(II)-DTPA (Fig. 4). Together, these data strongly indicate that the P*-cluster is the catalytically active species in Eu(II)-DTPA–based reactions.
Fig. 4.
Activities of cofactor-deficient MoFe proteins in hydrocarbon formation. Time-dependent formation of hydrocarbon products from the reduction of CN− (A) and CO (B) by ΔnifH (●), ΔnifBΔnifZ (▼), and ΔnifB (○) MoFe proteins. Activities were expressed in nanomole total reduced carbon/micromole protein, which were calculated based on the data in Fig. S2.
Discussion
A previous small angle X-ray scattering study has shown that the ΔnifH MoFe protein exists in a more extended conformation than the ΔnifB MoFe protein and that the increase in the size of ΔnifH MoFe protein is correlated with an increase in the solvent accessibility of its P*-cluster Fe atoms (22). Such a conformational change of ΔnifH MoFe protein can be best modeled by having a 6 Å gap at its α/β-subunit interface, which is absent from the structure of ΔnifB MoFe protein (22). The gap between the α- and β-subunits of ΔnifH MoFe protein not only results in a “split” of the P-cluster ([Fe8S7] structure) into a P*-cluster (paired [Fe4S4]-like structure), but also renders the P*-cluster easily accessible to both substrates and electrons, an advantage not shared by the well-shielded, intact P-cluster in ΔnifB MoFe protein in the Eu(II)-DTPA–driven reactions. It should be noted, however, that during ATP-dependent substrate turnover, MoFe protein normally undergoes a conformational change upon the docking of Fe protein (1, 5), which may render the P-cluster more accessible to substrates. Moreover, the redox change of P-cluster in this process could be accompanied by a structural change of the cluster toward a more “open” conformation, such as that observed in the case of the two-electron oxidized P-cluster (P2+). Interestingly, theoretical calculations have used a split [Fe4S4] cluster model to explain the spin state of P2+ (23), implying that a P*-cluster-like intermediate could be generated during the process of substrate turnover.
Facilitated by its unique location, the ΔnifH MoFe protein-associated P*-cluster takes up a resemblance to a surface-exposed Fe/S cluster that is capable of substrate reduction when a proper reducing agent is supplied. Like some of the synthetic Fe/S-based clusters, which can reduce C2H2 (24), H+ (25), and N2H4 (26) in the presence of strong reductants, the P*-cluster can catalyze the simple, two-electron reduction of the same set of substrates in reactions driven by Eu(II)-DTPA. However, this cluster can also reduce CN− and CO to hydrocarbons under ambient conditions, a catalytic property that has been attributed so far only to the isolated and protein-bound cofactors of nitrogenase (16, 27, 28). Consistent with its surface location, the P*-cluster of ΔnifH MoFe protein behaves more like the isolated cofactors in the Eu(II)-DTPA–driven reactions of CN−- and CO-reduction (16), because all these clusters (i) produce CH4 as the major product and (ii) show a clear and consistent preference for the formation of 1-alkenes over n-alkanes at alkene/alkane ratios of as high as 6.7/1 (Fig. 3). These observations point to a structural-functional analogy between the P-cluster and cofactor species of nitrogenases; moreover, they strengthen the link between nitrogenase- and Fischer–Tropsch-based reactions of hydrocarbon formation, as the latter reaction also has a well-documented tendency toward excess methanation and high ratios of alkene-to-alkane formation (29).
The catalytic properties of P*-cluster originate from its unique structural arrangement. Previous XAS/EXAFS studies of ΔnifH MoFe protein have shown that the P*-cluster consists of one standard [Fe4S4] subcubane and one atypical [Fe4S4]-like subcubane (8, 12). Thus, the P*-cluster in ΔnifH MoFe protein may resemble the catalytic modules in Ni/Fe-containing carbon monoxide dehydrogenase/acetyl-CoA synthase (30), siroheme-type sulfite reductase (31), and iron-only hydrogenase (32), all of which comprise a [Fe4S4] cluster that is either positioned near or bridged to a second metal-containing cluster, thereby permitting the transfer of electrons from the former to the latter for the subsequent reduction of substrates at the latter site. By analogy, the P*-cluster could follow the same mode of action as these two enzymes, transferring electrons from the standard [Fe4S4] cluster to the atypical [Fe4S4]-like cluster, where substrate reduction takes place. Given the capacity of standard, ferredoxin-type [Fe4S4] clusters in electron transfer (33, 34), such a hypothesis could account for the ability of P*-cluster to reduce a variety of substrates by two electrons. On the other hand, a different and perhaps more intricate mechanism may be used by the P*-cluster during catalysis, as this cluster is also capable of reducing more “complex” substrates, such as CN−, CO, and CO2, than those reduced by only two electrons.
Regardless, the fact that the surface-exposed P*-cluster and solvent-isolated cofactors can catalyze the ATP-independent reduction of CN−, CO, and CO2 points to a vivid possibility of developing biomimetic catalysts for hydrocarbon production under ambient conditions. Moreover, the observation that the biosynthetic precursors of P-cluster (e.g., the P*-clusters in ΔnifH MoFe protein) and cofactor [e.g., the L-cluster in NifEN (35–37) (see below)] show catalytic activities similar to, yet distinct from those of their mature counterparts implies that nitrogenase clusters might have evolved through several primordial intermediate stages, where they took on different functions, such as detoxification (e.g., reduction of C2H2 and N3−), hydrogenation (i.e., reduction of H+), carbon fixation (e.g., reduction of CN−, CO and CO2), and nitrogen fixation (i.e., reduction of N2).‡ Finally, given the topological analogy between the P-cluster and cofactor, it is chemically plausible that these clusters evolved from a common ancestral precursor (18, 38). This hypothesis raises the provocative question of whether the P*-cluster in ΔnifH MoFe protein represents a structural homolog of such a common precursor, particularly in light of the observed parallelism between the CO-reducing abilities of the P*-cluster and cofactor. Studies of the biosynthetic intermediates of nitrogenase, therefore, may not only shed light on the evolution of nitrogen fixation, but also unearth other lost functions—such as the reduction of CO to hydrocarbons—of early biological systems.
Materials and Methods
Cell Growth and Protein Purification.
A. vinelandii strains DJ1143, YM6A, and DJ1165 were grown in 180 L batches in a 200 L New Brunswick fermentor (New Brunswick Scientific) in Burke’s minimal medium supplemented with 2 mM ammonium acetate. The growth rate was measured by cell density at 436 nm using a Spectronic 20 Genesys (Spectronic Instruments). Cells were harvested in the late exponential phase using a flow-through centrifugal harvester (Cepa). The cell paste was washed with a buffer containing 50 mM Tris-HCl (pH 8.0). Previously published methods were used for the purification of His-tagged ΔnifB, ΔnifBΔnifZ, and ΔnifH MoFe proteins (7, 10).
EPR Spectroscopy.
All EPR samples were prepared in a Vacuum Atmospheres dry box at an oxygen level of less than 4 ppm. The ΔnifB, ΔnifBΔnifZ, and ΔnifH MoFe proteins were incubated with 5 mM Eu(II)-DTPA for 15 min, followed by the removal of Eu(II)-DTPA with a G25 column. Subsequently, these proteins were incubated with 2 mM sodium dithionite (Na2S2O4) before being transferred to and frozen in the EPR sample tubes. All protein samples contained 25 mM Tris-HCl (pH 8.0), 10% (vol/vol) glycerol, and 2 mM Na2S2O4. Perpendicular-mode EPR spectra of these samples were recorded by a Bruker ESP 300 Ez spectrometer (Bruker) interfaced with an Oxford Instruments ESR-9002 liquid helium continuous-flow cryostat (Oxford Instruments). All spectra were recorded at 10 K using a microwave frequency of 9.62 GHz, a microwave power of 50 mW, a gain of 5 × 104, a modulation frequency of 100 kHz, and a modulation amplitude of 5 G. A total of 10 scans were collected for each sample.
Activity Assays in Eu(II)-DTPA.
Eu(II)-DTPA was prepared as described previously (14). All assays contained, in a total volume of 25 mL, 25 mM Tris-HCl (pH 7.8), 5 mM Eu-DTPA, and 80 mg of ΔnifH, ΔnifBΔnifZ, or ΔnifB MoFe protein. The reaction of C2H2-, C2H4-, or CO-reduction was carried out in a gas atmosphere of 100% (vol/vol) C2H2, C2H4, or CO; whereas the reaction of CO2-reduction was carried out in a gas atmosphere of 0.05% CO2/99.95% (vol/vol) Ar and maintained at pH 7.8, with the addition of 0.5 M NaHCO3 to the assay. All other assays were carried out in a gas atmosphere of 100% (vol/vol) Ar, with the addition of 120 mM NaCN and 100 mM NaN3, respectively, to the reactions of CN−- and N3−-reduction. Production of H2 was analyzed as described previously (39), and formation of NH3 was determined by an HPLC fluorescence method (40). Alkene and alkane products CH4, C2H4, C2H6, C3H6, C3H8, 1-C4H8, n-C4H10, 1-C5H10, n-C5H12, 1-C6H12, n-C6H14, and n-C7H16 were analyzed by GC-FID on an activated alumina column (Grace), which was held at 40 °C for 2 min, increased to 200 °C at a rate of 10 °C/min, and held for an additional 2 min at 200 °C. These 12 hydrocarbons were quantified as described previously (16) and their detection thresholds were (in nanomole product/micromole protein): 1.1 (CH4), 1.3 (C2H4), 1.3 (C2H6), 1.5 (C3H6), 1.3 (C3H8), 1.4 (1-C4H8), 1.4 (n-C4H10), 3.1 (1-C5H10), 3.7 (n-C5H12), 6.1 (1-C6H12), 5.8 (n-C6H14), and 7.0 (n-C7H16), respectively. In control assays (i.e., without the ΔnifH MoFe protein), H2 (under Ar) and C2H4 (under C2H2) were produced at 27% and 9%, respectively, of those in the complete assays (i.e., with the ΔnifH MoFe protein). The specific activities of H2 and C2H4 formation were reported after subtracting the background activities of the control assays from those of the complete assays. No activities were observed in the cases of other substrates in the absence of ΔnifH MoFe protein.
GC-MS Analysis.
Hydrocarbon products were identified by GC-MS using a Hewlett-Packard 5890 GC and a 5972 MSD. The identities of CH4, C2H4, C2H6, C3H6, C3H8, 1-C4H8, n-C4H10, 1-C5H10, n-C5H12, 1-C6H12, n-C6H14, and n-C7H16 were confirmed by using a Scott standard gas mixture of n-alkanes and 1-alkenes. A total of 50 μL of gas was injected into a split/splitless injector operated at 125 °C in splitless mode. A 1-mm ID liner was used to optimize the sensitivity of gas detection. The separation of gaseous products was achieved by using a Restek PLOT-QS capillary column (0.320 mm ID × 30 m length), which was held at 40 °C for one min, heated to 220 °C at a rate of 10 °C/min, and held for another 3 min at 220 °C. Carrier gas (He) was passed through the column at a rate of 1.0 mL/min. The mass spectrometer was operated in electron impact ionization (EII) and selected ion monitoring mode.
Supplementary Material
Acknowledgments
We thank Prof. Douglas Rees [California Institute of Technology(CIT)] and Dr. Nathan Dalleska (CIT) for their kind help with GC-MS analysis. This work was supported by Herman Frasch Foundation Grant 617-HF07 and National Institutes of Health Grant GM-67626 (to M.W.R.).
Footnotes
The authors declare no conflict of interest.
†These calculations were based on the activities of the same molar amount of wild-type MoFe protein under conditions identical to those of the ΔnifH MoFe protein-based reactions, except that Eu(II)-DTPA was replaced by excess Fe protein and MgATP.
‡The L-cluster is a precursor form of FeMoco, which can be converted to a mature cofactor on NifEN before it is delivered to its target location in the MoFe protein. XAS/EXAFS and X-ray crystallographic studies show that the L-cluster is an 8Fe homolog to FeMoco, which is free of Mo and homocitrate (35, 36). Moreover, biochemical analyses reveal that the NifEN-bound L-cluster is catalytically active and capable of reducing C2H2 and N3− to C2H4 and NH3, respectively, in a Fe protein/ATP-dependent reaction (37).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202429109/-/DCSupplemental.
References
- 1.Burgess BK, Lowe DJ. Mechanism of molybdenum nitrogenase. Chem Rev. 1996;96:2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Einsle O, et al. Nitrogenase MoFe-protein at 1.16 A resolution: A central ligand in the FeMo-cofactor. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 3.Lancaster KM, et al. X-ray emission spectroscopy evidences a central carbon in the nitrogenase iron-molybdenum cofactor. Science. 2011;334:974–977. doi: 10.1126/science.1206445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spatzal T, et al. Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science. 2011;334:940. doi: 10.1126/science.1214025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction. Nature. 1997;387:370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- 6.Schmid B, et al. Structure of a cofactor-deficient nitrogenase MoFe protein. Science. 2002;296:352–356. doi: 10.1126/science.1070010. [DOI] [PubMed] [Google Scholar]
- 7.Ribbe MW, Hu Y, Guo M, Schmid B, Burgess BK. The FeMoco-deficient MoFe protein produced by a nifH deletion strain of Azotobacter vinelandii shows unusual P-cluster features. J Biol Chem. 2002;277:23469–23476. doi: 10.1074/jbc.M202061200. [DOI] [PubMed] [Google Scholar]
- 8.Corbett MC, et al. Comparison of iron-molybdenum cofactor-deficient nitrogenase MoFe proteins by X-ray absorption spectroscopy: implications for P-cluster biosynthesis. J Biol Chem. 2004;279:28276–28282. doi: 10.1074/jbc.M403156200. [DOI] [PubMed] [Google Scholar]
- 9.Broach RB, et al. Variable-temperature, variable-field magnetic circular dichroism spectroscopic study of the metal clusters in the DeltanifB and DeltanifH mofe proteins of nitrogenase from Azotobacter vinelandii. Biochemistry. 2006;45:15039–15048. doi: 10.1021/bi061697p. [DOI] [PubMed] [Google Scholar]
- 10.Hu Y, Fay AW, Dos Santos PC, Naderi F, Ribbe MW. Characterization of Azotobacter vinelandii nifZ deletion strains. Indication of stepwise MoFe protein assembly. J Biol Chem. 2004;279:54963–54971. doi: 10.1074/jbc.M408983200. [DOI] [PubMed] [Google Scholar]
- 11.Cotton MS, et al. VTVH-MCD study of the Δ nifB Δ nifZ MoFe protein from Azotobacter vinelandii. J Am Chem Soc. 2009;131:4558–4559. doi: 10.1021/ja807525m. [DOI] [PubMed] [Google Scholar]
- 12.Lee CC, et al. Stepwise formation of P-cluster in nitrogenase MoFe protein. Proc Natl Acad Sci USA. 2009;106:18474–18478. doi: 10.1073/pnas.0909149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Fay AW, Lee CC, Ribbe MW. P-cluster maturation on nitrogenase MoFe protein. Proc Natl Acad Sci USA. 2007;104:10424–10429. doi: 10.1073/pnas.0704297104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vincent KA, et al. Instantaneous, stoichiometric generation of powerfully reducing states of protein active sites using Eu(II) and polyaminocarboxylate ligands. Chem Commun (Camb) 2003;21:2590–2591. doi: 10.1039/b308188e. [DOI] [PubMed] [Google Scholar]
- 15.Danyal K, et al. Uncoupling nitrogenase: Catalytic reduction of hydrazine to ammonia by a MoFe protein in the absence of Fe protein-ATP. J Am Chem Soc. 2010;132:13197–13199. doi: 10.1021/ja1067178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee CC, Hu Y, Ribbe MW. ATP-independent formation of hydrocarbons catalyzed by isolated nitrogenase cofactors. Angew Chem Int Ed Engl. 2012;51:1947–1949. doi: 10.1002/anie.201108916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Holm RH. Structural conversions of molybdenum-iron-sulfur edge-bridged double cubanes and P(n)-type clusters topologically related to the nitrogenase P-cluster. Inorg Chem. 2004;43:674–682. doi: 10.1021/ic030259t. [DOI] [PubMed] [Google Scholar]
- 18.Ohki Y, Ikagawa Y, Tatsumi K. Synthesis of new [8Fe-7S] clusters: A topological link between the core structures of P-cluster, FeMo-co, and FeFe-co of nitrogenases. J Am Chem Soc. 2007;129:10457–10465. doi: 10.1021/ja072256b. [DOI] [PubMed] [Google Scholar]
- 19.Pickett CJ, et al. Synergic binding of carbon monoxide and cyanide to the FeMo cofactor of nitrogenase: Relic chemistry of an ancient enzyme? Chemistry. 2004;10:4770–4776. doi: 10.1002/chem.200400382. [DOI] [PubMed] [Google Scholar]
- 20.Carnahan EM, Protasiewicz JD, Lippard SJ. 15 years of reductive coupling: What have we learned? Acc Chem Res. 1993;26:90–97. [Google Scholar]
- 21.Seefeldt LC, Rasche ME, Ensign SA. Carbonyl sulfide and carbon dioxide as new substrates, and carbon disulfide as a new inhibitor, of nitrogenase. Biochemistry. 1995;34:5382–5389. doi: 10.1021/bi00016a009. [DOI] [PubMed] [Google Scholar]
- 22.Corbett MC, et al. Conformational differences between Azotobacter vinelandii nitrogenase MoFe proteins as studied by small-angle X-ray scattering. Biochemistry. 2007;46:8066–8074. doi: 10.1021/bi7005064. [DOI] [PubMed] [Google Scholar]
- 23.Torres RA, Lovell T, Noodleman L, Case DA. Density functional and reduction potential calculations of Fe4S4 clusters. J Am Chem Soc. 2003;125:1923–1936. doi: 10.1021/ja0211104. [DOI] [PubMed] [Google Scholar]
- 24.McMillan RS, Renaud J, Reynolds JG, Holm RH. Biologically related iron-sulfur clusters as reaction centers. Reduction of acetylene to ethylene in systems based on [Fe4S4(SR)4]3- J Inorg Biochem. 1979;11:213–227. doi: 10.1016/s0162-0134(00)80019-7. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh S, et al. Bio-inspired hydrogenase models: Mixed-valence triion complexes as proton reduction catalysts. Chem Commun (Camb) 2011;47:11222–11224. doi: 10.1039/c1cc13249k. [DOI] [PubMed] [Google Scholar]
- 26.Malinak SM, Demadis KD, Coucouvanis D. Catalytic reduction of hydrazine of ammonia by the VFe3S4 cubanes—Further evidence for the direct involvement of the heterometal in the reduction of nitrogenase substrates and possible relevance of the vanadium nitrogenase. J Am Chem Soc. 1995;117:3126–3133. [Google Scholar]
- 27.Lee CC, Hu Y, Ribbe MW. Vanadium nitrogenase reduces CO. Science. 2010;329:642. doi: 10.1126/science.1191455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, Lee CC, Ribbe MW. Extending the carbon chain: Hydrocarbon formation catalyzed by vanadium/molybdenum nitrogenases. Science. 2011;333:753–755. doi: 10.1126/science.1206883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santilli DS, Castner DG. Mechanism of chain growth and product formation for the Fischer–Tropsch reaction over iron catalysts. Energy Fuels. 1989;3:8–15. [Google Scholar]
- 30.Doukov TI, Iverson TM, Seravalli J, Ragsdale SW, Drennan CL. A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Science. 2002;298:567–572. doi: 10.1126/science.1075843. [DOI] [PubMed] [Google Scholar]
- 31.Oliveira TF, et al. The crystal structure of Desulfovibrio vulgaris dissimilatory sulfite reductase bound to DsrC provides novel insights into the mechanism of sulfate respiration. J Biol Chem. 2008;283:34141–34149. doi: 10.1074/jbc.M805643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 33.Beinert H. Iron-sulfur proteins: Ancient structures, still full of surprises. J Biol Inorg Chem. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- 34.Beinert H, Holm RH, Münck E. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 35.Corbett MC, et al. Structural insights into a protein-bound iron-molybdenum cofactor precursor. Proc Natl Acad Sci USA. 2006;103:1238–1243. doi: 10.1073/pnas.0507853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaiser JT, Hu Y, Wiig JA, Rees DC, Ribbe MW. Structure of precursor-bound NifEN: A nitrogenase FeMo cofactor maturase/insertase. Science. 2011;331:91–94. doi: 10.1126/science.1196954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y, et al. Catalytic activities of NifEN: Implications for nitrogenase evolution and mechanism. Proc Natl Acad Sci USA. 2009;106:16962–16966. doi: 10.1073/pnas.0907872106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Zuo JL, Zhou HC, Holm RH. Rearrangement of symmetrical dicubane clusters into topological analogues of the P cluster of nitrogenase: Nature’s choice? J Am Chem Soc. 2002;124:14292–14293. doi: 10.1021/ja0279702. [DOI] [PubMed] [Google Scholar]
- 39.Gavini N, Burgess BK. FeMo cofactor synthesis by a nifH mutant with altered MgATP reactivity. J Biol Chem. 1992;267:21179–21186. [PubMed] [Google Scholar]
- 40.Corbin JL. Liquid chromatographic-fluorescence determination of ammonia from nitrogenase reactions: A 2-min assay. Appl Environ Microbiol. 1984;47:1027–1030. doi: 10.1128/aem.47.5.1027-1030.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.