Abstract
The key regulators of intracellular trafficking, Ypt/Rab GTPases, are stimulated by specific upstream activators and, when activated, recruit specific downstream effectors to mediate membrane-transport events. The yeast Ypt1 and its human functional homolog hRab1 regulate both endoplasmic reticulum (ER)-to-Golgi transport and autophagy. However, it is not clear whether the mechanism by which these GTPases regulate autophagy depends on their well-documented function in ER-to-Golgi transport. Here, we identify Atg11, the preautophagosomal structure (PAS) organizer, as a downstream effector of Ypt1 and show that the Ypt1–Atg11 interaction is required for PAS assembly under normal growth conditions. Moreover, we show that Ypt1 and Atg11 colocalize with Trs85, a Ypt1 activator subunit, and together they regulate selective autophagy. Finally, we show that Ypt1 and Trs85 interact on Atg9-containing membranes, which serve as a source for the membrane component of the PAS. Together our results define a Ypt/Rab module—comprising an activator, GTPase, and effector—that orchestrates the onset of selective autophagy, a process vital for cell homeostasis. Furthermore, because Atg11 does not play a role in ER-to-Golgi transport, we demonstrate here that Ypt/Rabs can regulate two independent membrane-transport processes by recruiting process-specific effectors.
The conserved Ypt/Rab GTPases act as membrane organizers to regulate intracellular trafficking pathways. When stimulated by exchange factors termed “guanine nucleotide exchange factors” (GEFs), they interact with multiple downstream effectors, which mediate the different steps of vesicular trafficking (1, 2). In yeast, Ypt1 is required for endoplasmic reticulum (ER)-to-Golgi transport (3–5), and the TRAPP I complex acts as its GEF (6, 7). Rab1, the human functional homolog of Ypt1, also plays a role in ER-to-Golgi transport (8, 9). Conserved tethering factors, such as Uso1/p115, have been identified as downstream effectors of Ypt1 and hRab1 in ER-to-Golgi transport (10, 11).
Autophagy is a cellular recycling process. In this process, a double membrane surrounds parts of the cytoplasm, including cellular organelles, to form the autophagosome, which fuses with the lysosome (the vacuole in yeast), where macromolecules are degraded. Under stress conditions, such as starvation, nonselective autophagy is induced (12). In contrast, selective autophagy, in which specific cellular components are recycled, plays a role in cell homeostasis and therefore is important for human development and disease (13). The best-characterized type of selective autophagy is the cytosol-to-vacuole (CVT) pathway, which delivers specific enzymes from the cytoplasm to the vacuole under normal growth conditions. A conserved set of autophagy-specific proteins, Atgs, is required for the different types of autophagy. All types of autophagy start with the formation of the preautophagosomal structure (PAS), which originally was defined as a conserved multiprotein complex. More recently it was suggested that Atg9, an integral-membrane protein required for all types of autophagy, supplies the membrane component to the PAS (14). At present, it is not clear how the autophagy-specific and the membrane-trafficking machinery intersect to generate the autophagosome.
Although several Ypt/Rabs have been implicated in autophagy, the molecular mechanisms that underlie their function in this process are mostly unknown. Ypt1 and its mammalian homolog Rab1 play a role in autophagy (15, 16), and Trs85, in the context of the TRAPP III complex, can act as a Ypt1 GEF in this process (17). However, the molecular mechanism by which Ypt1 and Rab1 regulate autophagy is unknown, and it is not clear whether it is dependent on their well-documented function in ER-to-Golgi transport.
Atg11 is a PAS scaffold protein required for different types of selective autophagy including CVT (18, 19). Here, we used a combination of biochemistry, genetics, and imaging approaches to identify Atg11 as a downstream effector of Ypt1 and to show that the Ypt1–Atg11 interaction is required for PAS assembly under normal growth conditions. Moreover, we show that Trs85, Ypt1, and Atg11 function as one module and interact on Atg9-containing membranes and on the PAS. These results define a module comprising a GEF, Trs85-containing TRAPP III, Ypt/Rab GTPase, Ypt1, and an effector, Atg11, that plays a role at the onset of autophagy. Because Ypt1 and TRAPP complexes are involved in both ER-to-Golgi and autophagy, we propose that they coordinate the divergence of these processes by recruiting process-specific effectors.
Results
Atg11 Is a Downstream Effector of Ypt1.
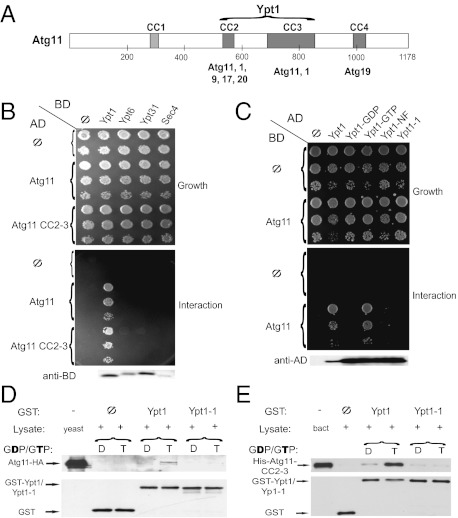
Atg11, which interacts with multiple Atg proteins through three of its coiled-coil (CC) domains (Fig. 1A) (19), was identified as a Ypt1 interactor in two independent yeast-two hybrid screens. We verified this interaction in both plasmid orientations (Fig. 1 B and C) and showed that it is specific to Ypt1, because Atg11 does not interact with Ypt6, Ypt31, or Sec4 (Fig. 1B); the last two play a role in autophagy (20). Furthermore, Atg11 interaction with Ypt1 is nucleotide specific, because it interacts with the GTP but not the GDP or nucleotide-free form of Ypt1 (Fig. 1C). CC 2 and 3 of Atg11 are required and sufficient for the interaction with Ypt1-GTP (Fig. 1B and Fig. S1). These results suggest that Atg11 is a Ypt1 effector, with the middle region of Atg11 mediating the interaction. This region is involved in multiple Atg11 interactions and is required for its function in selective autophagy (19).
Fig. 1.
Atg11 is a Ypt1 effector. (A) Schematic diagram of Atg11, its coiled-coil domains (CC), and interactors. CC2–4 positioning is based on COILS (http://www.ch.embnet.org/software/COILS_form.html); CC1 was suggested in ref. 19); interactions shown under CC2–4 were previously reported (19, 23); and the interaction with Ypt1 is reported here. (B) Ypt1, but not other Ypts, interacts with Atg11 in the yeast-two hybrid (Y2H) assay. Interaction of Atg11 and Atg11-CC2-3 with the GTP-restricted forms of the Ypts (Ypt1-Q67L, Ypt6-Q69L, Ypt31-Q72L, and Sec4-Q79L) was determined. (C) The Y2H interaction of Ypt1 with Atg11 is nucleotide specific. Only the wild type (Ypt1) and Ypt1-GTP (Q67L) interact with Atg11, whereas Ypt1-GDP (S22N), the nucleotide-free form (Ypt1-NF, D124N), and Ypt1-1 (T40K) do not. Immunoblot analysis (lower panels in B and C) shows expression of the different Ypt proteins. (D) Atg11-HA from yeast cell lysates coprecipitates with purified Ypt1-GTP but not with Ypt1-GDP or Ypt1-1 mutant protein (GTP or GDP). Atg11-HA (10% of lysate) coprecipitated preferentially with GST-Ypt1 loaded with GTP (T) (0.49 ± 0.02% of lysate above the background), and not with GST-Ypt1 loaded with GDP (D), GST-Ypt1-1 loaded with GTP or with GDP or GST (Φ). (E) Recombinant Ypt1, but not Ypt1-1, interacts with Atg11-CC2-3. The experiment was done as described in D, except that coprecipitation was done with bacterial lysates expressing His6-tagged Atg11-CC2-3 (10% loaded). His6-Atg11-CC2-3 coprecipitates preferentially with GST-Ypt1-GTP and not with GST-Ypt1-GDP or GST-Ypt1-1 GDP or with GTP, or GST (Φ). The level of precipitated GST-tagged proteins is shown at the bottom of panels D and E. Results are representative of at least two independent experiments.
To determine whether the Ypt1–Atg11 interaction occurs in vitro, we tested the coprecipitation of HA-tagged Atg11 from yeast lysates with purified recombinant GST-Ypt1. Atg11-HA coprecipitates preferentially with GST-Ypt1 loaded with GTP but not with GST-Ypt1-GDP or GST (Fig. 1D). The low level of the coprecipitation from yeast lysates can be attributed to the transient nature of the interaction or to competition with other yeast proteins interacting with Atg11. To determine whether recombinant Ypt1 and Atg11 proteins interact, the CC2-3 domain of Atg11 (amino acids 321–859), which interacts with Ypt1 in the yeast-two hybrid assay, was expressed in bacteria as a His6-tagged protein. Coprecipitation of His-Atg11-CC2-3 with purified Ypt1 showed that this protein interacts preferentially with Ypt1-GTP (Fig. 1E), suggesting that the Ypt1–Atg11 interaction is direct.
Cells carrying the ypt1-1 mutation, T40K, in the effector-binding domain of Ypt1 are defective in autophagy (15, 17). Therefore, we tested whether the Ypt1-1 mutant protein is defective in the interaction with Atg11 using the three interaction assays mentioned above: yeast-two hybrid, coprecipitation with Atg11-HA from yeast lysates and coprecipitation with bacterially expressed His-Atg11-CC2-3 (Fig. 1 C–E). Compared with the wild-type Ypt1 protein, the Ypt1-1 mutant protein is defective in the interaction with Atg11 or with Atg11-CC2-3 in all three assays.
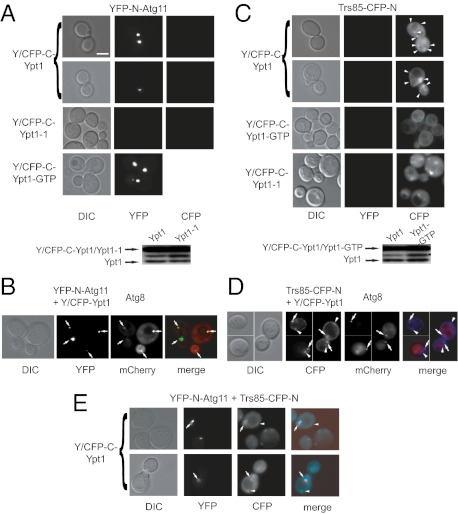
To determine whether Ypt1 and Atg11 interact in vivo, we used the bimolecular fluorescence complementation (BiFC) assay. BiFC is a protein-fragment complementation assay (PCA) in which two fragments of a fluorophore tagged to two different proteins are coexpressed in cells. Fluorescence is observed only if the two proteins interact to bring the two fluorophore fragments in close enough proximity (21). We constructed plasmids in which yeast-optimized YFP or CFP was split into N and C termini: The C termini of YFP and CFP are identical (Y/CFP-C), and the N-terminal domains, YFP-N and CFP-N, determine whether the interacting complex fluoresces in the YFP or CFP channel, respectively (21). Split YFP was used to determine the Ypt1–Atg11 interaction in vivo. Only in cells coexpressing YFP-N-Atg11 and Y/CFP-C-Ypt1 or Ypt1-GTP, but not Y/CFP-C-Ypt1-1, there is one dot per cell in the YFP channel (Fig. 2A), suggesting that Ypt1, but not Ypt1-1, interacts with Atg11 in vivo.
Fig. 2.
PCA for Ypt1, Atg11, and Trs85 using multicolor BiFC. (A) Positive PCA for Atg11 with Ypt1 and Ypt1-GTP (Ypt1-Q67L) but not with Ypt1-1. YFP fluorescence is seen in cells coexpressing YFP-N-Atg11 with C/YFP-C-Ypt1 or Ypt1-GTP but not C/YFP-C-Ypt1-1. No fluorescence is seen in the CFP channel. (Scale bar, 5 μm.) DIC, differential interference contrast. (B) Atg11 and Ypt1 interact in the Atg8-marked PAS. The experiment was done as described in A except that cells also express mCherry-Atg8. Overlap of YFP and mCherry fluorescence (merge) indicates that Atg11 and Ypt1 interact on the PAS (arrows). (C) Positive PCA for Trs85 with Ypt1 and Ypt1-1 but not Ypt1-GTP. CFP fluorescence is seen in cells coexpressing Trs85-CFP-N with C/YFP-C-Ypt1 (arrowheads) or C/YFP-C-Ypt1-1 but not C/YFP-C-Ypt1-GTP. No fluorescence is seen in the YFP channel. (D) Trs85 and Ypt1 interact in the Atg8-marked PAS. The experiment was done as described in C except that cells also express mCherry-Atg8. At least one CFP punctum per cell overlaps with mCherry (merge), indicating Trs85-Ypt1 interaction on the PAS (arrows), but the rest do not (arrowheads). (E) Multicolor BiFC of Trs85, Ypt1, and Atg11. Cells coexpress Trs85-CFP-N, YFP-N-Atg11, and C/YFP-C-Ypt1. Fluorescence in the CFP channel shows the Ypt1-Trs85 interaction, YFP shows the Ypt1-Atg11 interaction, and overlap of the Ypt1-Trs85 and Ypt1-Atg11 interactions is shown in the merged image. Arrows indicate puncta where all three proteins are present; arrowheads indicate dots where Ypt1 interacts with Trs85 but not with Atg11. Immunoblots in A and C show similar Ypt1 protein levels. Results are representative of at least two independent experiments.
Two pieces of evidence support the use of Ypt1-1 as a negative control in the BiFC assay. First, immunofluorescence microscopy shows a similar Ypt1 pattern in wild type and ypt1-1 (Fig. S2A). Second, Y/CFP-C-Ypt1-1 interacts with Trs85-CFP-N (see below, Fig. 2C). The Atg11-interacting PAS protein Atg1 (18) was used to verify the specificity of BiFC. Fluorescence was seen in cells coexpressing Y/CFP-C-Atg1 and YFP-N-Atg11 but not YFP-N-Atg1 and Y/CFP-C-Ypt1 (Fig. S2B). BiFC between Atg1 and Atg11, but not between Atg1 and Ypt1, provides support for the specificity of this assay and its relevance to protein interaction.
The importance of the CC2 and CC3 domains of Atg11 for its interaction with Ypt1 was confirmed using BiFC. Fluorescence was seen only in cells coexpressing Y/CFP-C-Ypt1 and YFP-N-Atg11, but not Atg11ΔCC2 or Atg11ΔCC3. All three YFP-N-Atg11 proteins show a BiFC interaction with Atg19 (Fig. S2C), which interacts with Atg11 through CC4 (19). These results show that the CC2 and CC3 domains of Atg11 are required for its interaction with Ypt1 in vivo.
When combined with markers, BiFC can be used for intracellular localization of protein interactions (21). To determine whether the Ypt1–Atg11 interaction occurs on the PAS, we tested the colocalization of the Ypt1-Atg11 BiFC puncta with the PAS marker Atg8 (22) tagged with mCherry. In all cells that show both the BiFC (YFP) and mCherry puncta, the fluorescence overlaps (40/40 cells; Fig. 2B). This result supports the idea that the Ypt1–Atg11 interaction occurs in the PAS.
In summary, using in vitro and in vivo approaches, Atg11 was identified as a Ypt1 effector candidate. Moreover, the Ypt1-1 mutant protein, in which one residue in the effector domain is changed, is defective in the interaction with Atg11.
Ypt1–Atg11 Interaction Is Required for PAS Assembly.
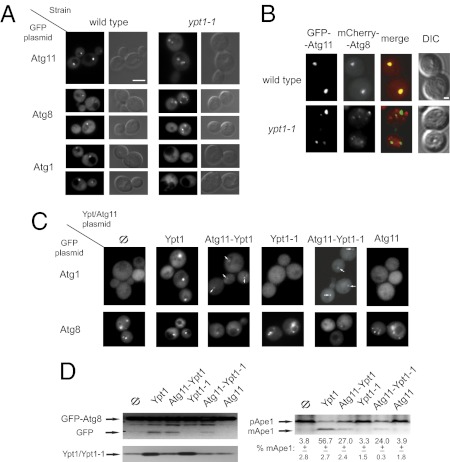
Ypt/Rab GTPases exert their function by recruiting their effectors to the proper location (1). To test whether Ypt1 regulates the localization of Atg11, the effect of the ypt1-1 mutation, which disrupts the Ypt1–Atg11 interaction, on the localization of GFP-Atg11 was determined. As previously shown, in wild-type cells, GFP-Atg11 localizes to a single dot per cell, which represents the PAS (18, 23). In contrast, in ypt1-1 mutant cells, GFP-Atg11 is seen as multiple puncta (Fig. 3A and Fig. S3A). This observation supports the idea that Atg11 is a downstream effector of Ypt1.
Fig. 3.
Ypt1 interaction with Atg11 is required for PAS assembly. (A) The localization of three GFP-tagged PAS components, Atg11, Atg8, and Atg1, is altered in ypt1-1 mutant cells. In wild-type cells each of the three proteins localizes to one dot, whereas in ypt1-1 cells Atg11 and Atg8 localize to multiple puncta, and Atg1 is diffuse. (Scale bar, 5 μm.) (See Fig. S3A for quantification and Fig. S3B for protein levels). (B) GFP-Atg11 and mCherry-Atg8 do not colocalize in ypt1-1 cells. In wild-type cells Atg11 and Atg8 colocalize in one dot, but in ypt1-1 cells the multiple puncta of Atg11 and Atg8 do not colocalize. (Scale bar, 1 μm.) (C) The Atg11-Ypt1-1 fusion protein can partially restore PAS formation in ypt1-1 cells. Like ypt1-1 cells (Φ), cells expressing Ypt1-1 or Atg11 exhibit diffuse Atg1 and multiple puncta of Atg8. The ypt1-1 Atg-localization defects are fully suppressed by Ypt1 and are partially suppressed by one of the fusion proteins, Atg11-Ypt1 or Atg11-Ypt1-1 (See Fig. S3C for quantification). (D) (Upper) The Ypt1-1-Atg11 fusion protein partially suppresses the maturation defects of GFP-Atg8 and Ape1 in ypt1-1 cells. The premature forms of GFP-Atg8 and Ape1 (pApe1) are present in all cells. As in ypt1-1 cells (Φ), in cells expressing Ypt1-1 or Atg11, there is no GFP (processed from GFP-Atg8) or mature Ape1 (mApe1). These processing defects are fully suppressed by Ypt1 and are partially suppressed by Atg11-Ypt1-1 or Atg11-Ypt1. (Lower Left) Ypt1 and Ypt1-1 protein levels. (Lower Right) Quantification of Ape1 maturation. Results are representative of at least two independent experiments.
The effect of the ypt1-1 mutation on two other PAS components, Atg8 and Atg1, was determined. Like GFP-Atg11, GFP-Atg8 localizes to a single dot in wild-type cells and to multiple dots in ypt1-1 mutant cells (Fig. 3A and Fig. S3A). Colocalization of Atg11 with Atg8 in wild-type and ypt1-1 mutant cells was tested using GFP-Atg11 and mCherry-Atg8. Although GFP-Atg11 and mCherry-Atg8 colocalize to one dot per cell in wild-type cells, the multiple dots of the two proteins do not overlap in ypt1-1 mutant cells (colocalization: 95% of 40 red dots in 39 wild-type cells and 2.5% of 81 dots in 25 ypt1-1 cells) (Fig. 3B). The appearance of Atg8 as multiple dots in several atg mutant cells, including atg9Δ (24), and of Atg11 in atg1Δ mutant cells (18) was reported previously. However, the nature of these dots is not clear. Atg1 is required for an early step of PAS assembly (14). In wild-type cells GFP-Atg1 localizes to a single dot, but in ypt1-1 mutant cells it is diffuse, even though its steady-state level is unchanged (Fig. 3A and Fig. S3 A and B). Together these results show that PAS assembly is defective in ypt1-1 mutant cells.
To support the idea that the inability of the Ypt1-1 mutant protein to interact with Atg11 results in a PAS-assembly defect, we tested the ability of an Atg11-Ypt1-1 fusion protein to bypass the mutant defect. In most ypt1-1 mutant cells transformed with an empty plasmid or plasmids expressing Ypt1-1 or Atg11, Atg1 is diffuse, and Atg8 is seen as multiple puncta. In cells expressing the wild-type Ypt1 protein, PAS assembly is restored, and Atg1 and Atg8 localize to a single dot in most cells. Importantly, in cells expressing the Atg11-Ypt1 and Atg11-Ypt1-1 fusion proteins, there is partial suppression of the PAS-assembly defect (Fig. 3C and Fig. S3C). Partial suppression of the ypt1-1 PAS-assembly defect by the Atg11-Ypt1-1 fusion protein also should restore PAS function. We followed two cargo proteins whose processing depends on delivery to the vacuole through the PAS, GFP-Atg8 and Ape1 (25). In ypt1-1 cells transformed with empty plasmid or plasmids expressing Ypt1-1 or Atg11, the processing of both GFP-Atg8 and Ape1 is defective. This defect is fully restored in cells expressing Ypt1 and is partially restored in cells expressing one of the fusion proteins, Atg11-Ypt1 or Atg11-Ypt1-1 (Fig. 3D). These observations support the idea that the Ypt1–Atg11 interaction is required for PAS assembly and function under normal growth conditions.
Interaction of Ypt1, Trs85, and Atg11 in the PAS.
We hypothesized that Trs85-containing TRAPP III functions together with Ypt1 and its effector Atg11 in a GEF-GTPase-effector module that regulates autophagy. Split CFP was used to determine the Trs85–Ypt1 BiFC interaction in vivo. Multiple fluorescent dots per cell are seen in the CFP channel only in cells expressing Trs85-CFP-N and Y/CFP-C-Ypt1 or Y/CFP-C-Ypt1-1 but not in cells expressing Y/CFP-C-Ypt1-GTP (Fig. 2C). GFP-Ypt1 localizes to multiple puncta per cell (26). Previously published studies of intracellular localization of Trs85 tagged with GFP or 3×GFP were inconclusive (17, 27). We tagged endogenous Trs85 with yeast-optimized EGFP and demonstrated that it is functional and localizes to multiple puncta per cell (Fig. S2D). Therefore, both live-cell microscopy and BiFC show that Ypt1 and Trs85 localize to and interact in more than one place in the cell. The BiFC interaction of Ypt1-1 with Trs85 shows that Ypt1-1 is not defective in the interaction with its activator. The fact that Ypt1-GTP does not show interaction with Trs85 in this BiFC assay serves as a negative control.
If the Ypt1–Trs85 interaction occurs in the PAS, we expect that one of the BiFC puncta in each cell would localize to the PAS. Cells expressing Trs85-CFP-N and Y/CFP-C-Ypt1 were transformed with a third plasmid expressing the PAS marker Atg8 tagged with mCherry. In each cell that shows both CFP and red mCherry puncta, at least one blue punctum overlaps with the red punctum (25/25 cells; Fig. 2D). This result indicates that Ypt1 and Trs85 interact in the PAS.
The colocalization of all three proteins, Trs85, Ypt1, and Atg11, was determined using multicolor BiFC. This assay allows simultaneous visualization of multiple protein interactions in the same cell (21). Cells were transformed with three plasmids expressing Y/CFP-C-Ypt1, Trs85-CFP-N, and YFP-N-Atg11. Fluorescence was determined in both the CFP and the YFP channels. As in the single-color BiFC described above, a few fluorescent puncta, showing the Ypt1–Trs85 interaction, are seen in the CFP channel, and only one dot per cell, reflecting the Ypt1–Atg11 interaction, is seen in the YFP channel. The merged images demonstrate that in each cell that shows YFP and CFP fluorescence there is a single dot in which all three proteins colocalize (in 50/50 cells with YFP and CFP fluorescence) (Fig. 2E). Because Atg11 is a component of the PAS, and because we showed that both the Ypt1-Atg11 and the Ypt1-Trs85 BiFC puncta colocalize with the PAS marker Atg8, these results indicates that all three proteins, Ypt1, Trs85, and Atg11, colocalize and interact in the PAS.
Role for the Trs85-Ypt1-Atg11 Module in Autophagy.
To support the idea that Trs85, Ypt1, and Atg11 function as a GEF-GTPase-effector module in autophagy, we used overexpression and double-mutant analyses.
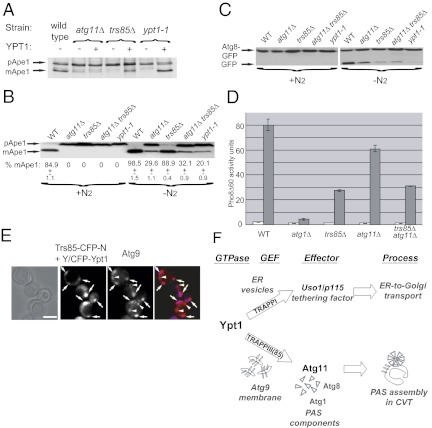
Usually, overexpression of a protein can suppress defects caused by mutant proteins that act upstream, but not downstream, in the same pathway (28). Therefore, overexpression of Ypt1 is expected to suppress the cargo-processing defect in cells in which its upstream regulator, Trs85, is deleted but not in cells in which its downstream effector, Atg11, is deleted. Suppression of the Ape1-processing phenotype of trs85Δ was shown previously when the GTP-restricted form, but not wild-type Ypt1, was expressed from the GAL1 promoter (17). We observed that overexpression of Ypt1 from its own promoter suppresses the Ape1-processing defect of ypt1-1 and trs85Δ but not that of atg11Δ (Fig. 4A). This suppression is specific to Ypt1, because overexpression of Ypt31 does not suppress this defect (Fig. S4A). Thus, overexpression analysis supports the idea that PAS function is regulated by a module in which Ypt1 functions downstream of Trs85 and upstream of Atg11.
Fig. 4.
A role for the Trs85-Ypt1-Atg11 module in autophagy. (A) Overexpression of Ypt1 suppresses the Ape1-processing defects of ypt1-1 and trs85Δ but not of atg11Δ. All three mutants exhibit an Ape1-processing defect (accumulated pApe1). mApe1 is seen in ypt1-1 and trs85Δ but not in atg11Δ overexpressing Ypt1. (B) The Ape1-processing defect of the atg11Δ trs85Δ double mutant under nitrogen starvation is not more severe than that of the single-deletion mutants. All mutants exhibit a complete Ape-processing defect during normal growth (+N2). Under nitrogen starvation (−N2, 4 h), Ape1 is processed to the mature form in wild-type cells, but it remains unprocessed in ypt1-1 mutant cells. Atg11Δ and atg11Δ trs85Δ exhibit a partial processing defect; trs85Δ exhibits a less severe defect. The percent of mApe1 is shown below the lanes. (C) The Atg8-processing defect of the atg11Δ trs85Δ double mutant under nitrogen starvation is not more severe than that of the single deletions. No strains process Atg8 during normal growth. Under nitrogen starvation (4 h), most of the Atg8-GFP protein in wild-type cells is processed to GFP, but in ypt1-1 mutant cells it remains unprocessed as Atg8-GFP. Trs85Δ and atg11Δtrs85Δ mutants exhibit a processing defect similar to but less severe than that of ypt1-1; atg11Δ exhibits a less severe defect. (D) The Pho8Δ60 alkaline phosphatase (ALP)-activity defect of atg11Δ trs85Δ is not more severe than in single-deletion mutants. ALP activity was determined in lysates prepared from cells grown in yeast extract-peptone-glucose medium (white bars) and after 6 h of nitrogen starvation (gray bars); atg1Δ mutant cells serve as a negative control. Trs85Δ and atg11Δtrs85Δ mutant cells exhibit a similar partial defect; atg11Δ mutant cells exhibit a less severe defect. Activity units represent nM nitrophenol/mg protein. Error bars represent SD. (E) Trs85 and Ypt1 BiFC puncta overlap with Atg9. The experiment was done as in Fig. 2D, except that Atg9 was tagged on the chromosome with mCherry. All CFP puncta overlap with mCherry (merge), indicating that Trs85 and Ypt1 interact on Atg9-marked compartments (arrows). There are more Atg9 than Ypt1-Trs85 puncta (arrowheads). (Scale bar, 5 μm.) Results shown in A–E are representative of at least two independent experiments. (F) A model showing how one Ypt/Rab GTPase, Ypt1, regulates two processes, ER-to-Golgi transport (Upper) and PAS assembly (Lower), by recruiting process-specific effectors. In ER-to-Golgi transport, Ypt1 activated by TRAPP I (6, 7) recruits the conserved tethering factor Uso1/p115 (10, 11) to stimulate ER vesicle tethering to the Golgi. In selective autophagy, a GEF-GTPase-effector module composed of Trs85-containing TRAPP III-Ypt1-Atg11 regulates the first step of both selective and nonselective autophagy, PAS assembly. We propose that Trs85, in the context of TRAPP III, and Ypt1 are localized to Atg9-containing membranes. Subsequently, activated Ypt1-GTP interacts with Atg11 to mediate PAS assembly on these membranes.
If two proteins function in the same pathway, a double-deletion mutant should confer a phenotype that is not more severe than the phenotypes of the single deletions. When grown in rich medium, the trs85Δ, ypt1-1, and atg11Δ mutations confer a complete block in Ape1 processing, and Atg8-GFP is not processed even in wild-type cells (Fig. 4 B and C). Therefore, we tested these processing phenotypes plus the growth and Pho8Δ60 activity phenotypes of the mutants under nitrogen starvation. Cells carrying single deletions of trs85Δ and atg11Δ exhibit intermediate growth and Pho8Δ60 defects compared with ypt1-1 and atg1Δ cells, respectively (Fig. S4B and Fig. 4D, respectively). In both assays, under nitrogen starvation trs85Δ confers a more severe phenotype than atg11Δ. These results are in agreement with the idea that Ypt1 and Trs85 play a role in both selective and nonselective autophagy (15, 17). The observation that atg11Δ mutant cells also exhibit mild growth and Pho860 defects under nitrogen starvation is in agreement with the idea that the PAS assembled during normal growth can persist and help cells survive under starvation conditions (29).
Because the single-deletion mutants trs85Δ and atg11Δ exhibit partial autophagy defects, it was possible to determine whether the double-deletion phenotypes are more severe than those of the single deletions. The double mutant trs85Δ atg11Δ exhibits starvation-induced growth and Pho8Δ60 defects similar to, but not more severe than, those of the single deletions (Fig. 4D and Fig. S4B). The cargo-processing phenotypes of the single- and double-mutant cells also were compared under nitrogen starvation. All mutant strains exhibit varying degrees of Ape1- and Atg8-processing defects, with ypt1-1 exhibiting the most severe defects. Importantly, the processing defects of the atg11Δ trs85Δ double deletion are not more severe than those of the single deletions (Fig. 4 B and C). Together, these results support the idea that the two Ypt1 interactors, Trs85 and Atg11, function in one module that regulates autophagy.
Localization of Trs85–Ypt1 Interaction to Atg9-Containing Membranes.
As shown above, one punctum of the Trs85–Ypt1 BiFC interaction in each cell localizes to the PAS (Fig. 2D). To determine the localization of the rest of the Ypt1–Trs85 interaction puncta, cells expressing RFP-tagged compartmental markers (27) were transformed with plasmids coexpressing Trs85-CFP-N and Y/CFP-C-Ypt1. We did not observe obvious colocalization of the CFP and RFP fluorescence for ER exit sites, cis Golgi, Golgi, trans Golgi, or endosomes (Fig. S5). Thus, the bulk of the Ypt1-Trs85 interaction does not occur on exocytic or endocytic compartments. However, whether some interaction occurs on those compartments is still an open question.
Atg9 is an integral membrane protein, and Atg9-containing membranes were proposed as a source for the autophagosome biogenesis. Like Ypt1 and Trs85, Atg9 localizes to multiple puncta per cell (30). Partial colocalization of Ypt1 and Atg9 was reported recently (17). To determine whether the Ypt1–Trs85 interaction occurs on Atg9-marked membranes, the Trs85-Ypt1 BiFC puncta were colocalized with Atg9-mCherry. Although there are more Atg9 mCherry puncta in each cell, all the CFP puncta representing the Ypt1–Trs85 interaction colocalize with Atg9 (multiple puncta in 25/25 cells) (Fig. 4E). If the BiFC puncta representing the Trs85–Ypt1 interaction sites colocalize with Atg9, we expected that Trs85 itself also colocalizes with Atg9. Like the BiFC puncta, all the Trs85 puncta overlap with Atg9 puncta (75/75 puncta in 30 cells), but there are additional Atg9 puncta in each cell (Fig. S4C). This result provides a BiFC-independent confirmation for the colocalization of Trs85 with Atg9. Because Atg11 interacts with Atg9 and affects its cellular localization (23), we wished to determine whether Ypt1 or Trs85 affects this localization as well. The number of Atg9-mCherry puncta is reduced in both trs85Δ and ypt1-1 mutant cells compared with wild-type cells (Fig. S4D). Thus, Ypt1 and Trs85 affect the localization of Atg9, an Atg11 interactor. Together, these results suggest that Trs85 and Ypt1 interact on Atg9-containing membranes, which serve as source for the membrane on which the PAS assembles. In addition, proper function of Trs85 and Ypt1 is important for Atg9 localization.
Discussion
PAS assembly is the first step of the selective and nonselective autophagy pathways, and Atg11 is a PAS organizer in selective autophagy (14). Here we show that Atg11 is a downstream effector of Ypt1 based on the following evidence: In vitro and in vivo analyses show that Atg11 interacts specifically with the GTP-bound form of Ypt1, and the localization of Atg11 to the PAS is regulated by Ypt1. Using ypt1-1, a mutant defective in the interaction with Atg11, we show that the Ypt1–Atg11 interaction is important for PAS assembly and function. Moreover, multicolor BiFC analysis shows that Trs85, an autophagy-specific subunit of the Ypt1 activator complex, interacts with Ypt1 on Atg9-containing membranes and with Ypt1 and Atg11 in the PAS. Finally, genetic analyses support the idea that the three proteins function as a GEF-GTPase-effector module to regulate PAS assembly (Fig. 4F). Because PAS assembly is the first known step of autophagy, this Ypt/Rab GTPase module regulates the onset of autophagy.
Our observation that under nitrogen starvation the ypt1-1 mutation confers more severe autophagy defects than those exhibited by trs85Δ and atg11Δ suggests that alternative Ypt1 activators and effectors function in nonselective autophagy. Atg11 and Atg17 seem to play similar roles in specific and nonspecific autophagy, respectively (29), including the recruitment of Atg9 to the PAS (23, 31). Because Ypt1 is involved in both nonselective and selective autophagy, and Atg11 is involved mainly in the former, Atg17 is a potential candidate for an alternative Ypt1 effector in nonselective autophagy.
Is the role of the Ypt1 module in PAS assembly conserved from yeast to human cells? Rab1 is a functional homolog of Ypt1 (8), and a role for Rab1 in autophagy has been shown in mammalian cells (16). A recent proteomic study suggests that KIAA1012, a human Trs85 homolog, plays a role in autophagy (32). Atg11 is conserved among yeast and fungi, but there is no clear human homolog for Atg11 (33). However, RB1CC1/FIP200 (KIAA0203) has been suggested as a candidate for a human homolog of the yeast Atg11 or Atg17 (34, 35). In addition, like the effect of the ypt1-1 mutation on Atg9 localization in yeast, inhibition of Rab1a function has been shown to alter the localization of Atg9 in human cells (36). Therefore, it is tempting to propose that the role of the Trs85-Ypt1-Atg11 module in autophagy is conserved.
An open question in the autophagy field is the identity of the membrane that serves as a source for autophagosomes. Using BiFC combined with colocalization analysis, we show that Ypt1 and Trs85 interact on Atg9-marked membranes. Because Atg9-containing membranes are considered a source for autophagosomal membrane (14), we propose that Ypt1 and Trs85 are recruited to these membranes to initiate PAS assembly (Fig. 4F). Ypt1 shows two different BiFC patterns: multiple puncta for the Trs85–Ypt1 interaction on the Atg9-containing membrane and a single punctum of the Trs85-Ypt1-Atg11 on the Atg8/Atg9-marked PAS. Because Atg9-containing reservoirs were shown to generate the PAS (30), we propose that Ypt1 interacts first with Trs85 on Atg9-containing membranes and later with Atg11 to facilitate PAS assembly (Fig. 4F). Interestingly, interaction with Atg11 is required for targeting Atg9 to the PAS (23), and here we show that Ypt1 also plays a role in Atg9 localization. Because one mechanism suggested for the Ypt/Rab mechanism of action is enhancement of interactions between effectors and effector-binding proteins (37), it is possible that Ypt1 enhances the Atg11–Atg9 interaction.
GTPases have been implicated in the coordination of vesicular transport substeps and in the integration of transport steps into whole pathways (38). Here, we propose that GTPases also can coordinate two different processes. How can one GTPase, Ypt1, function in two different processes, ER-to-Golgi transport and autophagy? Each Ypt/Rab GTPase can recruit multiple effectors in a timely and spatially regulated manner. We propose that two Ypt1 effectors exhibit process specificity: The conserved tethering factor Uso1/p115 acts as an effector of Ypt1/Rab1 in the ER-to-Golgi transport step (10, 11), whereas Atg11 is an autophagy-specific Ypt1 effector. Therefore, our results imply that Ypt/Rab GTPases can regulate two different processes by recruiting process-specific effectors (Fig. 4F). A future challenge is to determine the cues that allow Ypt1 to recruit a specific set of effectors to a specific membrane. One example of such discrimination is a Rab5 effector that can be recruited specifically to phosphatidylinositol 3-phosphate membranes (39).
Materials and Methods
All strains, plasmids, and reagents, yeast culture conditions and viability, protein level, coprecipitation and alkaline phosphatase activity analyses, and immunofluorescence and live-cell microscopy are detailed in SI Materials and Methods. Strains used in this paper are summarized in Table S1. Plasmids used in this study are summarized in Table S2.
Supplementary Material
Acknowledgments
We thank C. Richardson and F. Liu for performing early experiments, A. Shah for critical reading, and Y. Liang and D. Klionsky for providing strains, plasmids, and antibodies. This research was supported by National Institutes of Health Grant GM-45444 (to N.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121299109/-/DCSupplemental.
References
- 1.Segev N. Ypt and Rab GTPases: Insight into functions through novel interactions. Curr Opin Cell Biol. 2001;13:500–511. doi: 10.1016/s0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- 2.Segev N. Ypt/rab GTPases: Regulators of protein trafficking. Sci STKE. 2001;2001:re11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- 3.Jedd G, Richardson C, Litt R, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J Cell Biol. 1995;131:583–590. doi: 10.1083/jcb.131.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segev N. Mediation of the attachment or fusion step in vesicular transport by the GTP-binding Ypt1 protein. Science. 1991;252:1553–1556. doi: 10.1126/science.1904626. [DOI] [PubMed] [Google Scholar]
- 5.Segev N, Mulholland J, Botstein D. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell. 1988;52:915–924. doi: 10.1016/0092-8674(88)90433-3. [DOI] [PubMed] [Google Scholar]
- 6.Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell. 2000;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morozova N, et al. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat Cell Biol. 2006;8:1263–1269. doi: 10.1038/ncb1489. [DOI] [PubMed] [Google Scholar]
- 8.Haubruck H, Prange R, Vorgias C, Gallwitz D. The ras-related mouse ypt1 protein can functionally replace the YPT1 gene product in yeast. EMBO J. 1989;8:1427–1432. doi: 10.1002/j.1460-2075.1989.tb03524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pind SN, et al. Rab1 and Ca2+ are required for the fusion of carrier vesicles mediating endoplasmic reticulum to Golgi transport. J Cell Biol. 1994;125:239–252. doi: 10.1083/jcb.125.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: Programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- 11.Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraft C, Reggiori F, Peter M. Selective types of autophagy in yeast. Biochim Biophys Acta. 2009;1793:1404–1412. doi: 10.1016/j.bbamcr.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 15.Segev N, Botstein D. The ras-like yeast YPT1 gene is itself essential for growth, sporulation, and starvation response. Mol Cell Biol. 1987;7:2367–2377. doi: 10.1128/mcb.7.7.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 17.Lynch-Day MA, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–2269. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Huang WP, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He C, et al. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki K, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–26. doi: 10.1016/S0076-6879(08)03201-1. [DOI] [PubMed] [Google Scholar]
- 26.Calero M, et al. Dual prenylation is required for Rab protein localization and function. Mol Biol Cell. 2003;14:1852–1867. doi: 10.1091/mbc.E02-11-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 28.Jones S, et al. Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics. 1999;152:1543–1556. doi: 10.1093/genetics/152.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mari M, et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 2009;14:525–538. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- 32.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–116. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- 34.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21:3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winslow AR, et al. α-Synuclein impairs macroautophagy: Implications for Parkinson’s disease. J Cell Biol. 2010;190:1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segev N. Cell biology. A TIP about Rabs. Science. 2001;292:1313–1314. doi: 10.1126/science.1061114. [DOI] [PubMed] [Google Scholar]
- 38.Segev N. Coordination of intracellular transport steps by GTPases. Semin Cell Dev Biol. 2011;22:33–38. doi: 10.1016/j.semcdb.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simonsen A, et al. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.