Abstract
Despite the fact that most cancer cells display high glycolytic activity, cancer cells selectively express the less active M2 isoform of pyruvate kinase (PKM2). Here we demonstrate that PKM2 expression makes a critical regulatory contribution to the serine synthetic pathway. In the absence of serine, an allosteric activator of PKM2, glycolytic efflux to lactate is significantly reduced in PKM2-expressing cells. This inhibition of PKM2 results in the accumulation of glycolytic intermediates that feed into serine synthesis. As a consequence, PKM2-expressing cells can maintain mammalian target of rapamycin complex 1 activity and proliferate in serine-depleted medium, but PKM1-expressing cells cannot. Cellular detection of serine depletion depends on general control nonderepressible 2 kinase-activating transcription factor 4 (GCN2-ATF4) pathway activation and results in increased expression of enzymes required for serine synthesis from the accumulating glycolytic precursors. These findings suggest that tumor cells use serine-dependent regulation of PKM2 and GCN2 to modulate the flux of glycolytic intermediates in support of cell proliferation.
Keywords: amino acid synthesis, glucose, metabolism, nucleotide biosynthesis
One of the important metabolic signatures of tumor cells is the expression of the pyruvate kinase isoenzyme type-M2 (M2-PK or PKM2). Pyruvate kinase catalyzes the last reaction of glycolysis, converting phosphoenolpyruvate (PEP) to pyruvate and producing ATP. There are four pyruvate kinase isoenzymes: PKL (L-PK, liver isoform) (1), PKR (R-PK, red blood cell isoform) (2, 3), PKM1 (M1-PK, muscle isoform), and PKM2 (embryonic and tumor isoform) (4). PKM1 and PKM2 are encoded by the same but differentially spliced gene (5). Compared with PKM1, which is primarily expressed in normal tissues, such as brain and muscle (6, 7), which demonstrate high oxidative phosphorylation, PKM2 is expressed in proliferating tissues (i.e., embryonic cells, adult stem cells, and all tumor cells reported to date) (8–11). Because of this difference, it is believed that PKM2 could be used as a clinical cancer marker (12). It has been proposed that the oncogenic transcription factor c-Myc up-regulates nuclear RNA binding proteins PTB, hnRNAP1, and hnRNAP2 to promote the selective splicing of PKM mRNA precursor to PKM2 mRNA (13, 14). PKM2 is also induced by hypoxia-inducible factor 1 (15).
Unlike PKM1, which exists only as a tetramer with high affinity for PEP, PKM2 forms both tetramers (high affinity, low Km for PEP) and dimers (low affinity, high Km for PEP) (16). PKM2 requires fructose-1,6-bisphosphate (FBP) to form the active tetramer, but PKM1 does not (17, 18). Recent findings indicated that PKM2 can be phosphorylated at Y105 by tyrosine kinases in cancer cells (19, 20). The PKM2 conformational change caused by phosphorylation leads to FBP release and conversion of the enzyme from the tetramer to the less active dimer form. In fact, most of PKM2 exists as less active dimers with low catalytic rate in tumors (21, 22), yet PKM2 is predominantly a tetramer in nontransformed cells (9, 23). This interesting phenomenon has raised the question of what the benefit of reducing PKM activity is for tumor cells. One hypothesis is that the lower activity of PKM2 promotes the accumulation of upstream glycolytic intermediates, which are precursors for biosynthesis of nucleotides, amino acids, and lipids required for proliferation (21).
In addition to FBP, the nonessential amino acid serine is the only other known activator of PKM2 (24). Serine is used in proliferating cells for protein synthesis as well as the synthesis of other amino acids, such as glycine and cysteine. Furthermore, serine-derived glycine is used in nucleotide synthesis. Serine is also a precursor for the synthesis of lipids, such as phosphatidylserine and sphingolipids (25). Thus, serine is important for the synthesis of nucleotides, proteins, and lipids required for cell proliferation. The studies herein were undertaken to determine whether PKM2 contributes to the ability of cancer cells to undertake de novo serine production from glycolytic intermediates. Serine synthesis starts from the glycolytic intermediate 3-phosphoglycerate (3-PG). We hypothesized that reduced pyruvate kinase activity in PKM2-expressing cells would lead to accumulation of 3-PG, which could then be converted to serine. As serine levels increase, serine-dependent PKM2 activation could provide a feedback loop that restores glycolytic flux in growing cells. Here we demonstrate that PKM2 expression contributes to endogenous serine synthesis and to maintaining mammalian target of rapamycin complex 1 (mTORC1) activity in the absence of exogenous serine. To support these effects, we find that all three enzymes in the serine synthetic pathway are up-regulated upon nonessential amino acid starvation as a result of activation of the general control nonderepressible 2 kinase-activating transcription factor 4 (GCN2-ATF4) pathway.
Results
Serine Increases PKM2 Activity and Glycolysis Rate.
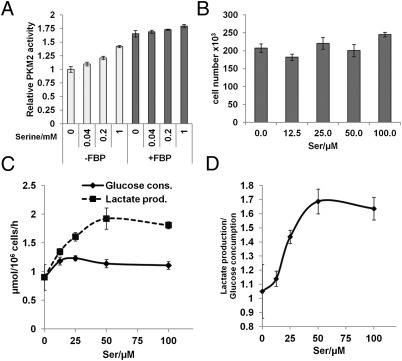
Although serine has previously been shown to be an activator of purified PKM2 in vitro (24), how this serine activation is affected by the presence of PKM2’s other allosteric activator FBP, and whether altered serine levels affect the rate of glycolysis in cells, remains unknown. To examine the effect that serine has on PKM2 enzyme activity in relation to FBP, we performed in vitro pyruvate kinase activity assays using recombinant human PKM2 protein incubated with increasing concentrations of serine in the absence or presence of FBP. In the absence of FBP, serine caused 10–40% increase in PKM2 activity in a dose-dependent manner, whereas only a slight increase of PKM2 activity was observed with increasing serine in the presence of 10 μM FBP (Fig. 1A). These data suggest that although FBP is the dominant activator of PKM2, serine may become important when the FBP concentration is low. To examine the effect of serine on modulating glycolysis in cells, we cultured H1299 cells (human lung carcinoma cell line) with various concentrations of serine for 16 h, and subsequently measured cellular glucose consumption and lactate production. The variation of serine levels over 16 h did not affect cell proliferation significantly (Fig. 1B). Interestingly, the lactate production of H1299 cells started to decrease when the serine concentration dropped below 50 μM. However, the glucose consumption did not decrease until the serine concentration was reduced to less than 12.5 μM (Fig. 1C). Accordingly, the ratio of lactate production/glucose consumption decreased from 1.69 to 1.13 when serine levels were reduced from 50 μM to 12.5 μM (Fig. 1D). The reduction of this ratio suggests that these tumor cells are able to adjust the glycolytic flux to lactate according to serine concentration, saving glycolytic intermediates for other needs when serine levels are low. Taken together, these data indicate that serine is a critical modulator of glycolysis in these tumor cells.
Fig. 1.
Serine increases PKM2 activity and glycolytic rate. (A) In vitro PKM2 activity increases in the presence of serine. Assay performed with a series of serine concentrations as indicated with and without 10 μM FBP (data represent mean ± SEM, n = 3). (B) Equal numbers of H1299 cells were incubated with various concentrations of serine for 16 h, cells were trypsinized, stained with Trypan blue, and counted (data represent mean ± SEM, n = 3). (C) Glucose consumption and lactate production in B were measured as described in Materials and Methods and normalized to cell number (data represent mean ± SEM, n = 3). (D) The lactate production/glucose consumption ratio increases with the serine concentration (data represent mean ± SEM, n = 3).
PKM2 Confers Resistance to Proliferation Arrest Under Serine Starvation.
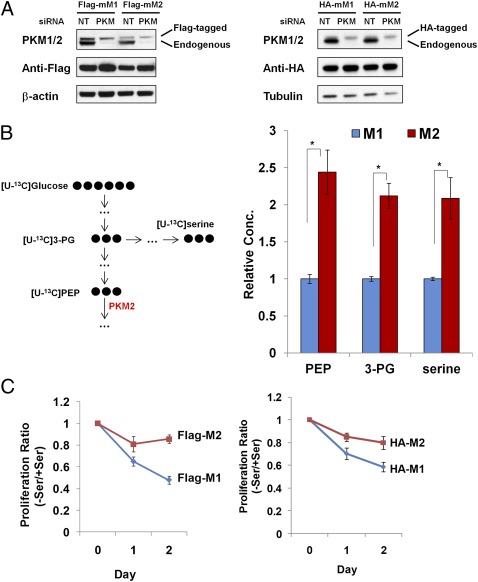
To distinguish the role of PKM2 from PKM1 in tumor cell metabolism, we developed matched tumor cells expressing either PKM1 or PKM2 using a similar strategy to that described previously (11). First, we generated stable H1299 cell lines expressing Flag/HA-tagged mouse PKM1 (Flag/HA-mPKM1) or PKM2 (Flag/HA-mPKM2). siRNA targeting human PKM but not mouse PKM was then used to knock down endogenous PKM expression, including both M1 and M2 isoforms, because they share the same mRNA precursor. As a result, the cells predominantly expressed mPKM1 or mPKM2 at comparable levels (referred to hereafter as M1 or M2 cells) (Fig. 2A).
Fig. 2.
PKM2 expressing cells are more resistant to serine starvation than PKM1-expressing cells. (A) To generate cell lines expressing PKM1 or PKM2, H1299 cells expressing Flag-tagged (Left) or HA-tagged (Right) mouse PKM1 or PKM2 were transiently transfected with 20 nM nontargeting siRNA (NT) or PKM siRNA. After 48 h, PKM1/2 expression was measured using immunoblot. (B) The levels of [U-13C] PEP, [U-13C] 3-PG, and [U-13C] serine in Flag-M1/M2 cells were measured using LC-MS. Flag-M1/M2 cells were incubated in serine-free medium for 24 h and then labeled with [U-13C6] glucose for 3 h. This process results in essentially complete labeling of PEP and 3-PG; thus, the data for these metabolites reflect total cellular concentrations. For serine, labeling is incomplete and the enhanced labeling reflects increased flux (data represent mean ± SEM, n = 3). *P < 0.05, two-tailed Student t test. (C) PKM2 confers resistance to serine starvation. Flag-M1/M2 (Left) or HA-M1/M2 (Right) cells were incubated with or without serine for 24 or 48 h, and cell proliferation was measured using an MTT assay. The ratios of proliferation between (−) serine and (+) serine media were calculated and plotted (data represent mean ± SEM, n = 3).
The cell lysates of M1 and M2 cells were then used in a PKM activity assay, and M2 cells showed significantly reduced pyruvate kinase activity compared with M1 cells, as reported previously (Fig. S1) (26). To test whether reduced pyruvate kinase activity in M2 cells promotes accumulation of metabolites related to the glycolytic pathway and diverts glycolytic intermediates into serine synthesis, Flag-M1/M2 cells were starved in serine-free medium for 24 h and then labeled with [U-13C6] glucose for 3 h. The cellular metabolites were subsequently measured using liquid chromatography-mass spectrometry (LC-MS). Consistent with our hypothesis, we found significantly higher 13C-labeled PEP (the substrate of pyruvate kinase), 3-PG (the precursor of serine synthesis), and serine in M2 cells than in M1 cells (Fig. 2B). These data demonstrate that PKM2 does contribute to the accumulation of downstream glycolytic intermediates and reroutes glycolysis into serine synthesis.
It has been published that M1 and M2 cells have the same proliferation rate under regular in vitro culture conditions, but in vivo M2 cells form much larger xenograft tumors than M1 cells (11). Similar to previous findings, we did not observe a significant difference in cell proliferation between M1 and M2 cells in complete medium (Fig. S2). Based on the accumulation of glycolytic intermediates that could contribute to de novo synthesis, we next tested whether M2 cells exhibit a proliferative advantage upon serine starvation. Indeed, M1 cells exhibited greater reduction in proliferation upon serine withdrawal than M2 cells, suggesting that M2 cells are less dependent on exogenous serine (Fig. 2C).
PKM2 Contributes to Preserved mTORC1 Activity upon Serine Depletion.
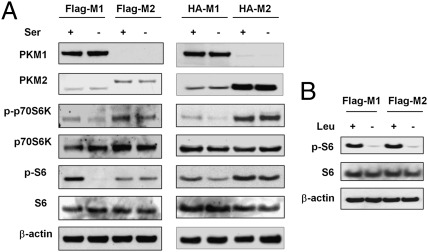
The impairment of growth in M1 cells in the absence of serine suggests that the cellular growth signaling was regulated by serine availability. mTOR is a key molecular sensor for nutrient availability and a regulator of cell growth and proliferation. The activation of mTORC1 requires glutamine and essential amino acids such as leucine (27, 28), but whether other nonessential amino acids, such as serine, are involved in mTORC1 activation remains unclear. To test whether low serine levels in M1 cells leads to mTORC1 inactivation and reduced proliferation, M1 and M2 cells were cultured with and without serine and immunoblots for the mTORC1 downstream effectors phospho-S6 kinase and phospho-S6 were performed. M1 cells, but not M2 cells, showed reduced mTORC1 activities in the absence of exogenous serine (Fig. 3A). To exclude the possibility that PKM2 may affect mTORC1 activity directly, we cultured M1 and M2 cells in medium depleted of the essential amino acid leucine, which is required for mTORC1 activation. As expected, leucine depletion eliminated mTORC1 activity in both M1 and M2 cells (Fig. 3B), indicating it is unlikely that PKM2 regulates mTORC1 activity directly. Taken together, these data suggest that PKM2-expressing cells are able to maintain serine synthesis that directly or indirectly contributes to mTORC1 activation upon serine deprivation.
Fig. 3.
PKM2 maintains mTORC1 activity upon serine depletion. (A) M2 cells, but not M1 cells, maintain mTORC1 activity upon serine deprivation. Flag-M1/M2 (Left) or HA-M1/M2 (Right) cells were incubated with/without serine for 16 h, and mTOR activities were measured by immunoblotting for p-S6K and p-S6. Data are representative of three independent experiments. (B) Leucine withdrawal reduces mTORC1 activity in both M1 and M2 cells. Flag-M1/M2 cells were incubated with/without leucine for 16 h.
Proliferative Advantage of M2 Cells Under Serine Withdrawal Depends on the Serine Synthetic Pathway.
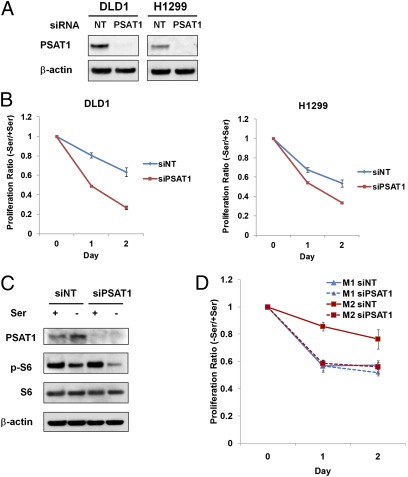
If the proliferative advantage of M2 cells under serine deprivation is a result of enhanced serine synthesis, we should be able to eliminate this advantage by blocking the serine synthetic pathway. There are three enzymes, 3-phospho-glycerate dehydrogenase (PHGDH), phosphoserine aminotransferase 1 (PSAT1), and phosphoserine phosphatase (PSPH), which are required for the three-step conversion of the glycolytic intermediate 3-PG to serine. All three enzymes have previously been found to be overexpressed or demonstrate increased activity in tumor tissue (29–34), suggesting this serine synthetic pathway may play an important role in tumorigenesis. To test this idea, we blocked this pathway by suppressing PSAT1 with RNAi. Upon PSAT1 knockdown, the cell proliferation of both DLD1 (human colon carcinoma cell line) and H1299 cells was significantly reduced under serine deprivation (Fig. 4 A and B). In addition, siRNA targeting PSAT1 inhibited mTORC1 activity more upon serine withdrawal than control siRNA (Fig. 4C). Moreover, under serine starvation, knocking down PSAT1 reduced the proliferation of M2 cells dramatically, but did not further reduce the proliferation of M1 cells (Fig. 4D). These data provide additional evidence that the proliferative advantage of M2 cells under serine starvation depends on elevated endogenous serine synthesis.
Fig. 4.
Blocking endogenous serine synthetic pathway by knocking down PSAT1. (A) Knocking down PSAT1 in tumor cells. DLD1 and H1299 cells were transfected with 50 nM nontargeting siRNA or PSAT1 siRNA. After 48 h, PSAT1 expression was measured using immunoblot. (B) Knocking down PSAT1 reduces tumor cell proliferation under serine starvation. Cells were transfected with PSAT1 siRNA as described earlier. After 48 h, transfected cells were incubated in media with or without serine for 24 or 48 h. Cell proliferation was measured using an MTT assay. The ratios of proliferation between (−) serine and (+) serine media were calculated and plotted (data represent mean ± SEM, n = 3). (C) Knocking down PSAT1 inhibits S6 phosphorylation upon serine depletion. H1299 cells were transfected with PSAT1 siRNA as described earlier. After 48 h, transfected cells were incubated with/without serine for 16 h. (D) Knocking down PSAT1 reduces proliferation of M2 cells significantly, but not M1 cells. HA-M1/M2 H1299 cells were transfected with PSAT1 siRNA as described before. After 48 h, transfected cells were incubated with or without serine for 24 or 48 h and cell proliferation was measured using an MTT assay. The ratios of proliferation between (−) serine and (+) serine media were calculated and plotted (data represent mean ± SEM, n = 3).
GCN2-ATF4 Pathway Up-Regulates Serine Synthetic Enzymes in Response to Amino Acid Deprivation.
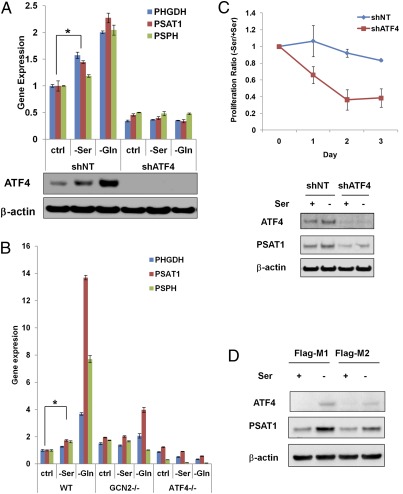
Not only was PSAT1 activity required for proliferation in serine-depleted medium, but we observed that PSAT1 expression was increased upon serine starvation (Fig. 4C). It has been reported that PHGDH, PSAT1, and PSPH are transcriptional targets of ATF4 (35, 36). ATF4 is translationally up-regulated upon amino acid depletion through GCN2-dependent eIF2α phosphorylation (37), and many ATF4 transcription targets are involved in nonessential amino acid synthesis and transport (38). To test whether these enzymes in the serine synthetic pathway are induced in an ATF4-dependent manner upon amino acid deprivation, we cultured DLD1 cells expressing nontargeting shRNA (shNT) or shRNA targeting ATF4 (shATF4) in serine-free or glutamine-free medium for 6 h. The mRNA levels of all three enzymes in the serine synthetic pathway were up-regulated under serine or glutamine starvation in control cells associated with ATF4 induction, but in ATF4-deficient cells the basal mRNA levels of the three enzymes were lower, and no induction was observed upon amino acid starvation (Fig. 5A), indicating that all three enzymes are up-regulated by ATF4 to compensate for amino acid starvation. The stronger mRNA induction upon glutamine starvation is probably because of the fact that cultured cancer cells are adapted to a high concentration of glutamine. The fact that glutamine withdrawal also induces enzymes in the serine synthetic pathway suggests that this induction was a result of a general amino acid response and not specific to serine withdrawal. We also confirmed that the induction depends on GCN2 because GCN2−/− mouse embryonic fibroblasts (MEFs) fail to up-regulate the enzymes under serine or glutamine starvation (Fig. 5B).
Fig. 5.
The induction of enzymes in the serine synthetic pathway under amino acid starvation depends on GCN2-ATF4 pathway. (A) DLD1 cells expressing nontargeting shRNA or ATF4 shRNA were starved in media without serine or glutamine for 6 h, and mRNA levels were measured using real-time PCR (data represent mean ± SD, n = 3). *PPHGDH = 0.0011, PPSAT1 = 0.0106, PPSPH = 0.0038. (B) Wild-type, GCN2−/− and ATF4−/− MEFs were starved in the media without serine or glutamine for 6 h, and mRNA levels were measured using real-time PCR (data represent mean ± SD, n = 3). *PPHGDH = 0.0124, PPSAT1 = 0.0001, PPSPH = 0.0004. (C) ATF4 is necessary for cell proliferation under serine starvation. (Upper) DLD1 cells expressing ATF4 shRNA (shATF4) or nontargeting shRNA (shNT) were incubated in media with or without serine for 24, 48, and 72 h. Cell proliferation was measured using an MTT assay. The ratios of proliferation between (−) serine and (+) serine media were calculated and plotted (data represent mean ± SEM, n = 3). (Lower) The protein levels of ATF4 and PSAT1 after 24-h serine withdrawal were measured using immunoblot. (D) Flag-M1/M2 cells were incubated with or without serine for 24 h. ATF4 and PSAT1 expression were measured using immunoblot. Data are representative of three independent experiments.
The above data suggest that the GCN2-ATF4 pathway collaborates with PKM2-dependent alterations in glycolytic metabolism to coordinate serine synthesis. To confirm this theory, we tested whether ATF4, like PKM2, was critical for cell proliferation in the absence of serine. Consistent with the observation that shATF4 cells have significantly lower PSAT1 expression than control cells, the proliferation of shATF4 cells was inhibited upon serine starvation, but the proliferation of the control cells was not (Fig. 5C). However, ATF4 induction of serine synthetic enzymes alone is not sufficient to promote serine-independent proliferation because the impaired proliferation of M1 cells in the absence of exogenous serine was associated with significantly higher ATF4 levels and higher expression of serine synthetic enzymes, but the induction of ATF4 and ATF4 target genes were less in serine-depleted M2 cells (Fig. 5D and Fig. S3).
Discussion
Although the preferred expression of PKM2 in tumor cells has been known for decades, how PKM2 contributes to tumorigenesis is still incompletely understood. Here we report that conversion of PKM1 to PKM2 contributes to the shunting of glycolytic precursors into the serine synthetic pathway (Fig. S4). Similar to FBP, serine positively regulates PKM2 enzyme activity. A low level of intracellular serine leads to a reduction in aerobic glycolysis, and the low pyruvate kinase activity of PKM2 facilitates accumulation of glycolytic intermediates that serve as substrates for endogenous serine synthesis. In turn a low level of serine leads to activation of GCN2 and the enhanced translation of ATF4. The ATF4 induction increases the transcription of the genes necessary for serine biosynthesis from the accumulating glycolytic intermediates upstream of PKM2. The combined functions of GCN2-ATF4 and PKM2 are necessary for cells to maintain cell proliferation when deprived of extracellular serine.
Essential amino acids and glutamine are well-established activators of mTORC1, which plays an important role in protein translation, cell growth, and proliferation (27, 28). However, it is not clear whether other nonessential amino acids besides glutamine can contribute to mTORC1 activity. Here we establish that serine also contributes to mTORC1 regulation. In serine-deficient medium, PKM1-expressing cells display reduced mTORC1 activity and impaired proliferation. In contrast, PKM2-expressing cells maintain mTORC1 activity and cell proliferation in serine-deficient medium. The maintenance of mTORC1 activity depends on an intact serine synthetic pathway because mTORC1 activity is lost when PKM2-expressing cells are transfected with PSAT1 siRNA in serine-deficient medium.
GCN2 is a kinase activated by uncharged tRNA, so it is a direct sensor of amino acid depletion. The GCN2-ATF4 pathway plays a critical role in preserving intracellular amino acid homeostasis (38, 39). Here we found that serine and glutamine deprivation activates the GCN2-ATF4 pathway to up-regulate three enzymes needed for serine synthesis. Together with the increased substrate availability provided by PKM2 expression, this up-regulation of the serine synthetic pathway provides tumor cells with an enhanced ability to produce serine when exogenous serine is not sufficient to support proliferation. Two recent reports suggest that the genomic regions containing PHGDH are amplified in breast cancer and melanomas (33, 34), diverting glycolysis intermediates to serine synthesis. Our data suggest that activation of the GCN2-ATF4 signaling pathway represents a physiologic mechanism to up-regulate PHGDH and the other two enzymes in the serine synthetic pathway independent of changes at their genomic loci.
Based on the present finding there may be two distinct ways to exploit the selective expression of PKM2 by tumor cells to enhance cancer therapies. One way is using PKM2 activators to increase its enzyme activity to restore glycolytic flux from PEP to pyruvate so that less 3-PG accumulates. Currently there are PKM2 activators under development (40, 41). The present studies also suggest that an alternative strategy might be to develop drugs that inhibit one of the enzymes in the serine synthetic pathway.
Materials and Methods
PKM1 and PKM2 cDNA Transfection and Establishment of Stable Cell Lines.
The cDNAs encoding the complete coding region of mouse PKM1 and PKM2 genes were purchased from Open Biosystems and then subcloned into the modified pIRESpuro vector (Clontech), which contains a double HA or FLAG tag at the N terminus of the multiple cloning sites. cDNAs were transfected in the H1299 cell line using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The empty modified pIRESpuro vector was used as a control for transfection. Stable cell lines were selected following puromycin treatment (2.5 mg/mL; Invitrogen) followed by limiting dilution.
RNAi.
siRNA targeting endogenous human PKM and PSAT1 was ordered from Invitrogen. Next, 20 nM siRNA was used to suppress endogenous PKM1/2 and 50 nM was used for PSAT1. Transfection of siRNA was performed using Lipofectamine RNAiMAX (Invitrogen).
Pyruvate Kinase Activity Assay.
Pyruvate kinase activity assay was performed as previously reported (11, 20). Recombinant PKM2 was purchased from GenWay Biotech.
LC-MS Analysis.
For measurement of metabolite levels, Flag-tagged M1/M2 cells were transfected with human PKM1/2 siRNA. At 48 h posttransfection, cells were switched to culture media without serine. After 24 h, this media was replaced by otherwise equivalent media containing 25 mM [U-13C6] glucose. After 3 h of labeling, metabolism was quenched and metabolites were extracted by quickly aspirating the media and immediately adding −80 °C 80:20 methanol:water extraction solution. The extraction process was allowed to continue for 15 min at −80 °C, and then the cells were scraped from the plate. The cell suspension was then centrifuged at 5,300 × g for 10 min, and the supernatant was kept and the debris was re-extracted with −80 °C 80:20 methanol:water. The resulting suspension was centrifuged again and the supernatant was combined with first supernatant. Each sample was then divided into two identical portions, which were dried under nitrogen flow. One portion was resuspended in HPLC-grade water. The other portion was resuspended in 100 μL 80:20 methanol:water and derivatized by adding 10 μL triethylamine and 2 μL benzyl chloroformate, to enhance the measurement sensitivity of amino acids. Samples were analyzed by multiple LC-MS systems (each from Thermo Scientific and fed by electrospray ionization), as described previously (42–44). Briefly, a stand-alone orbitrap mass spectrometer (Exactive) operating in negative-ion mode was coupled to reversed-phase ion-pairing chromatography and used to scan from m/z 85–1,000 at 1 Hz and 100,000 resolution; A TSQ Quantum Discovery triple-quadrupole mass spectrometer operating in negative-ion mode was coupled to reverse-phase ion-pairing chromatography and used to analyze selected compounds by multiple reaction monitoring. Data were analyzed using the MAVEN software suite (45). The results are normalized by protein concentration.
Immunoblot.
Cells were harvested in 1×RIPA buffer (Cell Signaling). The following antibodies were used: p-S6 kinase, total S6K, S6, total S6, PKM2, total PKM (Cell Signaling), PKM1 (Abgent), PSAT1 (Abnova), and ATF4 (Santa Cruz).
Reverse-Transcription and Real-Time PCR.
Total RNA was extracted following the TRIzol Reagent (Invitrogen) protocol. One to three micrograms total RNA were used in reverse transcription following SuperScript II (Invitrogen) protocol. Quantitative PCR was performed on a 7900HT Sequence Detection System (Applied Biosystems) using Taqman Gene Expression Assays (Applied Biosystems). Gene expression data were normalized to 18S rRNA.
Measurements of Metabolites.
Glucose uptake and lactate production was measured using the YSI 7100 MBS (YSI Life Science). After 16-h cell culture, glucose and lactate concentrations in the medium were determined; the amount consumed or produced by cells was determined by subtracting the concentration in the sample medium from the concentration in medium incubated without cells, then normalized to cell number.
Supplementary Material
Acknowledgments
We thank Dr. Constantinos Koumenis (University of Pennsylvania) for the shATF4 DLD1 cells and ATF4−/− mouse embryonic fibroblasts, and Dr. David Ron (University of Cambridge) for the GCN2−/− mouse embryonic fibroblasts. This work was supported in part by grants from the National Institutes of Health and the National Cancer Institute.
Footnotes
Conflict of interest statement: C.B.T. is a cofounder of and consultant at Agios Pharmaceuticals.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204176109/-/DCSupplemental.
References
- 1.Carbonell J, Feliu JE, Marco R, Sols A. Pyruvate kinase. Classes of regulatory isoenzymes in mammalian tissues. Eur J Biochem. 1973;37(1):148–156. doi: 10.1111/j.1432-1033.1973.tb02969.x. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Horche P, Luque J, Perez-Artes E, Pineda M, Pinilla M. Comparative kinetic behaviour and regulation by fructose-1,6-bisphosphate and ATP of pyruvate kinase from erythrocytes, reticulocytes and bone marrow cells. Comp Biochem Physiol B. 1987;87:553–557. doi: 10.1016/0305-0491(87)90051-4. [DOI] [PubMed] [Google Scholar]
- 3.Kahn A, Marie J, Boivin P. Pyruvate kinase isozymes in man. II. L type and erythrocyte-type isozymes. Electrofocusing and immunologic studies. Hum Genet. 1976;33:35–46. doi: 10.1007/BF00447284. [DOI] [PubMed] [Google Scholar]
- 4.Marie J, Levin MJ, Simon MP, Kahn A. Genetic and epigenetic control of the pyruvate kinase isozymes in mammals. Isozymes Curr Top Biol Med Res. 1983;7:221–240. [PubMed] [Google Scholar]
- 5.Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–13812. [PubMed] [Google Scholar]
- 6.Marie J, Kahn A, Boivin P. Pyruvate kinase isozymes in man. I. M type isozymes in adult and foetal tissues, electrofocusing and immunological studies. Hum Genet. 1976;31:35–45. doi: 10.1007/BF00270397. [DOI] [PubMed] [Google Scholar]
- 7.Guguen-Guillouzo C, Szajnert MF, Marie J, Delain D, Schapira F. Differentiation in vivo and in vitro of pyruvate kinase isozymes in rat muscle. Biochimie. 1977;59:65–71. doi: 10.1016/s0300-9084(77)80087-4. [DOI] [PubMed] [Google Scholar]
- 8.Gali P, Bourdin M. Ontogenesis of pyruvate kinase in the brain and liver tissues of the rat. Biochimie. 1978;60:1253–1260. doi: 10.1016/s0300-9084(79)80442-3. [DOI] [PubMed] [Google Scholar]
- 9.Reinacher M, Eigenbrodt E. Immunohistological demonstration of the same type of pyruvate kinase isoenzyme (M2-Pk) in tumors of chicken and rat. Virchows Arch B Cell Pathol Incl Mol Pathol. 1981;37:79–88. doi: 10.1007/BF02892557. [DOI] [PubMed] [Google Scholar]
- 10.Max-Audit I, et al. Pattern of pyruvate kinase isozymes in erythroleukemia cell lines and in normal human erythroblasts. Blood. 1984;64:930–936. [PubMed] [Google Scholar]
- 11.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 12.Eigenbrodt E, Basenau D, Holthusen S, Mazurek S, Fischer G. Quantification of tumor type M2 pyruvate kinase (Tu M2-PK) in human carcinomas. Anticancer Res. 1997;17(4B):3153–3156. [PubMed] [Google Scholar]
- 13.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clower CV, et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci USA. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Berkel TJ, de Jonge HR, Koster JF, Hülsmann WC. Kinetic evidence for the presence of two forms of M2-type pyruvate kinase in rat small intestine. Biochem Biophys Res Commun. 1974;60:398–405. doi: 10.1016/0006-291x(74)90218-6. [DOI] [PubMed] [Google Scholar]
- 17.Muroya N, Nagao Y, Miyazaki K, Nishikawa K, Horio T. Pyruvate kinase isozymes in various tissues of rat, and increase of spleen-type pyruvate kinase in liver by injecting chromatins from spleen and tumor. J Biochem. 1976;79:203–215. doi: 10.1093/oxfordjournals.jbchem.a131048. [DOI] [PubMed] [Google Scholar]
- 18.Eigenbrodt E, Schoner W. Modification of interconversion of pyruvate kinase type M2 from chicken liver by fructose 1,6-bisphosphate and l-alanine. Hoppe Seylers Z Physiol Chem. 1977;358:1057–1067. doi: 10.1515/bchm2.1977.358.2.1057. [DOI] [PubMed] [Google Scholar]
- 19.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 20.Hitosugi T, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eigenbrodt E, Reinacher M, Scheefers-Borchel U, Scheefers H, Friis R. Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells. Crit Rev Oncog. 1992;3:91–115. [PubMed] [Google Scholar]
- 22.Schneider J, et al. Tumor M2-pyruvate kinase in lung cancer patients: Immunohistochemical detection and disease monitoring. Anticancer Res. 2002;22(1A):311–318. [PubMed] [Google Scholar]
- 23.Mazurek S, Zwerschke W, Jansen-Dürr P, Eigenbrodt E. Metabolic cooperation between different oncogenes during cell transformation: Interaction between activated ras and HPV-16 E7. Oncogene. 2001;20:6891–6898. doi: 10.1038/sj.onc.1204792. [DOI] [PubMed] [Google Scholar]
- 24.Eigenbrodt E, Leib S, Krămer W, Friis RR, Schoner W. Structural and kinetic differences between the M2 type pyruvate kinases from lung and various tumors. Biomed Biochim Acta. 1983;42:S278–S282. [PubMed] [Google Scholar]
- 25.Mazurek S. Pyruvate kinase type M2: A key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43:969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Vander Heiden MG, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicklin P, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara K, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 29.Ojala P, et al. mRNA differential display of gene expression in colonic carcinoma. Electrophoresis. 2002;23:1667–1676. doi: 10.1002/1522-2683(200206)23:11<1667::AID-ELPS1667>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 30.Pollari S, et al. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. 2011;125:421–430. doi: 10.1007/s10549-010-0848-5. [DOI] [PubMed] [Google Scholar]
- 31.Snell K, Natsumeda Y, Eble JN, Glover JL, Weber G. Enzymic imbalance in serine metabolism in human colon carcinoma and rat sarcoma. Br J Cancer. 1988;57:87–90. doi: 10.1038/bjc.1988.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herzfeld A, Legg MA, Greengard O. Human colon tumors: Enzymic and histological characteristics. Cancer. 1978;42:1280–1283. doi: 10.1002/1097-0142(197809)42:3<1280::aid-cncr2820420337>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 33.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo J, et al. Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes. 2009;58:2565–2573. doi: 10.2337/db09-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams CM. Role of the transcription factor ATF4 in the anabolic actions of insulin and the anti-anabolic actions of glucocorticoids. J Biol Chem. 2007;282:16744–16753. doi: 10.1074/jbc.M610510200. [DOI] [PubMed] [Google Scholar]
- 37.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 38.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 39.Ye J, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang JK, et al. Evaluation of thieno[3,2-b]pyrrole[3,2-d]pyridazinones as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2010;20:3387–3393. doi: 10.1016/j.bmcl.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boxer MB, et al. Evaluation of substituted N,N’-diarylsulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. J Med Chem. 2010;53:1048–1055. doi: 10.1021/jm901577g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munger J, et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemons JM, et al. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 2010;8:e1000514. doi: 10.1371/journal.pbio.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu W, et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.