Abstract
Polycomb Group (PcG) proteins mediate heritable gene silencing by modifying chromatin structure. An essential PcG complex, PRC1, compacts chromatin and inhibits chromatin remodeling. In Drosophila melanogaster, the intrinsically disordered C-terminal region of PSC (PSC-CTR) mediates these noncovalent effects on chromatin, and is essential for viability. Because the PSC-CTR sequence is poorly conserved, the significance of its effects on chromatin outside of Drosophila was unclear. The absence of folded domains also made it difficult to understand how the sequence of PSC-CTR encodes its function. To determine the mechanistic basis and extent of conservation of PSC-CTR activity, we identified 17 metazoan PSC-CTRs spanning chordates to arthropods, and examined their sequence features and biochemical properties. PSC-CTR sequences are poorly conserved, but are all highly charged and structurally disordered. We show that active PSC-CTRs—which bind DNA tightly and inhibit chromatin remodeling efficiently—are distinguished from less active ones by the absence of extended negatively charged stretches. PSC-CTR activity can be increased by dispersing its contiguous negative charge, confirming the importance of this property. Using the sequence properties defined as important for PSC-CTR activity, we predicted the presence of active PSC-CTRs in additional diverse genomes. Our analysis reveals broad conservation of PSC-CTR activity across metazoans. This conclusion could not have been determined from sequence alignments. We further find that plants that lack active PSC-CTRs instead possess a functionally analogous PcG protein, EMF1. Thus, our study suggests that a disordered domain with dispersed negative charges underlies PRC1 activity, and is conserved across metazoans and plants.
Keywords: intrinsically disordered protein, protein evolution
Regulation of chromatin structure occurs through covalent modification of histone proteins, and noncovalent effects on chromatin structure and folding (1, 2). Chromatin modifying enzymes add or remove posttranslational modifications of histone proteins through structured domains, which are readily identified through sequence alignments and predicted structure comparisons. On the other hand, proteins that alter chromatin folding and compaction frequently use intrinsically disordered regions. These domains tend to evolve more rapidly, making it difficult to track their activity across evolution using sequence alignments (1).
An essential set of chromatin-based regulators are the Polycomb Group (PcG) proteins, which mediate heritable gene silencing through the modification of chromatin structure (2). They have been implicated in a wide variety of biological processes, ranging from genome imprinting (3), cell cycle regulation (4), mammalian X-chromosome inactivation (5), and the maintenance of stem cell identity (6). PcG proteins are also misregulated in cancer and contribute to cancer progression (7). PcG proteins assemble into multiprotein complexes, with different biochemical activities toward chromatin (2). One PcG complex, PRC2, methylates histone H3 on lysine 27 through the SET domain of one of its subunits, Enhancer of Zeste [E(Z)] (8). PRC2 and its enzymatic activity are clearly conserved across metazoans and plants (9), as reflected by sequence alignments of the SET domain in diverse genomes (10). Another PcG complex, Polycomb Repressive Complex 1 (PRC1), consists of four “core” subunits: RING, Posterior sex combs (PSC), Polycomb (PC), and Polyhomeotic (PH), all of which are essential in Drosophila (11, 12). Several in vitro activities have been described for PRC1, all of which entail the noncovalent modification of chromatin structure. These include chromatin compaction (13), inhibition of chromatin remodeling (14), and repression of transcription from DNA and chromatin templates in vitro (14). Two PRC1 subunits—RING and PSC—are also present in a distinct complex, dRAF, which functions as an E3 ligase to stimulate the covalent modification of chromatin through H2A ubiquitination (15).
The PRC1 subunit PSC is sufficient for its noncovalent effects on chromatin structure (11, 13, 16). PSC is a large protein with a conserved motif near its N terminus containing a RING and RAWUL domain (17–19). This region is important for assembly of PSC into PRC1 and likely its activity in Polycomb complexes (17, 20). The C-terminal region (CTR) of the protein is necessary and sufficient for PSC effects on chromatin structure. Nonsense mutations that encode truncations of most of the CTR are severe hypomorphs, and disrupt PcG-dependent gene silencing in vivo (16, 21). The truncated proteins also lack PSC’s effects on chromatin in vitro, indicating that these biochemical activities are central to its biological function.
The primary sequence of PSC-CTR is poorly conserved even within the dipterans (20), rendering it difficult to identify conserved sequence features that encode its biochemical activities. In addition, a paralogue of PSC in Drosophila melanogaster—Suppressor 2 of zeste [Su(z)2]—has a CTR of similar biochemical function to PSC-CTR, even though their primary sequences are poorly conserved, according to Lo et al. (20). Su(z)2 shares high sequence similarity with PSC only in the N-terminal domain (20, 22, 23). The CTRs of PSC and Su(z)2 share unusual amino acid compositions, and are both predicted to be disordered, leading to the hypothesis that these properties underlie their conserved activity. The poor sequence conservation of the PSC-CTR has also made it difficult to determine the extent of its evolutionary conservation beyond Drosophila (20).
To determine essential sequence properties encoding PSC-CTR activity, and to assess the extent of its evolutionary conservation, we identified 17 metazoan PSC-CTRs and compared their sequences and biochemical activities. We find that biochemically active PSC-CTRs are present in widely diverged species despite the lack of sequence similarity, indicating they are broadly conserved in function but not primary sequence. We identified sequence properties shared by active PSC-CTRs, which bind DNA tightly and inhibit chromatin remodeling efficiently. Importantly, we determined that the presence of extended contiguous negative charge impairs PSC-CTR activity. Our work illustrates the importance of using empirical measurements, rather than sequence alignments, to assess the functional properties of PSC-like proteins. The mechanistic and evolutionary features of PSC-CTR uncovered in this study may be generally relevant to chromatin-binding proteins, which commonly possess similar intrinsically disordered regions (24).
Results
Large, Intrinsically Disordered PSC-CTRs Are Present in a Wide Sampling of Invertebrate Taxa.
To assess the conservation of PSC-CTR sequence and function, we assembled a comprehensive set of PSC-CTRs by querying 30 diverse metazoan and plant genomes for PSC homologues using the conserved N terminus of D. melanogaster PSC. This region contains a RING-finger domain (InterPro domain IPR001841). Our search encompassed both deuterostome and protostome lineages within the bilaterian clade, yielding 154 PSC-like genes (Table S1). The Su(z)2 gene was also classified as a PSC-like gene, as it is a paralogue of PSC. The amino acid sequence downstream of the RING-finger domain was designated as the CTR for each PSC-like gene (see Materials and Methods for annotation procedure).
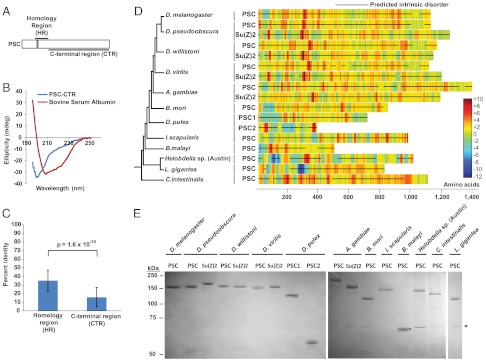
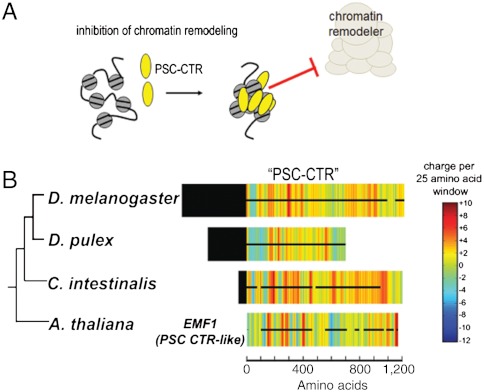
We selected 17 PSC-CTRs for biochemical analysis. These proteins exhibit gross features similar to the CTRs of D. melanogaster PSC and Su(z)2. In particular, they are large (> 350 amino acids) and the majority of each sequence is predicted to be structurally disordered (Fig. 1D). The species from which we selected PSC-CTRs range from closely related homologues within the Drosophilids, to distant homologues in annelids, mollusks, and chordates (Fig. 1D and Table S1). ClustalW alignments of PSC homologues yielded significantly higher alignment scores for the N-terminal “homology region” than the CTR (P < 10-29, two-sample t-test with unequal variance; Fig. 1C), indicating that CTR sequences are poorly conserved. Epitope-tagged PSC-CTRs were expressed in Sf9 insect cells, and purified by immunoaffinity chromatography (Fig. 1E). We measured two properties of PSC-CTRs, which are well characterized for D. melanogaster PSC-CTR: DNA binding, and inhibition of chromatin remodeling.
Fig. 1.
Sequences of diverse metazoan PSC-CTRs are highly charged, intrinsically disordered and poorly conserved. (A) Schematic representation of the homology region (HR) (amino acids 264–463) and the large, disordered C-terminal region (CTR) (amino acids 456–1603) of PSC. Gray box (amino acids 263–302) represents the conserved RING-finger domain. (B) Far-UV circular dichroism spectrum of the intrinsically disordered D. melanogaster PSC-CTR. A minimum of ellipticity at 200 nm is observed, consistent with a disordered structure. The CD spectrum of the well-folded globular protein bovine serum albumin is shown for comparison. (C) PSC-CTR sequences are significantly less conserved than PSC-HR sequences (P = 1.6 × 10-35, two sample t-test with unequal variance). (D) Charge density plots of experimentally tested PSC-CTRs (named PSC, Su(z)2, PSC1, and PSC2). The accompanying cladogram is based on published metazoan phylogenies (45–47). (E) Coomassie-strained gel of PSC-CTRs purified from Baculovirus-infected Sf9 cells. Black circle denotes the band corresponding to B. malayi PSC-CTR. Asterisk on this and all gels indicates HSC70 contaminant. Errors in all figures and tables represent standard deviation. A similar contaminant was purified away from D. melanogaster PSC through gel filtration chromatography and shown not to be active (15). The region in each “parent” PSC protein corresponding to PSC-CTR is listed in Table S2B.
Diverse Metazoan PSC-CTRs Bind DNA Tightly and Inhibit Chromatin Remodeling.
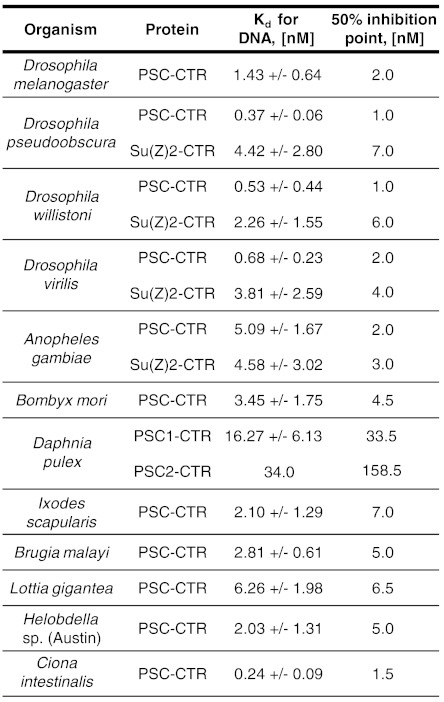
We measured the apparent Kd of each PSC-CTR for dsDNA by double filter binding (25). All of the tested PSC-CTRs bind DNA. Most bind with high affinity (Kd less than 8 nM); two PSC-CTRs, Daphnia pulex PSC1-CTR and PSC2-CTR bind more weakly (Kd of 16.27 ± 6.13 nM and 34 nM, respectively) (Fig. 2 and Fig. S1A). As a control, we measured the apparent Kd of D. melanogaster PSC-CTR as 1.43 ± 0.64 nM (Fig. 2 and Fig. S1A), similar to that of full-length PSC (11).
Fig. 2.
Most PSC-CTRs bind DNA tightly and inhibit chromatin remodeling. Summary of measured Kd from double-filter binding assays and 50% inhibition points from Restriction Enzyme Accessibility (REA) assays for all experimentally tested PSC-CTRs. Kd values represent the affinity of PSC-CTRs for free DNA, while 50% inhibition points represent the ability of PSC-CTRs to inhibit chromatin remodeling.
We then measured the ability of each PSC-CTR to inhibit chromatin remodeling using a restriction enzyme accessibility (REA) assay. In this assay, the chromatin remodeling factor human SWI/SNF uses the energy of ATP hydrolysis to expose a nucleosome-occluded restriction enzyme site in an array of nucleosomes, increasing its digestion (26, 27). Preincubation of the nucleosomal template with D. melanogaster PSC-CTR blocks the activity of SWI/SNF, resulting in decreased digestion of nucleosomal restriction sites (16). The inhibition of chromatin remodeling can thus be measured by the extent of digestion at a nucleosome-occluded restriction site. D. melanogaster PSC-CTR inhibited remodeling by 50% at a concentration of 2 nM (Fig. 2 and Fig. S1B), similar to the previously reported value (16).
PSC-CTRs inhibited chromatin remodeling in a concentration-dependent manner, but with varying efficiencies (Fig. 2 and Fig. S1B). Importantly, they did not alter the level of background restriction enzyme digestion by HhaI when incubated with chromatin template in the absence of chromatin remodeler (compare first and second lanes in Fig. S1B). This indicates that chromatin remodeling, and not restriction enzyme digestion, is specifically inhibited. The 15 PSC-CTRs that bound DNA most tightly also inhibited chromatin remodeling with high efficiency, with 50% inhibition points from 1 to 7 nM. In contrast, D. pulex PSC1-CTR and PSC2-CTR required concentrations of 34 nM and 159 nM, respectively, to achieve 50% inhibition of remodeling, corresponding to an approximately 17 and approximately 80-fold lower efficiency than D. melanogaster PSC-CTR. A modest correlation was observed between the affinity for DNA and the efficiency of inhibition of chromatin remodeling among repressive PSC-CTRs (r = 0.532, P = 0.041). Intuitively, this correlation is expected since chromatin binding is likely mediated at least in part by DNA binding, and must be important for inhibition of chromatin remodeling. However, the weak correlation suggests that the two assays reflect at least partially distinct properties of the PSC-CTR, and that DNA binding alone is likely not sufficient for inhibition of chromatin remodeling, which is supported by previous analysis of D. melanogaster PSC (28). We conclude that most of the large and disordered PSC-CTRs that we tested experimentally indeed share similar biochemical activities with D. melanogaster PSC-CTR.
Repressive PSC-CTRs Do Not Share Conserved Sequence Motifs.
Having identified and experimentally characterized 17 PSC-CTRs, we performed a comparative sequence analysis to identify shared properties that might explain their activity. Because D. pulex PSC1-CTR and PSC2-CTR both bind DNA and inhibit chromatin remodeling less well than any of the others, we designated these two as “nonrepressive” and the remaining 15 as “repressive.” This allowed us to look for distinguishing sequence features that are specific to repressive CTRs, as well as common properties that are shared between both repressive and nonrepressive PSC-CTRs. Using the MEME suite, we queried all repressive PSC-CTRs for conserved sequence motifs, but were unable to find any. These data are consistent with the poor sequence conservation of the CTR (Fig. 1C), and indicate that repressive PSC-CTR sequences are highly divergent in sequence despite their functional similarity in vitro.
Analysis of Sequence Properties in PSC-CTRs.
Because repressive PSC-CTRs are poorly conserved in primary sequence, we hypothesized that “architectural” sequence features, such as the distribution of charged amino acids, may give rise to their similar biochemical properties. We further predicted that at least one such architectural property should distinguish repressive and nonrepressive PSC-CTRs.
Because it is likely that electrostatic forces contribute to the interaction between PSC-CTR and negatively charged chromatin, we examined the charge features of repressive and nonrepressive PSC-CTRs to identify shared and distinguishing properties of PSC-CTRs. Indeed, almost all PSC-CTRs exhibit higher overall charge than the average protein in the UniProt Knowledgebase (Fig. 3A). However, net charge did not distinguish repressive from nonrepressive PSC-CTRs (0 of 4 nonrepressive PSC-CTRs separated) (Fig. 3A). We were also unable to fully distinguish repressive and nonrepressive PSC-CTRs by percent positively charged residues, percent negatively charged, percent polar amino acids, or percent nonpolar amino acids (Fig. S2). Both repressive and nonrepressive PSC-CTRs also have similar overall compositions of polar, nonpolar, and negatively charged amino acids and similar levels of structural disorder that deviate from the UniProt average, regardless of their repressive activity (Fig. S2 and Table S1). We conclude that all experimentally tested PSC-CTRs share overall charge, amino acid composition, and predicted intrinsic disorder.
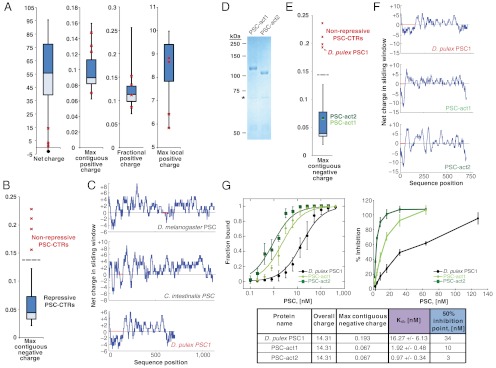
Fig. 3.
The extent of contiguous negative charge in PSC-CTR determines repressive activity. Repressive PSC-CTRs are depicted with box and whisker plots, while nonrepressive PSC-CTRs are represented by red crosses. The ends of the whiskers respectively represent the maximum and minimum data points; upper and lower bounds of the box represent the upper and lower quartiles; horizontal line through the box is the median. Nonrepressive PSC-CTRs are D. pulex PSC1 and PSC2, which were experimentally tested in this study, and M. musculus BMI1 and X. laevis PCGF2, which were previously tested (43). (A) Overall charge and local positive charge properties cannot distinguish the repressive activities of PSC-CTR. “Net charge” is calculated from each full-length PSC-CTR sequence. The average of the UniProt Knowledgebase is represented as a filled circle. “Max contiguous positive charge” represents the length of the longest positively charged stretch in the protein, normalized by total protein length. “Fractional positive charge” represents the fraction of 25-amino acid windows with charge greater than +3.5 in each PSC-CTR. “Max local positive charge” represents the maximum charge attained amongst all 25-amino acid windows in each PSC-CTR. (B) Maximum contiguous negative charge distinguishes repressive from nonrepressive PSC-CTRs. This metric represents the length of the longest negatively charged domain within PSC-CTR, normalized by the length of the protein. (C) Representative charge plots of repressive PSC-CTRs (D. melanogaster and C. intestinalis) and nonrepressive PSC-CTRs (D. pulex), with red horizontal lines denoting the longest stretch of contiguous negative charge. Charge plots were generated from consecutive 25-amino acid sliding windows. (D) Coomassie-strained gel of PSC-act1 and PSC-act2 purified from Baculovirus-infected Sf9 cells. Although PSC1 and PSC2 have identical molecular weights, they migrate slightly differently. (E) Box and whisker plot depicts maximum contiguous negative charge of PSC-CTRs, PSC-act1 and PSC-act2 are indicated by green circles and the nonrepressive ‘parent’ D. pulex PSC1-CTR by a red cross. (F) Charge plots of the ‘parent’ D. pulex PSC1-CTR and the ‘activated’ PSC-act1 and PSC-act2. (G) Measurements of Kd and inhibition of chromatin remodeling for PSC-act1, PSC-act2, and the nonrepressive ‘parent’ D. pulex PSC1-CTR. All error bars denote standard deviation.
Local Charge Properties Distinguish Repressive and Nonrepressive PSC-CTRs.
The local distribution of charged residues—rather than overall charge—could define PSC-CTR activity. We measured the local charge distribution of all PSC-CTR sequences by calculating the net charge within each window of size 25 amino acids in each protein sequence. We then devised a number of quantitative metrics to characterize this local charge distribution. We quantified the fraction of 25 amino acid windows in each PSC-CTR with charge greater than +3.5, but this metric could not distinguish active from inactive PSC-CTRs (Fig. 3A). The fraction of windows in each PSC-CTR with charge less than -3.5 also failed to distinguish activity (Fig. S2). We then tested whether the maximum positive or negative charge attained in a window for each protein was able to distinguish activity (Fig. 3A and Fig. S2), and found that they were unable to do so. However, the maximum number of contiguous negatively charged windows that occur within PSC-CTR, normalized by the sequence length (henceforth abbreviated as “maximum contiguous negative charge”) distinguishes repressive PSC-CTRs (Fig. 3 B and C). This result is robust for window sizes ranging from 15 to 40 sequence residues (Fig. S3). In contrast, the maximum number of contiguous positively charged windows, normalized by sequence length, does not differentiate the two groups (Fig. 3A).
To directly test the functional importance of contiguous negative charge, we sought to “activate” a nonrepressive PSC-CTR by lowering its maximum contiguous negative charge without changing its amino acid composition or net charge. The primary sequence of D. pulex PSC1-CTR was rearranged (as described in Materials and Methods) to decrease the maximum contiguous negative charge without creating domains of high positive charge or altering overall charge. The resulting protein was named PSC-act1 (Fig. 3 D–F; and protein sequence in Table S2B) and tested for biochemical activity. Strikingly, PSC-act1 exhibited an approximately 8.5-fold increase in affinity for DNA, with a measured Kd of 1.92 ± 0.48 nM (Fig. 3G). PSC-act1 also inhibited chromatin remodeling approximately 3.5-fold more efficiently, achieving 50% inhibition of remodeling at 10 nM (Fig. 3G). Thus, PSC-act1 is clearly more active than the nonrepressive D. pulex PSC1-CTR, which exhibited an apparent Kd of 16.27 ± 6.13 nM, and a 50% remodeling inhibition point at 34 nM. We conclude that an extended stretch of contiguous negative charge can disrupt PSC-CTR function, since the redistribution of negative charge away from this region clearly increases activity.
Intuitively, dense positive charge within PSC-CTR could facilitate its interaction with negatively charged chromatin (29). To test this directly, we rearranged charged residues within PSC-act1 (as described in Materials and Methods) to create dense positively charged patches without altering overall charge or maximum contiguous negative charge. The biochemical properties of the resulting protein—named PSC-act2 (Fig. 3 D–F; and protein sequence in Table S2B)—were assessed through filter binding and REA assays. Indeed, PSC-act2 was more active than PSC-act1, exhibiting an approximately twofold increase in affinity for DNA (Kd = 0.97 ± 0.34 nM) and an approximately 3.5-fold increase in efficiency of inhibition of chromatin remodeling (50% inhibition point = 3 nM) (Fig. 3G). We note that repressive and nonrepressive PSC-CTRs could not be distinguished by the size or number of positively charged domains, nor the magnitude of positive charge in these domains (see “Max contiguous positive charge,” “Fractional positive charge,” and “Max positive charge” metrics in Fig. S2). However, the number of domains with high positive charge and the magnitude of positive charge in these domains are significantly correlated with the ability of repressive PSC-CTRs to inhibit chromatin remodeling (Fig. S4 A–F). This correlation is robust over a range of thresholds used to quantify charged patches. Together, these data suggest that multiple patches of high positive charge contribute to PSC-CTR activity.
Predicting PSC-CTR Activity Through Sequence-Based Criteria.
From the sequence analysis of PSC-CTRs, we were able to identify features shared by all PSC-CTRs (amino acid composition, length, intrinsic disorder) and one that distinguishes repressive and nonrepressive PSC-CTRs (extent of contiguous negative charge per length of protein). Using these features, we formulated a set of sequence criteria to identify proteins with PSC-CTR-like activity (see Materials and Methods).
To validate these criteria, we asked whether they could successfully predict PSC-CTR-like activity in other proteins. The ExPASy AACompSim tool (30, 31) was used to query the D. melanogaster proteome for candidates with amino acid composition similar to PSC-CTR. The closest match to PSC-CTR was Su(z)2, followed by an unknown protein (y5098_DROME), and the transcriptional regulator Jing. Previous studies indicate that Jing mediates the repression of proximal-distal patterning genes homothorax and teashirt during D. melanogaster development, reminiscent of the role of PSC in silencing developmental genes (32). Genetic interactions were also observed between jing and Polycomb (32). The N-terminal region (NTR) of Jing lies upstream of a series of C2H2-type zinc fingers (33) (Fig. S5A). It is predicted to be largely disordered, and exhibits low maximum contiguous negative charge, similar to repressive PSC-CTRs (Fig. S5B). We expressed and purified Jing-NTR from baculovirus-infected Sf9 cells (Fig. S5B), and assessed its biochemical properties. It bound to free DNA with a Kd of 3.0 ± 2.0 nM (Fig. S5C) and achieved 50% inhibition of chromatin remodeling at a concentration of 9 nM (Fig. S5D), indicating that Jing-NTR is biochemically equivalent to PSC-CTRs in these assays. Thus, our sequence-based criteria identified a protein with PSC-CTR-like activity, even though it exhibits low levels of sequence similarity to PSC-CTR.
PSC-CTR Activity Is Broadly but Not Universally Conserved Across Metazoan Evolution.
To understand the mode of PSC-CTR evolution, we analyzed the experimentally validated PSC-CTRs in the context of established metazoan phylogenies (34–36). Repressive PSC-CTRs are broadly conserved in invertebrates, spanning a diverse range of protostomes, including mollusks, annelids, nematodes, and arthropods (Fig. 4A), and the deuterostome/chordate Ciona intestinalis. In order to increase the resolution of phylogenetic sampling, we predicted repressive activity in all PSC-CTRs that were annotated in our initial search of 28 diverse metazoan genomes (Table S1). Each organism was classified as either possessing or lacking repressive PSC-CTR(s).
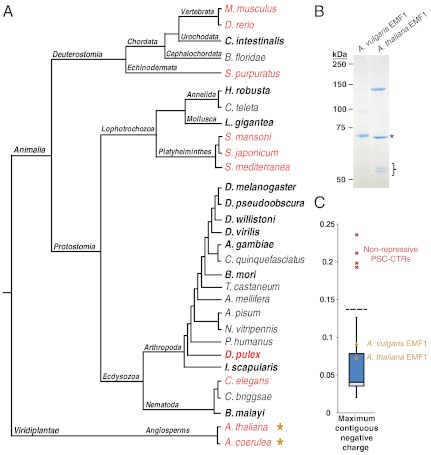
Fig. 4.
Broad conservation of PSC-CTR in metazoans and identification of plant EMF1 as a PSC-CTR-like protein. (A) PSC-CTR activity is superimposed upon an established metazoan phylogeny (34–36). Taxa with repressive PSC-CTRs are colored black, while taxa that lack repressive PSC-CTRs are colored red. PSC-CTR activity in bolded taxa was experimentally tested, while activity in nonbolded taxa was predicted from their maximum contiguous negative charge, intrinsic disorder, and amino acid composition. Yellow asterisks denote taxa that possess the PSC-like protein EMF. Data from M. musculus and D. rerio PSC-CTRs were obtained from (43). Note that the A. coerulea genome was queried for repressive PSC-CTRs, but subsequent affinity purification and biochemical analysis was performed on EMF1 from Aquilegia vulgaris (a very closely related species to A. coerulea). (B) Coomassie-strained gel of A. thaliana EMF1 and A. vulgaris EMF1 purified from Baculovirus-infected Sf9 cells. The asterisk denotes a an approximately 70 kDa band, likely to be HSC70. The closed bracket denotes approximately 55 kDa bands, likely to be β-tubulin. (C) Box and whisker plot depicting maximum contiguous negative charge of PSC-CTRs, relative to EMF1 proteins (yellow asterisks).
Notably, we find that independent evolutionary transitions in PSC-CTR activity have occurred in multiple metazoan lineages (Fig. 4A). Within the lophotrochozoans, repressive PSC-CTRs were predicted to be absent in the platyhelminthes Schistosoma japonicum and Schistosoma mansoni, while experimentally tested repressive PSC-CTRs were present in the mollusk Lottia gigantea and the annelid Helobdella sp. (Austin). Similar evolutionary transitions were observed in the deuterostomes, with the basal chordates Branchiostoma floridae and Ciona intestinalis—and not the more derived chordates Mus musculus and Danio rerio—possessing repressive PSC-CTRs. In addition, the echinoderm Strongylocentrotus purpuratus, which represents a sister taxon to the chordates, was predicted to lack repressive PSC-CTRs. Within the arthropods, D. pulex lacked repressive PSC-CTRs (as supported by the biochemical analysis of D. pulex PSC1-CTR and PSC2-CTR) while seven other taxa in this phylum (Drosophila melanogaster, Drosophila pseudoobscura, Drosophila willistoni, Drosophila virilis, Anopheles gambiae, Bombyx mori, Ixodes scapularis) were experimentally verified to possess repressive PSC-CTRs. It should be pointed out, however, that while the biochemical properties of repressive and nonrepressive PSC-CTRs are distinct, our confidence in prediction of repressive PSC-CTRs is higher than for nonrepressive ones since we do not know whether proteins with the reduced biochemical activity such as D. pulex PSC1-CTR might still contribute to gene regulation in vivo. Taken together, our comparative analysis suggests that PSC-CTR activity is not a static feature of PRC1, and may instead vary widely within independent lineages. This mode of evolutionary change is superimposed upon the broad conservation of repressive PSC across the metazoan phylogeny.
Plant PcG Protein EMF1 Has PSC-CTR-Like Properties.
We extended our analysis beyond metazoans, and asked whether plant genomes also encode proteins with PSC-CTR-like activity. However, repressive PSC-CTRs could not be found in the distantly related eudicots Arabidopsis thaliana and Aquilegia coerulea (Table S1), which are separated by 120–130 million years of divergence (9). This led us to wonder whether PRC1 function is conserved in plants. If so, a functionally similar PSC-CTR-like protein should be present. Previously, it was reported that the putative PRC1 subunit Arabidopsis thaliana EMBRYONIC FLOWER1 (EMF1) inhibits transcription from naked DNA templates in vitro (37), and interacts physically with other PcG proteins, including the PRC1 subunits AtBMI1A, AtBMI1B, AtRING1A, and AtRING1B (37, 38). We asked whether EMF1 has PSC-CTR-like properties. To this end, we cloned EMF1 from A. thaliana and Aquilegia vulgaris [a closely related species that is separated from A. coerulea by approximately 6 million years of divergence (39)] and expressed them in Sf9 cells for subsequent affinity purification (Fig. 4B). Both A. thaliana and A. vulgaris EMF1 are predicted to be largely disordered, and exhibit low contiguous negative charge, consistent with our criteria for repressive PSC-CTRs (Fig. 4C). The cloned A. vulgaris EMF1 was substantially shorter than the predicted sequence obtained from the Phyotozome database, suggesting that it could be a short isoform of the full-length protein. The shorter protein still fits the criteria for PSC-CTR-like repressive activity. We found that both A. thaliana and A. vulgaris EMF1 bind tightly to free DNA with Kd of 1.5 nM and 2.5 nM, respectively (Fig. S6A), similar to that observed for repressive PSC-CTRs. In addition, A. thaliana and A. vulgaris EMF1 inhibit chromatin remodeling with similar efficiency as D. melanogaster PSC-CTR, achieving 50% inhibition at a concentration of 3 nM and 8 nM, respectively (Fig. S6B), similar to the repressive PSC-CTRs characterized in this study. Thus, EMF1 and D. melanogaster PSC-CTR could be considered as being functionally equivalent in these assays. To sum, we are unable to detect repressive PSC-CTRs in A. thaliana and A. vulgaris, and instead find that a PSC-like protein, EMF1, is present in these genomes. These data suggest that plant PRC1 and D. melanogaster PRC1 may be functionally conserved, except that a different subunit in each complex is responsible for PSC-like activity.
Discussion
We set out to determine the sequence properties that confer the biochemical activities of the PSC-CTR, and the extent to which the PSC-CTR is evolutionarily conserved. Our key findings are: i) PSC-CTR activity is determined (and can be predicted) by physical properties rather than sequence similarity. PSC-CTRs are large, intrinsically disordered, and have patches of positive charge interspersed with local negatively charged regions. High numbers of positively charged windows correlate with repressive activity, while an extended region of negative charge can disrupt PSC-CTR activity. ii) The repressive activity of the PSC-CTR (involving nonenzymatic modification of chromatin structure) is broadly conserved across the metazoan phylogeny. iii) The plant protein EMF1 may functionally replace the PSC-CTR in PRC1.
How Can the Sequence Properties of the PSC-CTR Explain Its Activities?
Our data suggest the electrostatic properties of PSC-CTRs are important for their activity. Sections of the PSC-CTR that have local negative charge will be repelled from the chromatin. Because the intracellular screening length is small, only electrostatic interactions between charges in close proximity will contribute significantly to the PSC-CTR-chromatin-binding energy. Regions of the protein that have net negative local charge (or are uncharged) can loop away from the chromatin to reduce the electrostatic repulsion. Positively charged residues within a section of the PSC-CTR with local negative charge are unlikely to be in close proximity to the nucleosome array, and so may not contribute to the PSC-CTR-chromatin-binding energy. This may explain why local charge distribution rather than global net charge predicts PSC-CTR activity (Fig. 3). We suggest that the presence of multiple regions of local positive charge, separated by regions of local negative charge, allows the PSC-CTR to make multiple contacts over a long stretch of DNA or chromatin. Indeed, previous footprinting experiments with PRC1 indicate two complexes can protect close to 200 base pairs of DNA (40). This could lead to bending of the DNA around the protein, which has been observed in scanning force microscopy experiments carried out with PRC1 bound to DNA (41), It could also lead to chromatin compaction if PSC bends the linker DNA, or contacts nonadjacent segments of chromatin, either of which would bring nucleosomes closer together (Fig. S7). The distributed binding mode suggested by the properties of PSC-CTRs has been proposed for how other intrinsically disordered proteins interact with their binding partners, forming “binding clouds” through multiple low affinity, dynamic interactions (42).
Our data indicate a significant correlation between the biochemical activity of each PSC-CTR homologue, and the number of regions of high local positive charge (Fig. S4). This is consistent with the idea that PSC-CTRs make multiple contacts with chromatin or DNA. We observe that extended stretches of contiguous negative charge, but not net-negative charge, disrupt PSC-CTR activity [the repressive Helobdella sp. (Austin) PSC-CTR has a net charge of -2.9]. Extended stretches of net-negative charge may directly interfere with protein-DNA binding by preventing stretches of the protein from approaching the chromatin, or may compete with DNA for binding to positively charged patches on the protein. They may also interfere with CTR–CTR interactions which are likely important for activity (28, 41).
One question is whether PSC-CTRs contain functional subdomains. Our biochemical analysis of D. melanogaster PSC suggests that both chromatin binding and self-interaction are important for inhibition of chromatin remodeling (28). Structure-function analysis of PSC and phenotypic analysis of PSC mutant alleles reveal that the region between amino acid residues 456 and 909 is especially important for activity, since proteins lacking this region are not active in vitro or in vivo (16, 21). D. melanogaster PSC (456–909) also satisfies our predictive criteria for repressive activity: It is predicted to be intrinsically disordered, exhibits low maximum contiguous negative charge, and is very similar in amino acid composition to D. melanogaster PSC-CTR (456–1603) (Fig. S8). Furthermore, the magnitude and density of positively charged patches are greater in amino acids 456–909 than in any other region of D. melanogaster PSC (Fig. S8). This underscores the importance of D. melanogaster PSC (456–909), since localized positive charge is correlated with repressive activity (43) (Fig. S4).
Despite the importance of region 456–909 in contributing to PSC activity, the purified PSC 456–909 fragment binds chromatin but is impaired for inhibition of chromatin remodeling (28). On the other hand, D. melanogaster PSC (1–910) and PSC (456–1603) are fully active in vitro, suggesting that either region flanking PSC 456–909 contributes to the core biochemical properties of PSC (16, 28). We analyzed the charge properties of these truncated proteins and found that the maximum contiguous negative charge in D. melanogaster PSC 1–910, 456–1603, and 1–1603 is two- to threefold lower than PSC (456–909) (Fig. S8). These data suggest that lowering contiguous negative charge across the whole protein by increasing its length “counterbalances” negatively charged regions in the CTR through an unknown mechanism. Indeed, the extent of contiguous negative charge in repressive PSC-CTRs is inversely correlated with repressive activity (Fig. S9). Future analysis of PSC-CTRs from other species will include testing them for self-association activity, which is not addressed in our current study, and testing whether they contain positively charged regions that are especially critical for interactions with chromatin.
Taken together, these observations provide a framework for considering how PSC-CTRs interact with chromatin in more detail; for example, which parts of the protein actually contact chromatin. They provide an explanation for how PSC can bind an extended region of DNA, or possibly multiple segments of DNA or chromatin. Together with our previous data demonstrating the importance of PSC-PSC interactions, we suggest the mode of binding of the PSC-CTR to chromatin and self-association between chromatin-bound CTRs together mediate compaction. In addition to noncovalent modification of chromatin, PSC also contributes to E3 ligase activity toward histone H2A (15). This activity involves its RING finger, and is conserved in mammalian PSC homologues that lack a functional PSC-CTR (44, 45); mammalian E3 ligase complexes equivalent to the dRAF complex thus likely do not have sequences that noncovalently modify chromatin (46). It is possible that for PSC-like proteins, noncovalent modification of chromatin affects histone ubiquitylation, perhaps by anchoring PSC to its chromatin substrate. It would be interesting to determine if histone ubiquitylation by the dRAF complex (containing PSC and dRING) on multinucleosome substrates displays different parameters than that mediated by equivalent mammalian complexes.
Evolution and Conservation of the PSC-CTR.
Although PSC-CTR activity is broadly conserved across the metazoans, our phylogenetic analysis reveals that it may in fact vary widely in several independent lineages (Fig. 4A). At least three independent evolutionary transitions in activity are likely to have occurred. Firstly, repressive PSC-CTRs are not found in the crustacean D. pulex, but are present in eight other arthropods that occupy basal and derived phylogenetic positions relative to D. pulex. Secondly, within the lophotrochozoans, the platyhelminth S. japonicum is predicted not to have a repressive PSC-CTR, while the mollusk L. gigantea and the annelid Helobdella sp. (Austin) possess repressive PSC-CTRs. Thirdly, the basal chordate C. intestinalis possesses repressive PSC-CTRs, while the more derived chordates M. musculus and D. rerio and the echinoderm S. purpuratus are predicted to lack repressive PSC-CTRs.
Importantly, the loss of PSC-CTR activity is associated with the compensatory gain of a distinct but functionally analogous PRC1 subunit. For example, the vertebrates Danio rerio and Mus musculus lack repressive PSC-CTRs. Instead, they possess a distinct PRC1 subunit—M33—with analogous activities to D. melanogaster PSC-CTR (43). Similarly, the angiosperms A. thaliana and A. coerulea lack repressive PSC-CTRs, but instead possess a functionally analogous PRC1 subunit, EMF1 (Fig. 4 and Fig. S6). The evidence presented in our study is consistent with the hypothesis that PRC1 activity is conserved across eukaryotes, while the identity of the functional subunit may vary between taxa (43, 47).
What might contribute to the labile nature of PSC-CTR activity? Our comparative analysis indicates that there is low sequence conservation among repressive PSC-CTRs, and no conserved sequence motifs. Rather, the maximum extent of contiguous negative charge distinguishes between repressive and nonrepressive PSC-CTRs (Fig. 3B). It is theoretically possible to modulate this charge feature through many different mutational paths, rendering its repressive activity intrinsically amenable to evolutionary change. These include nonsynonymous substitutions that alter charge density, and insertions or deletions that change the length of charged patches.
Central to the argument that the PSC-CTR is evolutionarily labile is the presence of genomes that lack repressive PSC-CTRs. Although it is possible that repressive PSC-CTRs were not found because of inadequate genome annotation, we believe that our conclusion about the lability of PSC-CTRs is robust. First, for two of the monophyletic groups (the deuterostomes and lophotrochozoans) in which an independent gain/loss of repressive activity was observed, multiple genomes were predicted to lack repressive PSC-CTRs. It is less likely that all of these genomes possess repressive PSC-CTRs that were not annotated. Secondly, we comprehensively queried genomes for PSC homologues using an inclusive e-value cutoff of 10-5. The PSC genes with repressive CTRs in our study yielded an average e-value of 4.85 × 10-11 from our TBLASTN searches using the homology region of D. melanogaster PSC, well within the e-value cutoff of 10-5. The distant repressive PSC homologues outside of the arthropod clade yielded an average e-value of 2.06 × 10-10, still comfortably within the 10-5 cutoff. Thus, it is unlikely that PSC homologues with repressive CTRs were excluded because of their e-values returned from the TBLASTN searches.
Interestingly, Caenorhabditis elegans PSC-CTR is classified as nonrepressive (Fig. S10A). However, REA measurements indicate that full-length C. elegans PSC is repressive (43). Analysis of the sequence of the C. elegans PSC suggests that the repressive activity lies in the N-terminal region (NTR), upstream of the RING finger. The NTR—but not the CTR—exhibits several key characteristics of repressive proteins: i) C. elegans PSC-NTR is predicted to be intrinsically disordered (Fig. S10B). This stands in contrast to the CTR, which is predicted to be ordered (Fig. S10C); ii) The maximum contiguous negative charge of C. elegans PSC-NTR is lower than the CTR (whose value is very close to the cutoff for nonrepressive proteins) (Figure S10A); iii) C. elegans PSC-NTR is more similar in amino acid composition to the repressive D. melanogaster PSC-CTR (Fig. S10A). Indeed, C. elegans PSC-NTR—but not the CTR—satisfies the corresponding criteria for repressive activity (Fig. S10A). These data suggest that the NTR and not the CTR of C. elegans PSC harbors repressive activity. More broadly, the reallocation of activity away from C. elegans PSC-CTR is consistent with the hypothesis that the repressive activity of PSC-CTR is not a static feature of PRC1.
Recent experimental data shows that the murine Polycomb (Pc) homologue, M33, possesses analogous biochemical properties to the repressive D. melanogaster PSC-CTR (43), thus suggesting that M33 may functionally replace the PSC-CTR in mammalian PRC1. M33 satisfies our prediction criteria for repressive activity (Fig. S10A). It should be noted, though, that C. elegans PSC-NTR and M33 are shorter and possess higher local positive charge than most repressive PSC-CTRs (Fig. S10 D–F). C. elegans PSC-NTR and mouse M33 may thus constitute a distinct class of chromatin regulators that have both overlapping and distinct properties from the repressive PSC-CTRs we have studied, but retain similar biochemical activities.
In summary, we find that PSC-CTRs with little sequence similarity are, in fact, functionally equivalent because of their shared physical properties. Empirical measurements rather than sequence alignments must therefore be employed to determine PSC-CTR function, and allowed us to discover broad conservation of PSC-CTR function in metazoans and PSC-CTR-like function associated with PRC1 in plants. Our data show that primary sequence divergence cannot automatically be equated with functional divergence. This dichotomy may hold true more generally for intrinsically disordered regions, which also tend to evolve rapidly and exhibit poor sequence similarity (1).
Materials and Methods
Tissues, Cell Lines, and Nucleic Acid Samples.
Whole (unsexed) tissue used for nucleic acid isolation and subsequent amplification of PSC-CTR sequences are listed in Table S2A.
Circular Dichroism.
Far-UV circular dichroism (CD) analysis was performed at 25 °C using a J-815 circular dichroism spectrometer (Jasco). Samples were dialyzed in 50 mM phosphate buffer pH 7.4, 150 mM NaF prior to CD analysis.
Identification of PSC-CTRs and PSC-CTR-Like Proteins.
For all BLAST queries, the e-value cutoff was set to 10-5 and partial sequences were excluded. All protein sequences are listed in Table S1A. Metazoan and plant genomes were queried for PSC-like sequences in Genbank or the Joint Genome Institute database with TBLASTN, using the homology region of D. melanogaster PSC (see Table S2B for PSC homology region sequence). Sequences that lacked the RING-finger domain (InterPro domain IPR001841) were excluded. Drosophilid PSC-CTRs were designated from similarity to D. melanogaster PSC-CTR or Su(Z)2-CTR sequences. For non-Drosophilid PSC homologues, the sequence downstream of the RING-finger domain was designated as the PSC-CTR. Several amino acids immediately downstream of the RING-finger domain were omitted from some PSC-CTRs to optimize PCR amplification. PSC-CTRs selected for biochemical analysis are listed in Table S2B. All amino acids upstream of this conserved region were defined as the N-terminal region (NTR). Sequences that lacked the C2H2-type zinc finger domain (InterPro domain IPR007087) were excluded. The Aquilegia corerulea genome was queried for EMF1 homologues using full-length Arabidopsis thaliana EMF1 (Uniprot accession no. Q9LYD9) and BLASTP in the Phytozome v7.0 database.
Cloning.
PSC-CTR, EMF1, and Jing genes were amplified using iProof DNA polymerase (BioRad) from either genomic DNA (48), or from cDNA that was reverse transcribed (SuperScript II, Invitrogen; oligo(dT)12–18 primer) from Trizol-isolated total RNA. PCR products were cloned into pCR8/GW/TOPO TA (Invitrogen), and transferred into pFBacFG (a modified pFastBac1 vector containing a FLAG tag upstream of Gateway destination vector cassette) through LR recombination (Invitrogen). All cloned sequences, primers used for amplification, and tissue sources for gDNA or RNA are listed in Table S2.
Protein Expression and Purification.
FLAG-tagged proteins were expressed in Sf9 insect cells using the Bac-to-Bac expression system (Invitrogen) as previously described (11, 49).
Biochemical Assays.
At least three independent measurements of each biochemical activity were performed for every protein, using two independent protein preparations. Double filter binding assays were performed as previously described (11, 25) to measure the apparent affinity of proteins for 32P-labeled 157 base-pair DNA. Filters were quantified using a Typhoon Trio variable mode imager (GE Healthcare), and the fraction of bound DNA-over-protein titrations was fit to a single exponential curve to calculate the apparent dissociation constant (Kd). For binding data which did not fit a single exponential (R2 < 0.985), the apparent Kd was estimated from the graph of the points. Restriction enzyme accessibility (REA) assays were performed as previously described (20).
Chromatin Assembly.
A 12-nucleosomal G5E4 array (50) was prepared for REA assays by assembling HeLa core histones on chromatin templates through salt gradient dialysis, as previously described (51, 52). Assembly of the G5E4 array was verified by EMSA after EcoRI digestion to release mononuclesomes as previously described (53).
Motif Analysis.
Conserved motif searches within repressive PSC-CTRs were performed using the MEME suite web server (54). The width of allowable motifs was set to 2–50 amino acids, with no restriction on the number of copies of each motif within the PSC-CTR sequences.
Criteria for Prediction of Repressive Activity.
PSC-CTRs were classified as repressive if they fulfilled all of these criteria: i) Sequence length greater than 500 amino acids; ii) More than 60% of the protein sequence is predicted to be structurally disordered, predictions were carried out using the VL3-BA algorithm under the PONDR prediction server at http://www.pondr.com (55); iii) The amino acid composition of each potential PSC-CTR was within 0.18 of D. melanogaster PSC-CTR. The distance is calculated as the square root of the sum of squared frequency differences for each of the 20 amino acids; iv) the number of contiguous negatively charged windows within the protein must not exceed 15% of its sequence length. Each window spanned 25 amino acids, so the first window contains sequence residues 1∶25 and the last window amino acids N-25∶N-1, where N is the sequence length. For each window, the charge Q at pH 7.9 was calculated as  where Qx is the charge of each amino acid, given by
where Qx is the charge of each amino acid, given by  with the appropriate sign, and the pKX values were obtained from the emboss iep application (56).
with the appropriate sign, and the pKX values were obtained from the emboss iep application (56).
Design of Activated Charge Mutants.
PSC-act1 and PSC-act2 were constructed strictly by rearranging amino acids within D. pulex PSC1-CTR. Thus, the overall amino acid composition and net charge of the protein of PSC-act1 and PSC-act2 was unchanged relative to D. pulex PSC1-CTR. To construct PSC-act1, the longest stretch of contiguous negative charge in D. pulex PSC1-CTR (amino acids 12–150) was reduced in length by rearranging the negatively charged amino acids aspartic acid and glutamic acid. These residues were moved away from the long negatively charged patch, to regions of D. pulex PSC1-CTR that were positively charged (as computed by 25-amino acid sliding windows). In addition, negatively charged amino acids within the patch were rearranged to be in closer physical proximity to one another, in order to reduce the length of contiguous negative charge. In both cases, the negatively charged amino acids were only swapped with positively charged or uncharged polar amino acids to limit the disruption of the distribution of polarity across the protein. Together, these rearrangements reduced the length of maximum contiguous negative charge (normalized by protein length) in D. pulex PSC1-CTR from 0.193 to 0.067 (see Fig. 3G).
PSC-act2 was constructed from PSC-act1 by rearranging charged amino acids outside of the longest contiguous negative charged patch, while leaving this region unchanged. To create dense patches of positive charge, positively charged amino acids were rearranged to be in closer proximity to one another. Only swaps with negatively charged or polar uncharged amino acids were allowed. These rearrangements gave rise to dense patches of positive charge within PSC-act2, resulting in an uneven distribution of charge relative to PSC-act1.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Kyle McElroy, Nicole Follmer; and Drs. Angela DePace, Elena Kramer, Andrew Murray, Stanley Lo, Ajaz Ul Wani, Polina Kehayova, Bettina Lengsfeld; and members of the Francis laboratory for thoughtful discussions and comments on the manuscript. We thank Drs. Michael Brenner, Eric Siggia, David Weisblat, William Gelbart, and Jiangwen Zhang for helpful discussions and advice. We thank Daniel Grau for discussions and sharing of data prior to publication; and Drs. Robert Kingston, Ting Wu, and Amanda Lobell for encouragement and support. We gratefully acknowledge the laboratories that provided tissue samples for gene cloning, and help in animal rearing (listed in Table S2A). This work was supported by an Engineering and Physical Sciences Research Council Fellowship (EP/H028064/1) to L.J.C and a grant from the National Institute of General Medical Sciences GM078456-01 to N.J.F.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.L.W. is a guest editor invited by the Editorial Board.
See Author Summary on page 6798 (volume 109, number 18).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118678109/-/DCSupplemental.
References
- 1.Brown CJ, Johnson AK, Dunker AK, Daughdrill GW. Evolution and disorder. Curr Opin Struct Biol. 2011;21:441–446. doi: 10.1016/j.sbi.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 3.Mager J, Montgomery ND, de Villena FP, Magnuson T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat Genet. 2003;33:502–507. doi: 10.1038/ng1125. [DOI] [PubMed] [Google Scholar]
- 4.Martinez AM, Cavalli G. The role of polycomb group proteins in cell cycle regulation during development. Cell Cycle. 2006;5:1189–1197. doi: 10.4161/cc.5.11.2781. [DOI] [PubMed] [Google Scholar]
- 5.Lee JT. Lessons from X-chromosome inactivation: Long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su Y, Deng B, Xi R. Polycomb group genes in stem cell self-renewal: A double-edged sword. Epigenetics. 2011;6:16–19. doi: 10.4161/epi.6.1.13298. [DOI] [PubMed] [Google Scholar]
- 7.Bracken AP, Helin K. Polycomb group proteins: Navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 8.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitcomb SJ, Basu A, Allis CD, Bernstein E. Polycomb Group proteins: An evolutionary perspective. Trends Genet. 2007;23:494–502. doi: 10.1016/j.tig.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- 12.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 13.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 14.King IF, Francis NJ, Kingston RE. Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol Cell Biol. 2002;22:7919–7928. doi: 10.1128/MCB.22.22.7919-7928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagarou A, et al. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 2008;22:2799–2810. doi: 10.1101/gad.484208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King IF, et al. Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol Cell Biol. 2005;25:6578–6591. doi: 10.1128/MCB.25.15.6578-6591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunk BP, Martin EC, Adler PN. Drosophila genes Posterior Sex Combs and Suppressor two of zeste encode proteins with homology to the murine bmi-1 oncogene. Nature. 1991;353:351–353. doi: 10.1038/353351a0. [DOI] [PubMed] [Google Scholar]
- 18.Ishida A, et al. Cloning and chromosome mapping of the human Mel-18 gene which encodes a DNA-binding protein with a new ‘RING-finger’ motif. Gene. 1993;129:249–255. doi: 10.1016/0378-1119(93)90275-8. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Pulido L, Devos D, Sung ZR, Calonje M. RAWUL: A new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics. 2008;9:308. doi: 10.1186/1471-2164-9-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo SM, Ahuja NK, Francis NJ. Polycomb group protein Suppressor 2 of zeste is a functional homolog of Posterior Sex Combs. Mol Cell Biol. 2008;29:515–525. doi: 10.1128/MCB.01044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu CT, Howe M. A genetic analysis of the Suppressor 2 of zeste complex of Drosophila melanogaster. Genetics. 1995;140:139–181. doi: 10.1093/genetics/140.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunk BP, Martin EC, Adler PN. Molecular genetics of the Posterior sex combs/Suppressor 2 of zeste region of Drosophila: Aberrant expression of the Suppressor 2 of zeste gene results in abnormal bristle development. Genetics. 1991;128:119–132. doi: 10.1093/genetics/128.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emmons RB, et al. Molecular genetic analysis of Suppressor 2 of zeste identifies key functional domains. Genetics. 2009;182:999–1013. doi: 10.1534/genetics.108.097360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuxreiter M, et al. Malleable machines take shape in eukaryotic transcriptional regulation. Nat Chem Biol. 2008;4:728–737. doi: 10.1038/nchembio.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: Application to protein-nucleic acid interactions. Proc Natl Acad Sci USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Logie C, Peterson CL. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 1997;16:6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: A dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 28.Lo SM, Francis NJ. Inhibition of chromatin remodeling by polycomb group protein posterior sex combs is mechanistically distinct from nucleosome binding. Biochemistry. 2010;49:9438–9448. doi: 10.1021/bi100532a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grau DJ, Antao JM, Kingston RE. Functional dissection of Polycomb repressive complex 1 reveals the importance of a charged domain. Cold Spring Harb Symp Quant Biol. 2011;75:61–70. doi: 10.1101/sqb.2010.75.056. [DOI] [PubMed] [Google Scholar]
- 30.Wilkins MR, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 31.Wilkins MR, et al. From proteins to proteomes: Large scale protein identification by two-dimensional electrophoresis and amino acid analysis. Biotechnology (N Y) 1996;14:61–65. doi: 10.1038/nbt0196-61. [DOI] [PubMed] [Google Scholar]
- 32.Culi J, Aroca P, Modolell J, Mann RS. Jing is required for wing development and to establish the proximo-distal axis of the leg in Drosophila melanogaster. Genetics. 2006;173:255–266. doi: 10.1534/genetics.106.056341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Montell DJ. Jing: A downstream target of slbo required for developmental control of border cell migration. Development. 2001;128:321–330. doi: 10.1242/dev.128.3.321. [DOI] [PubMed] [Google Scholar]
- 34.Hejnol A, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philippe H, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 36.Regier JC, et al. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature. 2010;463:1079–1083. doi: 10.1038/nature08742. [DOI] [PubMed] [Google Scholar]
- 37.Calonje M, Sanchez R, Chen L, Sung ZR. EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell. 2008;20:277–291. doi: 10.1105/tpc.106.049957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bratzel F, Lopez-Torrejon G, Koch M, Del Pozo JC, Calonje M. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol. 2010;20:1853–1859. doi: 10.1016/j.cub.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 39.Bastida J, Alcantara J, Rey P, Vargas P, Herrera C. Extended phylogeny of Aquilegia: The biogeographical and ecological patterns of two simultaneous but contrasting radiations. Plant Syst Evol. 2009;284:171–185. [Google Scholar]
- 40.Mohd-Sarip A, Cleard F, Mishra RK, Karch F, Verrijzer CP. Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev. 2005;19:1755–1760. doi: 10.1101/gad.347005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohd-Sarip A, et al. Architecture of a polycomb nucleoprotein complex. Mol Cell. 2006;24:91–100. doi: 10.1016/j.molcel.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Uversky VN. Multitude of binding modes attainable by intrinsically disordered proteins: A portrait gallery of disorder-based complexes. Chem Soc Rev. 2011;40:1623–1634. doi: 10.1039/c0cs00057d. [DOI] [PubMed] [Google Scholar]
- 43.Grau DJ, et al. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011;25:2210–2221. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Elderkin S, et al. A phosphorylated form of Mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol Cell. 2007;28:107–120. doi: 10.1016/j.molcel.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 46.Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hennig L, Derkacheva M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009;25:414–423. doi: 10.1016/j.tig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 48.Strauss WM. Preparation of genomic DNA from mammalian tissue. Curr Protoc Mol Biol. 2001:2.2.1–2.2.3. doi: 10.1002/0471142727.mb0202s42. [DOI] [PubMed] [Google Scholar]
- 49.Abmayr SM, Yao T, Parmely T, Workman JL. Preparation of nuclear and cytoplasmic extracts from mammalian cells. Curr Protoc Mol Biol. 2006:12.1.1–12.1.10. doi: 10.1002/0471142727.mb1201s75. [DOI] [PubMed] [Google Scholar]
- 50.Utley RT, et al. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 51.Carruthers LM, Tse C, Walker KP, III, Hansen JC. Assembly of defined nucleosomal and chromatin arrays from pure components. Methods Enzymol. 1999;304:19–35. doi: 10.1016/s0076-6879(99)04004-5. [DOI] [PubMed] [Google Scholar]
- 52.Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee KM, Narlikar G. Assembly of nucleosomal templates by salt dialysis. Curr Protoc Mol Biol. 2001:21.6.1–21.6.16. doi: 10.1002/0471142727.mb2106s54. [DOI] [PubMed] [Google Scholar]
- 54.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Obradovic Z, et al. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53(Suppl 6):566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- 56.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]