Abstract
The metabolic genotype of an organism can change through loss and acquisition of enzyme-coding genes, while preserving its ability to survive and synthesize biomass in specific environments. This evolutionary plasticity allows pathogens to evolve resistance to antimetabolic drugs by acquiring new metabolic pathways that bypass an enzyme blocked by a drug. We here study quantitatively the extent to which individual metabolic reactions and enzymes can be bypassed. To this end, we use a recently developed computational approach to create large metabolic network ensembles that can synthesize all biomass components in a given environment but contain an otherwise random set of known biochemical reactions. Using this approach, we identify a small connected core of 124 reactions that are absolutely superessential (that is, required in all metabolic networks). Many of these reactions have been experimentally confirmed as essential in different organisms. We also report a superessentiality index for thousands of reactions. This index indicates how easily a reaction can be bypassed. We find that it correlates with the number of sequenced genomes that encode an enzyme for the reaction. Superessentiality can help choose an enzyme as a potential drug target, especially because the index is not highly sensitive to the chemical environment that a pathogen requires. Our work also shows how analyses of large network ensembles can help understand the evolution of complex and robust metabolic networks.
Keywords: drug resistance, drug target identification, essential reactions
The metabolic networks of free-living organisms are complex and comprise hundreds to thousands of chemical reactions. Most of these reactions are catalyzed by enzymes encoded in genes. A metabolic network’s most important function is to synthesize all small-molecule precursors of biomass that are necessary for the growth and survival of an organism. For well-studied free-living organisms, these comprise precursors some 50 different small molecules, including amino acids and nucleotides (1).
The metabolic genotype of an organism comprises all enzyme-coding genes. It determines the enzymatic reactions in a metabolic network. This genotype can change dramatically without affecting the metabolic phenotype (that is, the ability to synthesize biomass in a given environment). For instance, loss of function mutations in many enzyme-coding genes can leave the metabolic phenotype unaffected (2–8). In addition, reactions can get added to a metabolic network through horizontal gene transfer of enzyme-coding genes, a process that is especially frequent in prokaryotes. The deletion and addition of multiple reactions over time may lead to metabolic networks that differ in many reactions but can still sustain life in the same chemical environment.
This enormous genotypic plasticity has implications for the evolution of metabolism. It means that reactions or entire pathways necessary for life in one organism may be dispensable in another organism. For example, the isoprenoid pathway synthesizes isopentenyl diphosphate, which is important for synthesis of cell wall constituents. This pathway is essential in Bacillus subtilis, but it is replaced by the mevalonate pathway in Staphylococcus aureus, where the mevalonate pathway is essential (9, 10). Neither of the pathways would be essential in an organism possessing both of these metabolic routes. For the purpose of our work, we define a biochemical reaction as essential if its elimination abolishes the network’s ability to synthesize all biomass molecules in a given environment. A reaction is nonessential if an organism has the ability to bypass that reaction through alternate reactions or metabolic pathways or if the product of the reaction is not needed in a given environment. We emphasize that reaction essentiality depends on the environment. Earlier analyses (11–15) have explored the extent to which reaction essentiality varies among environments. However, these studies focused on a single metabolic network and its genotype. They did not take into account that metabolic networks with the same phenotype can vary in their genotype. Such genotypic variation can also lead to variation in reaction and gene essentiality. The reactions in the isoprenoid and mevalonate pathways mentioned above provide one example. Another example is the existence of essential genes unique to particular strains of S. cerevisiae (16).
Enormous investments are necessary to develop new antibiotic drugs that combat pathogens (17). The genotypic plasticity of metabolic networks has very practical implications for these efforts and the long-term success of the drugs that they produce. Multiple existing drugs target the metabolism of pathogens, such as sulfonamides, fosmidomycin, and isoniazid (18–21). An ideal enzymatic drug target has to fulfill several criteria, among them that the target is essential for the pathogen’s survival. Only in this case can the drug suppress the pathogen. However, as we pointed out, whether an enzyme is essential may depend on the metabolic network of which it is a part. The same enzyme can be essential in one metabolic network that can sustain life in a given environment but nonessential in a different network. Drugs targeting such enzymes are vulnerable to pathogens that evolve resistance against them (for example, through horizontal gene transfer).
Which reactions in a metabolic network may be most easily bypassed? Which reactions cannot be bypassed? We do not know the answer to these questions, which is not surprising. Answering them would require examining many different metabolic genotypes and evaluating the essentiality of reactions in each of them. This process cannot be done systematically with current experimental technology, and it requires new computational approaches. We have recently developed an approach that can answer these questions (22, 23). It uses flux balance analysis (FBA) to compute the phenotype of a network from its genotype. This phenotype is the ability of the network to sustain life in an environment or a set of environments. FBA has been shown to predict gene essentiality with an accuracy of nearly 90% (12, 13). Mismatches between FBA predictions and experimental data can often be attributed to enzyme misregulation, wherein regulatory constraints prevent enzymes from being expressed at optimal levels (24–26). Such constraints are easily broken in laboratory evolution experiments (24–26) and are of limited relevance to our work, because we are concerned with a more fundamental question, namely how the presence or absence of some reactions (enzyme-coding genes) affects the essentiality of other reactions.
Even more central to our approach than FBA is our current considerable knowledge of the universe of biochemical reactions. This known universe currently comprises more than 5,000 stoichiometrically defined reactions (27, 28) that are known to take place in some organisms. Based on this information, our approach can generate random samples of metabolic genotypes (metabolic networks) with a given phenotype (22, 23) (SI Appendix, SI Methods). We refer to such genotypes as random viable metabolic networks. Briefly, we here generate large samples of such networks, examine the reactions in them, and determine whether they are essential. We then use the concept of reaction superessentiality (23) to estimate a superessentiality index for each reaction. This index indicates the fraction of random metabolic networks with a given phenotype in which a reaction is essential. Reactions where this index is low are easily bypassed, reactions where this index is high are difficult to bypass, and reactions where this index is maximal are impossible to bypass based on our current knowledge. The word superessentiality is motivated by the fact that reactions can be more than just essential. They can be essential in many, most, or all metabolic networks with a given phenotype. Our analysis focuses on carbon metabolism, because carbon is central to life.
In this context, we ask fundamental questions about essential reactions and the extent to which they are also superessential. Which are the reactions that cannot be bypassed? How many reactions cannot be bypassed? To what extent does their essentiality depend on the environment? We relate the outcome of these and other analyses to metabolic evolution and the problem of finding drug targets with high superessentiality and thus, low propensity for resistance evolution.
Results
Core of Absolutely Superessential Reactions in Carbon Metabolism.
We begin our analysis with a single carbon source phenotype, an aerobic minimal environment that contains glucose as the only carbon source (SI Appendix shows all of the environmental metabolites that we study). Our point of departure is the biomass composition of Escherichia coli, because it is well-understood; additionally, its major components are representative of other free-living organisms (1, 12). Starting from the set of all currently known reactions, we generated a metabolic network that we call the universal network. This network comprises all known 5,906 biochemical reactions with well-defined stoichiometry (SI Appendix, SI Methods details construction of the universal network). Not surprisingly, this network can produce all biomass components in a glucose minimal environment. For any one metabolic reaction, the universal network contains all possible alternative pathways that could bypass a reaction. Because the reactions of any viable network (including the E. coli network) are a subset of this universe of reactions, an essential reaction in this network cannot be bypassed in any network that uses reactions from the known universe of reactions. That is, if a reaction is essential in the universal network, no known pathway can bypass this reaction and render it nonessential. We analyzed each reaction in the universal network for its essentiality, and thus, we identified 133 reactions essential for growth on glucose. This set of 133 essential reactions forms an irreducibly essential set of reactions. We call it the superessential core of metabolism for viability on glucose (Dataset S1).
Broad Distribution of Reaction Superessentiality.
As opposed to reactions in the superessential core, which are essential regardless of which other reactions occur in a network, the essentiality of many reactions may depend on other reactions. Although the universal network allowed us to identify absolutely superessential reactions, it does not allow us to understand how reaction essentiality depends on other reactions in a network. To this end, we took a different approach; we evaluated the essentiality of each reaction in a large number of genome-scale metabolic networks that contain an otherwise random assortment of known reactions but are viable on a given set of environments. Starting from the E. coli metabolic network, we used the approach detailed in SI Appendix, SI Methods to generate random samples of metabolic networks that can synthesize all E. coli biomass precursors in an aerobic minimal environment containing glucose as the only carbon source. Briefly, this approach relies on Markov Chain Monte Carlo sampling from the set of all metabolic networks that can be formed using 5,609 known biochemical reactions. Our method ensures that the resulting networks have the same number of reactions but are otherwise unrelated to the starting network; additionally, they have a randomized reaction content relative to each other. We refer to these networks as random viable networks. We generated 500 such random viable networks and identified all essential reactions in each such network. These reactions are reactions where removal abolishes a network’s ability to synthesize all biomass components in this environment. On average, only 283.59 reactions (20.3%) were essential in networks of our sample, with a low SD of 8.51 reactions.
To quantify a reaction’s superessentiality with this approach, we define its superessentiality index (ISE) as the fraction of networks in which the reaction is essential. The maximum value of ISE is one for reactions that are essential in all networks of the sample—we call such reactions absolutely superessential. The lowest value of ISE is zero for reactions nonessential in all networks. An ISE of 0.002 would indicate that a reaction was essential only in 1 of 500 random viable networks. Of the total number of 5,906 chemical reactions that occurred in at least 1 of our 500 random viable networks (SI Appendix, SI Methods), 1,400 (23.7%) reactions were essential in at least one network. Fig. 1A shows a rank plot in which reactions are ranked according to their superessentiality index. It indicates that different reactions can vary widely in their superessentiality.
Fig. 1.
Most essential reactions are environment-general. (A) Rank plot of the superessentiality index of 1,400 reactions essential for growth on glucose in 500 random viable metabolic networks. The plateau to the left of the plot corresponds to 139 absolutely superessential reactions (ISE = 1). (B) The distribution of environment-specific and environment-general essential reactions in 500 random metabolic networks viable on 54 carbon sources. The mean proportion of environment-general essential reactions for all 500 networks is 2.38 times higher than the mean proportion of environment-specific essential reactions. (C) The vertical axis shows the mean number of reactions whose elimination abolishes viability on the number of carbon sources shown on the horizontal axis. Error bars correspond to 1 SD. Data are based on 500 random networks viable on 54 carbon sources. Deletion of most essential reactions abolishes growth only on a very limited number of carbon sources (one to eight). (D) Superessentiality indices of reactions essential for growth on glucose (horizontal axis) and 54 carbon sources (vertical axis). The strong association shows that superessentiality does not depend strongly on the environment. One reaction with superessentiality that differs dramatically in glucose and 54 carbon sources is indicated with an arrow (TMDPP) (SI Appendix, SI Results). Data are based on essential reactions from 500 random metabolic networks viable on glucose and another set of 500 random metabolic networks viable on 54 carbon sources.
Comparing the results of this approach to our previous determination of the superessential core from the universal network allows us to validate the network sampling approach. Specifically, sampling identified 139 reactions (2.3%) as absolutely superessential (ISE = 1; that is, they cannot be bypassed in any of our 500 random networks viable on glucose). These reactions correspond to the plateau on the left side of Fig. 1A. Based on the universal network, we had found that 133 reactions formed the superessential core for viability on glucose. Most importantly, these 133 reactions are all contained in the set of 139 absolutely superessential reactions identified by sampling (Dataset S1). In other words, only six reactions that sampling identified as absolutely superessential are artifactually identified as absolutely superessential because of insufficient sampling. This observation shows that even modest samples of 500 random metabolic networks can provide good estimates of reaction superessentiality.
How many of the essential reactions in E. coli can potentially be bypassed by reactions from the reaction universe? Two hundred and forty reactions are essential in E. coli for growth in the glucose minimal environment; of these 240 reactions, 133 reactions are absolutely superessential in the universal network. This finding means that 55.4% (133 of 240) of the essential reactions from E. coli are, in fact, absolutely superessential and thus, irreplaceable. The remaining 44.6% of reactions have an ISE < 1, meaning that an organism could bypass such reactions by acquiring new metabolic genes through mechanisms such as horizontal gene transfer.
Examples of Superessential Reactions.
We next discuss a few examples of reactions in the superessential core. The first of them is phosphoglucosamine mutase (Blattner number b3176) (29), which catalyzes a reversible conversion between glucosamine-1-phosphate to glucosamine-6-phoshate. This enzyme plays an important role in the synthesis of UDP-N-acetyl-d-glucosamine, which is used in peptidoglycan and lipid IVA biosynthesis (30). Second, nicotinamide adenine dinucleotide (NAD) kinase (Blattner number b2615) is important in the generation of nicotinamide adenine dinucleotide phosphate from NAD in an ATP-dependent manner. NAD kinase, thus, may play an important role in determining the size of a cell’s nicotinamide adenine dinucleotide phosphate pool and its turnover in the cell (31). A third example is diaminopimelate decarboxylase (Blattner number b2838), which generates l-lysine from meso-diaminopimelate. This enzyme catalyzes the last reaction in the l-lysine biosynthesis pathway. It is essential if l-lysine is not supplied by the environment.
In addition to absolutely superessential reactions (ISE = 1), reactions with lower superessentiality index (ISE < 1) can also shed light on the structure of metabolism. For example, if a reaction is nonessential in a fraction (1 − ISE) of random viable networks, this fraction indicates how easily the reaction can be bypassed by alternate metabolic pathways based on known reactions. For instance, glucose-6-phosphate isomerase (Blattner number b4025), although not essential in E. coli (11), has an ISE of 0.314, indicating that it is essential in 31.4% of networks. This finding means that it is bypassed in 68.6% of the networks in our sample. Our analysis of reactions shows that reactions from central pathways such as glycolysis, citric acid cycle, or pyruvate metabolism tend to have low superessentiality indices, whereas reactions from amino acid synthesis, such as histidine metabolism, tend to have especially high superessentiality indices (Dataset S2).
Individual networks may contain reactions that do not contribute to biomass production, for example, because they are part of an isolated pathway fragment or a pathway that does not contribute to biomass synthesis in a given environment. Such reactions and pathway fragments do occur in well-annotated metabolic models like the model of E. coli (12). They are also inevitable consequences of unbiased Markov Chain Monte Carlo sampling of random viable networks (SI Appendix, SI Methods). We refer to such reactions as blocked reactions (23, 32). To identify them, we computed the maximum allowable flux through each reaction for viability on glucose for each network in our sample of 500 random viable networks (SI Appendix, SI Methods) (32). If this flux was below a threshold of 10−8, we consider the reaction blocked. When considered together, the number of networks in which a reaction occurs, its superessentiality index ISE, and the number of networks it is blocked in can indicate the extent to which a reaction and its alternate pathways coexist and are functional in a sample of random networks. As mentioned earlier, glucose-6-phosphate isomerase is essential in 31.4% of networks. However, it is present in 44.2% of networks (and blocked in none). Together, these proportions mean that 12.8% (44.2% − 31.4% − 0%) of the random networks in our sample have more than one functional route for this particular reaction. The penultimate reaction in histidine biosynthesis is carried out by histidinol dehydrogenase (Blattner number b2020). It is present in 87% of the networks, essential in 84.6% of networks, and blocked in 0.6% of networks, meaning that it coexists along with its alternate pathways only in 1.8% (87% − 84.6% − 0.6%) of networks. This measure of superessentiality is complementary to the ISE index in providing information on how easily a reaction is bypassed. We report it for all E. coli reactions in Dataset S2.
Metabolic Networks Have Many Environment-General Essential Reactions.
Thus far, we discussed reaction essentiality for a single carbon source phenotype. How do our observations generalize to multiple carbon source phenotypes? Our definition of phenotype regards the ability of a network to sustain life on a given number of sole carbon sources. Networks with a multiple carbon source phenotype can sustain life on many sole carbon sources and any subset of these carbon sources, such as sources that might occur in an environment that changes over time. The highest numbers of carbon sources for a multiple carbon source phenotype that we consider are the 54 different sole carbon sources in which E. coli is known to be viable from experiments (SI Appendix) (12). We can represent the phenotype of viability on 54 carbon sources as a binary string of length 54 in which all entries are equal to one. A deletion of a reaction that abolishes viability on carbon source i would change the value of entry i in this string to zero. We define a reaction as essential in this multiple carbon source phenotype if it abolishes viability on at least one carbon source. Deletion of some reactions abolishes viability in all 54 environments. We refer to such reactions as environment-general essential reactions. Deletion of other reactions abolishes viability only in a few environments. We refer to these reactions as environment-specific essential reactions.
We next revisit (for a multiple carbon phenotype) the concept of a superessential core of metabolism—the set of absolutely superessential reactions. As in our analysis with the universal network for a single carbon source phenotype, we identified absolutely superessential reactions for growth on all 54 carbon environments from our universal reaction network of 5,906 reactions. This approach yielded 148 absolutely superessential reactions. We note that only 15 additional reactions became absolutely superessential as our analysis moved from the single carbon source to the multiple carbon source phenotype (148 vs. 133 absolutely superessential reactions). This observation argues for a common core of superessential reactions that does not depend on the actual environment considered. Indeed, we find that 125 of 148 reactions in the superessential core are environment-general, meaning that deleting these reactions abolishes growth in all 54 different environments.
We next identified all essential reactions (as defined above) with our sampling approach (that is, in each of 500 random networks that were viable on 54 carbon sources) and determined each reaction’s superessentiality index (ISE; we note that this index disregards the number of environments in which a reaction is essential). The number (1,569) of reactions that were essential in at least one network was only 12% higher than the 1,400 reactions essential for growth on glucose that we discussed earlier. SI Appendix, Fig. S1 shows a rank plot of ISE for these 1,569 reactions. Its shape is very similar to the curve in Fig. 1A, and its left side also contains a plateau of 155 (9.9%) reactions that were absolutely superessential for growth on each of the 54 carbon sources. In sum, we identified only seven more reactions as absolutely superessential through the sampling approach compared with the 148 reactions from the universal network. The 148 true-positive reactions are shown in Dataset S3.
Although deletion of some reactions abolishes growth on one or few environments, deletion of other reactions abolishes growth on all 54 environments. Which of these two types of essential reaction is more prominent? Fig. 1B shows the distribution of the proportion of a metabolic network’s reactions that are environment-specific and -general. The data are based on 500 random networks viable on 54 different sole carbon sources. Fig. 1B clearly shows that many more essential reactions are environment-general (mean = 0.18 or 251 reactions) than -specific (mean = 0.0755 or 105 reactions). That is, most essential reactions abolish viability on all carbon sources. Fig. 1C shows the average number of environment-specific essential reactions per network that abolishes viability on a given range of carbon sources when deleted. The number of reactions that abolishes growth on fewer than eight sources when deleted is, by far, the highest (about 42 reactions per network; SEM = 4 reactions), whereas reactions that abolish growth on 45–53 sources are next, with 12 reactions per network (SEM = 3 reactions). Also, there are fewer reactions (one to three reactions per network, SEM = 1 reaction) that abolish growth on 9–45 environments. In sum, most essential reactions in a metabolic network fall into two categories: those reactions whose deletion abolishes viability in very few environments and the vast majority of reactions whose deletion abolishes viability in all environments.
Superessentiality Index of a Reaction Is Not Very Sensitive to the Environment.
Our analysis thus far also suggests that most essential reactions are essential, irrespective of the number of different environments and regardless of the specific carbon source examined. For example, SI Appendix, Fig. S1 shows that 1,569 reactions are essential in at least one network for viability on at least 1 of 54 alternative carbon sources, whereas Fig. 1A shows that 1,400 reactions are essential in at least one network for growth on glucose. Thus, of 1,655 unique reactions that are essential for life on either glucose or at least 1 of 54 carbon sources, 1,314 reactions (79.4%) are essential for both kinds of phenotypes. In addition, the superessentiality index of reactions is similar for both the single carbon source and the multiple carbon source phenotype. Fig. 1D shows that a strong correlation (Pearson’s r = 0.95, P value < 10−300, n = 1,314) exists between the superessentiality index ISE in the single and multiple carbon source phenotypes. A comparison between essential reactions of the phenotype requiring growth on 54 carbon sources and simpler phenotypes that require growth on 5, 10, 20, 30, and 40 alternative carbon sources yields correlations as high as those correlations seen in Fig. 1D (Pearson’s r > 0.92, P value < 10−300, n ≥ 1,409 in all five cases). Moreover, as we already discussed, most essential reactions in a network are essential for growth in more than one environment (Fig. 1 C and D). Taken together, this finding means that a reaction’s superessentiality index ISE does not depend strongly on the number of carbon sources on which it can support viability. The same finding holds, therefore, for how readily a reaction can be bypassed; it does not depend strongly on the environment for most reactions. In SI Appendix, SI Results, we discuss some notable exceptions, such as reactions that can have high superessentiality in 54 different minimal environments but low superessentiality in a glucose-minimal environment.
Absolutely Superessential Reactions Are Enriched in Anabolic Pathways.
As mentioned earlier, we found 133 reactions that are absolutely superessential for growth on glucose and 125 reactions that are absolutely superessential and environment-general for growth on 54 alternative sole carbon sources. We asked whether reactions in these two superessential cores preferentially derive from specific pathways (SI Appendix, SI Methods). We found that both cores were significantly enriched for reactions in pathways that synthesize several amino acids (histidine, valine, leucine, isoleucine, tyrosine, and tryptophan) and cell wall components. In contrast, the cores were not enriched for pathways that synthesize murein, threonine, lysine, and methionine as well as membrane lipids. The results are similar for both superessential cores (SI Appendix, Tables S2 and S3). Reactions from central metabolism such as glycolysis or the citric acid cycle are notably absent from the superessential cores (Datasets S1 and S3). Taken together, this finding means that most absolutely superessential reactions are anabolic in nature, whereas catabolic reactions from pathways such as glycolysis can be more easily bypassed. This observation agrees with experimental and computational studies of essential reactions in E. coli and S. cerevisiae (6, 11, 33, 34). It is also consistent with our earlier observation that reactions from these pathways generally have low superessentiality indices (Dataset S2). We speculate that the reticulate structure of some parts of metabolism may be the reason why one or more pathways, such as central carbon metabolism, are not enriched for superessential reactions, although these pathways are very important (35) (Datasets S1 and S3). In contrast, some amino acids, such as histidine or tryptophan, are synthesized through more linear pathways, which may, thus, not be as easy to bypass. In SI Appendix, SI Results, we show that the environment-general superessential core is a compact and highly connected part of metabolism.
Genes Responsible for Superessential Reactions Occur in Most Prokaryotic Genomes.
If a reaction is frequently essential for preserving a phenotype in random viable metabolic networks, then the corresponding enzyme-coding gene should also occur frequently in many prokaryotic genomes. This line of reasoning will fail if either our knowledge of the universe of metabolic reactions is incomplete or our understanding of an organism’s enzyme complement is partial. The extent to which it fails can shed light on the imperfection of our current metabolic knowledge. With these observations in mind, we analyzed the relationship between reaction superessentiality and reaction occurrence in prokaryotic genomes. To this end, we defined the genome occurrence index (IGO) of a reaction as the fraction of genomes that carry a gene with a product that is known to catalyze this reaction. For each reaction in the universe of reactions, we used Kegg orthology numbers (http://www.genome.jp/kegg/ko.html) (36) to estimate the fraction of prokaryotic genomes that encode an enzyme catalyzing the reaction (SI Appendix, SI Methods).
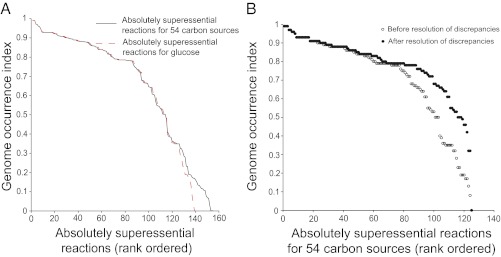
We first focused on absolutely superessential reactions (ISE = 1) for a single carbon source phenotype (viability on glucose) and for multiple carbon source phenotypes. Fig. 2A shows a rank plot of the genome occurrence index IGO for these reactions. If our current understanding of metabolism was perfect, we would expect that the IGO values of all absolutely superessential genes are equal to one. However, the rank plot shows that this expectation is not the case. We highlight two features of this plot. First, the plots for single carbon source and multiple carbon source phenotypes are very similar and nearly congruent. This finding corroborates our earlier observation that most superessential reactions are environment-general. It suggests that differences in the environment in which different species live cannot account for most differences in genome occurrences among absolutely superessential reactions. Second, of the 155 absolutely superessential reactions for growth in 54 carbon sources, 57% (88 reactions) occur in more than 75% of genomes (820 genomes); 73.5% (114 reactions) of 155 reactions occur in more than 50% of genomes (IGO ≥ 0.5). This finding means that a majority of absolutely superessential reactions occurs in most prokaryotic genomes sequenced to date. This association between superessentiality and genome occurrence (IGO) is highly significant with a P value smaller than 10−5 (n = 105, permutation test) (SI Appendix, Fig. S3). The 125 absolutely superessential and environment-general reactions also showed a highly significant association with genome occurrence. Specifically, of 125 reactions, 105 reactions (84%) occurred in more than 50% of genomes (P value < 10−5, n = 105). We next expanded our analysis to include reactions with lower than absolute superessentiality (ISE < 1) and determined whether there exists a statistical association between a reaction’s superessentiality index and its genome occurrence. Such an association indeed exists (Spearman’s ρ = 0.4, P value < 10−300, n = 5,609). SI Appendix, Fig. S4 shows that this correlation in the observed data is significantly different from the correlation in randomized data (P value < 10−5, n = 105), suggesting that the association that we see between superessentiality index and the occurrence of a reaction’s enzyme-coding genes does not occur by chance alone.
Fig. 2.
Absolutely superessential reactions occur in most prokaryotic genomes. (A) The vertical axis shows the genome occurrence index of absolutely superessential reactions for growth on glucose and 54 carbon sources. The curves are almost congruent, underscoring that most absolutely superessential reactions are environment-general. Furthermore, a majority (73.5%) of 155 absolutely superessential reactions for growth on 54 carbon sources has a high genome occurrence (IGO ≥ 0.5). (B) The plot shows the change in the genome occurrence index of 125 environment-general and absolutely superessential reactions when we manually improve the functional annotation of those 20 reactions that occur in fewer than 50% of genomes. Before the improvement, 105 of 125 reactions have a genome occurrence of more than 50% (○), whereas after improvement, this number increases to 118 (94.4%; ●). See Genes Responsible for Superessential Reactions Occur in Most Prokaryotic Genomes and Dataset S4 for details.
These observations indicate, on the one hand, that our approach of characterizing reaction superessentiality in random viable networks reveals biologically relevant information. On the other hand, they also show that our knowledge of metabolism and its enzymes is still incomplete. For example, as we discussed earlier, 114 of 155 absolutely superessential reactions occur in at least 50% of genomes. Of the remaining 41 reactions, about one-half are environment-specific reactions, whereas other reactions have low genome occurrences. The reasons for apparently low occurrence of highly superessential reactions highlight various limitations in existing genome annotation and database information, which a few examples will show.
Glycerol-3-phosphate acyltransferase, encoded by plsB, plays a role in phospholipid synthesis and is essential in E. coli (11) and S. typhirium (37) in the murine in vivo infection model. The enzyme uses fatty acyl-ACP or acyl-CoA thioesters to form acyl-glycerol-3-phosphate, and it is mainly limited to γ-proteobacteria (38), which explains its low genome occurrence index. Other prokaryotes contain a similar enzyme that uses acyl-phosphate to synthesize acyl-glycerol-3-phosphate (encoded by the gene plsY), but the Kegg reaction database does not distinguish between these reactions. It only contains the E. coli variant. If both variants were taken into account, the reaction would occur in a much larger fraction (0.91) of genomes. The glycerol-3-phosphate acyltransferase reaction seems superessential, because the other pathway (encoded by plsY) is absent from the set of known reactions that we used. Another example concerns the coaA gene encoding the enzyme pantothenate kinase. Pantothenate kinase catalyzes the first step of CoA biosynthesis, an essential and ubiquitous cofactor in almost all biological organisms. Prokaryotes can have two different types of pantothenate kinases encoded by the gene coaA or coaX. These genes do not share sequence similarity (39), but they, nonetheless, encode enzymes that catalyze the same reaction. The enzyme encoded by coaA is of type I, whereas the enzyme encoded by coaX is a type III enzyme (39, 40). The existence of the alternative coaX explains the low genome occurrence of coaA, but it also reinforces the essentiality of the reaction. If we take both variants into account, the reaction occurs in 83% of the genomes that we analyzed. Another case in point is reactions catalyzed by promiscuous enzymes, such as the enzyme pyrimidine phosphatase (PMDPHT). PMDPHT catalyzes the transformation of 5-amino-6-(5′-phosphoribitylamino) uracil to 4-(1-d-Ribitylamino)-5-aminouracil with the release of one inorganic phosphate (12), and it is essential in the riboflavin biosynthesis pathway (1). PMDPHT is absolutely superessential and environment-general, but the enzyme-coding gene responsible for PMDPHT has not yet been identified. It, therefore, has the minimum genome occurrence of zero.
In sum, missing information about relevant enzymes and genes can lead to low apparent genome counts, even for reactions with high superessentiality. We identified the reasons for low genome occurrence for those 20 of 125 environment-general reactions in the absolutely superessential core that occur in fewer than 50% of genomes. Fig. 2B shows a comparison of the genome occurrences before and after a correction for such discrepancies based on (limited) independent information. After correction, 94.4% (118 of 125) of absolutely superessential reactions occur in more than 50% genomes. The reasons for these discrepancies are similar to those listed in the above examples. They include misleading assignments of orthology, undiscovered enzymes, and nonorthologous gene displacement (41–45) (SI Appendix, SI Results and Dataset S4).
Most Absolutely Superessential Environment-General Reactions Remain Superessential in Complex in Vivo and Rich Environments.
So far, we have used 54 minimal environments distinguished by their sole carbon source to characterize reaction superessentiality. Although the use of minimal environments makes our analysis simpler, it raises the question of the extent that reaction superessentiality would be similar in the complex chemical environments that many pathogens need to survive. To answer this question, we used the universal network approach to identify absolutely superessential reactions in the complex environments known to sustain in vivo growth of Salmonella typhimirium LT2 (46), Mycobacterium tuberculosis H37Rv (47), Pseudomonas aeruginosa PAO1 (48), and Mycoplasma pneumoniae (49) as well as a synthetic complete medium (50) (SI Appendix, SI Methods). These environments consist of various nutrients such as amino acids, cofactors, fatty acids, and nucleotides. In addition, we supplemented each environment with all 54 carbon sources that we studied here to render our identification of absolutely superessential reactions conservative, because additional nutrients will lead to a reduction but never an increase of reaction superessentiality. Nonetheless, we found that, in each of the five supplemented environments, a majority (at least 77.6%) of the 125 absolutely superessential reactions that we had previously identified (Dataset S3) is still superessential (Dataset S5). Among the absolutely superessential reactions that this approach identified (101 reactions on average for the five environments), every single one is contained in our previously identified set of 125 absolutely superessential environment-general reactions. Furthermore, about 83% of 101 reactions are represented in more than 50% of prokaryotic genomes (P value < 10−5, n = 105 for each of the five environments) (SI Appendix, SI Results and Table S4). The latter observation indicates not only the importance of these reactions, but it also indicates that this importance is not restricted to organisms with a biomass composition similar to E. coli; the organisms in which these reactions occur are taxonomically diverse and may vary widely in their biomass composition. To validate this assertion (that is, that the superessentiality of reactions is not highly sensitive to biomass composition), we also studied reaction superessentiality for different biomass compositions (SI Appendix, SI Results). We found that the relative magnitude of superessentiality indices is highly correlated between different biomass compositions (Spearman’s r = 0.8, P value < 10−13, n = 1,561) (SI Appendix, SI Results).
Discussion
The metabolism of an organism can evolve through elimination of reactions because of loss of function mutations and by addition of new reactions by horizontal gene transfer. Several studies, experimental and computational alike, have identified essential reactions (3–6, 11–14, 51) in different organisms or genome-scale metabolic networks. Most such studies are organism-specific, and they do not address the question of how readily a reaction could be bypassed through alternate reactions or pathways based on our current knowledge of metabolism. This question is important not only to understand the evolutionary plasticity of metabolic networks but also to identify those enzymatic targets for antimetabolic drugs where the risk of evolving resistance is smallest. Our approach of universal network analysis allows us to identify absolutely superessential reactions that are essential in any metabolic network. Random viable network sampling, in addition, allows us to quantify superessentiality for many reactions and study its causes in individual networks. Both approaches are complementary and can be to used cross-validate each other.
One might argue that an analysis like ours should ideally use many reconstructed metabolic networks from diverse prokaryotes (52) instead of random viable network samples. However, the number of high-quality reconstructed networks suitable for FBA is currently still too low. In addition, all sets of such networks would be related through a common evolutionary history, which creates a bias in data that random viable network samples can avoid. Finally, random viable network samples can be directly used for statistical hypothesis testing (22, 23, 53). For our analysis, random viable metabolic networks are, thus, currently an indispensible tool.
We focused here on metabolic networks with a size that is identical to the size of E. coli and with a viability that is defined as the ability to synthesize all E. coli biomass precursors. We did so because the E. coli biomass composition is well-studied, and many of its components—amino acids, nucleotides, etc.—are representative of biomass precursors in most other free-living organisms. Moreover, E. coli is an environmental generalist and thus able to survive in multiple different environments. This feature allowed us to compare reaction superessentiality for networks viable in one and multiple environments. In this regard, we focused on minimal environments that vary in their sole carbon sources because carbon is life’s most central chemical element. Specifically, we compare networks viable in a minimal environment with glucose as its sole carbon source with networks that are viable on 54 different sole carbon sources. We refer to these two types of networks as networks with multiple and single carbon source phenotypes.
We began by identifying a core of absolutely superessential reactions (Datasets S1 and S3). These are reactions that occur and are essential in all networks that we study. This core comprises 133 reactions for the single carbon source phenotype and 148 reactions in the multiple carbon source phenotype. The vast majority of reactions in this core are not specific to a given carbon source, but they are required for viability on all carbon sources. They also form a statistically highly significant connected component in a graph-based representation of metabolism. The properties of this core show that the reactions that are most difficult to bypass do not allow the organism to survive in specific environments, but they are essential to life in multiple environments. Computational and experimental studies have tried to identify common essential reactions across a small number of organisms to develop antibiotics (37, 54). Our identification of an absolutely superessential core of reactions goes beyond these analyses. It carries important implications for drug target identification. It predicts that antimetabolic drugs targeted to enzymes that are most difficult to bypass will not come from pathways that mediate adaptation to specific environment but rather, from anabolic pathways responsible for the synthesis of cell wall components and amino acids.
The next step of our analysis focused on the superessentiality of reactions for single and multiple carbon source phenotypes. First, we showed that reaction superessentiality in single minimal and multiple minimal environments is highly correlated. This finding again showed that a reaction’s superessentiality derives mostly from reactions not specific to an environment. Second, we analyzed the relationship between a reaction’s superessentiality and how frequently genes encoding known enzymes for this reaction occur in 1,093 prokaryotic genomes; 94.4% (118 of 125) of reactions in the environment-general superessential core occur in more than 50% of prokaryotic genomes, a number much greater than expected by chance alone. In addition, the statistical association between the superessentiality of a reaction and the number of genomes encoding it is much higher than expected by chance alone. Not unexpectedly, this association explains only a modest fraction of the variance in genome occurrence, which reflects our incomplete knowledge about metabolism and cell biology. For example, a highly superessential reaction catalyzed by several nonorthologous enzymes may show a low genome count if genes encoding some of these enzymes have not yet been identified (41–43). Other reasons for high superessentiality and low genome count involve promiscuous enzymes that catalyze more than one reaction but are not known to do so (44, 45) or unknown biochemical pathways that can bypass a reaction (45). Conversely, an enzyme with high genome count and low superessentiality may have important nonmetabolic functions. Examples that we discuss in SI Appendix, SI Results include the glycolytic enzyme enolase (for its role in the RNA degradasome) or thioredoxin reductase (for its indirect but essential role in reducing important cytoplasmic enzymes and regulatory proteins).
We also studied how rich environments and environments known to support the life of pathogens inside a host affect the complement of absolutely superessential reactions. We found that a majority of reactions from a set of 125 absolutely superessential reactions needed for life on 54 carbon sources remained absolutely superessential in these environments. This observation is consistent with our earlier observations that most superessential reactions are environment-general. Moreover, enzyme-coding genes of 114 of 125 absolutely superessential reactions have experimentally been confirmed as essential in at least one of three well-studied organisms, namely S. enterica serovars (37, 55), M. tuberculosis H37Rv (56, 57), and P. aeruginosa (58, 59) (Dataset S6). Furthermore, a significantly larger number of absolutely superessential reactions than expected by chance alone (mean = 83%) is encoded by a majority of sequenced prokaryotic genomes. This finding indicates that these reactions are not just highly superessential, because they support synthesis of biomass molecules highly specific to one organism, such as E. coli. All of the above means that the superessential core is not highly sensitive to the chemical composition of an environment and of biomass.
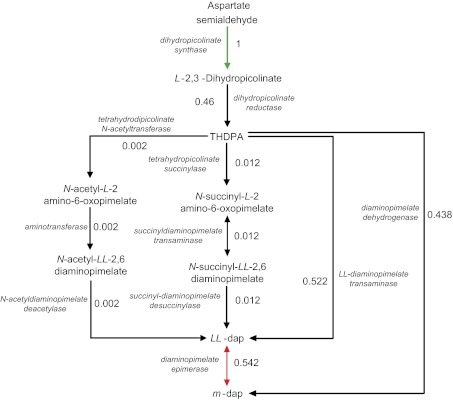
As knowledge about metabolism accumulates, our estimates of reaction superessentiality will become ever more accurate. Already at the present time, the estimates that we obtained can help guide the selection of drugs targeting metabolism. That is, although our observations do not answer the question of how to inhibit a specific enzyme, they can help answer which essential enzymes are the best candidates for inhibition based on how difficult it is to bypass the reactions that these enzymes catalyze. The main idea is that enzymes with reactions that have high superessentiality (large ISE) are good drug target candidates, because cells cannot easily evolve resistance by rerouting metabolism around these enzymes (60). This finding holds even more so for reactions with high superessentiality and high genome count. One example is methionine adenosyltransferase (Blattner number b2942), an absolutely superessential enzyme present in 97% of prokaryotic genomes. Other examples include shikimate kinase (ISE = 1, IGO = 0.86) and chorismate synthase (ISE = 1, IGO = 0.89), enzymes that are already being explored as possible targets (61, 62). As another example, Fig. 3 shows a more detailed analysis involving the diaminopimelate (DAP) pathway. The DAP pathway is responsible for meso-2,6-Diaminopimelate (m-DAP) synthesis. m-DAP is an important precursor to l-lysine and peptidoglycan synthesis in prokaryotes, both of which are not synthesized in humans (this characteristic is important, because the ideal drug targets the pathogen but not the host). DAP epimerase [enzyme commission number (EC) 5.1.1.7], the final enzyme involved in the production of m-DAP, is actively being explored as a drug target (63–66). Although DAP epimerase is essential for growth on all 54 carbon sources in E. coli, it is essential only in 54.2% of random viable networks, because many microbes can synthesize m-DAP through diaminopimelate dehydrogenase (EC 1.4.1.16) (67). In other words, inhibition of this enzyme could be ineffective in the long run, because there is a known route of resistance. A better target in the same pathway would be dihydrodipicolinate synthase (EC 4.2.1.52), which is superessential, environment-general, and present in 92% of prokaryotic genomes. In addition, its product, l-2,3-dihydropicolinate, is not essential in humans, making dihydrodipicolinate synthase an ideal drug target. In sum, the analysis of superessential reactions may be one of several worthy starting points to development of drug targets that block or slow down the evolution of antidrug resistance (68–70).
Fig. 3.
The figure indicates the superessentiality index of each reaction in the m-DAP synthesis pathway. Diaminopimelate epimerase (red), a drug target that is being actively pursued (63–66), has an intermediate superessentiality index of ISE = 0.542 because of an alternate pathway catalyzed by diaminopimelate dehydrogenase that also synthesizes m-DAP. In contrast, dihydropicolinate synthase (green) is absolutely superessential, making it a very attractive drug target from this perspective. Note that the majority of the reactions shown possess alternate metabolic routes signified by their low superessentiality. LL-dap, LL-2,6-diaminopimelate; m-DAP, meso-2,6-diaminopimelate; THDPA, L-2,3,4,5-tetradihydropicolinate.
We next discuss potential caveats to our study. First, as noted above, estimation of reaction superessentiality depends on our knowledge about the universe of all feasible enzyme-catalyzed metabolic reactions. If future work adds reactions and pathways to the known universe, then the superessentiality of individual reactions may decline. However, we expect that the ranking of superessentiality of many reactions will remain similar over time. If so, a reaction with superessentiality that is much higher than the superessentiality of another reaction would still be a better candidate drug target. Second, our comparison of essential reactions with enzyme occurrence in organisms with sequenced genomes depends on the quality of available metabolic genome annotation (28, 36). This annotation currently has numerous limitations, which we discussed. Third, completely unknown spontaneous (not enzyme-catalyzed) reactions could lower the superessentiality of enzyme-catalyzed reactions. They could, thus, contribute to some of the yet unresolved low genome occurrences of absolutely superessential reactions. However, based on our knowledge of known spontaneous reactions, which constitute a very small fraction (1.3%) (SI Appendix) of all known reactions, this effect may be minor. Fourth, there are uncertainties in biomass composition. We carried out our analysis with the biomass composition of a free-living organism in mind, and the majority of biomass molecules that we consider would be found in typical free-living organisms. However, some minor biomass components may be restricted to some organisms and may not be required in others. A potential example is siroheme, a molecule that is part of E. coli biomass but may not be needed in other organisms (46, 52, 71–73). In an organism that does not need this molecule, 1.6% (two reactions) of our absolutely superessential reactions lose this status. However, even when taking some variation in biomass composition into account, the relative order of superessentiality among reactions would be largely preserved. To show this, we used the biomass composition of SEED models, metabolic models of diverse organisms that are created with a semiautomatic procedure from genomic and other information (52). Specifically, we recalculated the superessentiality index (ISE) of reactions for a biomass composition that contains molecules found in a majority of these SEED models. The rank correlation coefficient between superessentiality indices for the E. coli biomass and this biomass composition exceeds 0.8. This finding means that the superessentiality index is not highly sensitive to biomass composition. Fifth, the generation of random viable networks, especially for multiple carbon source phenotypes, is computationally expensive (22, 74). Only limited sample sizes are currently feasible. However, we note that even these limited sample sizes yield results that are in agreement with complementary approaches (for example, the identification of absolutely superessential reactions from the universal network that we analyzed).
In sum, our analysis sheds light on the evolution and genotypic plasticity of metabolic networks. It shows that metabolic networks contain a core of absolutely superessential reactions regardless of their metabolic genotype. The composition of this core is not highly sensitive to the environment in which viability is required. More generally, reactions vary broadly in their superessentiality and thus, in how readily they can be bypassed by alternative pathways. A comprehensive evolutionary approach like ours may help identify putative drug targets and develop effective antibiotic therapies. We hope that our data on reaction superessentiality (Datasets S1, S3, S4, and S5) will become broadly useful resources to other researchers.
Methods
Monte Carlo Markov Chain Random Walk.
We generate random metabolic networks through long random walks that leave a metabolic network’s ability to synthesize biomass unchanged. Each step in a random walk consists of the addition of a randomly chosen reaction from the known universe of biochemical reactions followed by the deletion of another randomly chosen reaction. After each step, we use flux balance analysis to predict the viability of a network in one or more chemical environments. We accept the step if the network remains viable after deletion; otherwise, we reject the step and carry out another step. Sequential addition and deletion of reactions ensure that the size of the network remains constant throughout the random walk. We generated 500 random networks for each phenotype that we consider.
Essential and Absolutely Superessential Reactions.
We define a reaction as essential for a given phenotype if its elimination causes cessation of biomass synthesis. To identify essential reactions in a given network, we eliminate each reaction and use FBA to assess whether nonzero biomass growth flux is still achievable. If not, the reaction is called essential for this network and growth environment. We call a reaction absolutely superessential if it is essential in a given environment in all metabolic networks that we considered.
Methods are described in greater detail in SI Appendix.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 6810 (volume 109, number 18).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113065109/-/DCSupplemental.
References
- 1.Neidhardt FC, Ingraham JL. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 1st Ed. Washington, DC: American Society for Microbiology; 1987. [Google Scholar]
- 2.Almaas E, Oltvai ZN, Barabási AL. The activity reaction core and plasticity of metabolic networks. PLOS Comput Biol. 2005;1:e68. doi: 10.1371/journal.pcbi.0010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segrè D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci USA. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards JS, Palsson BO. Robustness analysis of the Escherichia coli metabolic network. Biotechnol Prog. 2000;16:927–939. doi: 10.1021/bp0000712. [DOI] [PubMed] [Google Scholar]
- 5.Price ND, Papin JA, Palsson BO. Determination of redundancy and systems properties of the metabolic network of Helicobacter pylori using genome-scale extreme pathway analysis. Genome Res. 2002;12:760–769. doi: 10.1101/gr.218002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerdes SY, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pál C, et al. Chance and necessity in the evolution of minimal metabolic networks. Nature. 2006;440:667–670. doi: 10.1038/nature04568. [DOI] [PubMed] [Google Scholar]
- 8.Shlomi T, Berkman O, Ruppin E. Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proc Natl Acad Sci USA. 2005;102:7695–7700. doi: 10.1073/pnas.0406346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilding EI, et al. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J Bacteriol. 2000;182:4319–4327. doi: 10.1128/jb.182.15.4319-4327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhuri RR, et al. Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH) BMC Genomics. 2009;10:291. doi: 10.1186/1471-2164-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feist AM, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Zhang J. Abundant indispensable redundancies in cellular metabolic networks. Genome Biol Evol. 2009;1:23–33. doi: 10.1093/gbe/evp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papp B, Pál C, Hurst LD. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature. 2004;429:661–664. doi: 10.1038/nature02636. [DOI] [PubMed] [Google Scholar]
- 15.Joyce AR, et al. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J Bacteriol. 2006;188:8259–8271. doi: 10.1128/JB.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowell RD, et al. Genotype to phenotype: A complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 18.Gadad AK, Mahajanshetti CS, Nimbalkar S, Raichurkar A. Synthesis and antibacterial activity of some 5-guanylhydrazone/thiocyanato-6-arylimidazo[2,1-b]-1,3, 4-thiadiazole-2-sulfonamide derivatives. Eur J Med Chem. 2000;35:853–857. doi: 10.1016/s0223-5234(00)00166-5. [DOI] [PubMed] [Google Scholar]
- 19.Wiesner J, Borrmann S, Jomaa H. Fosmidomycin for the treatment of malaria. Parasitol Res. 2003;90(Suppl 2):S71–S76. doi: 10.1007/s00436-002-0770-9. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee A, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 21.Timmins GS, Deretic V. Mechanisms of action of isoniazid. Mol Microbiol. 2006;62:1220–1227. doi: 10.1111/j.1365-2958.2006.05467.x. [DOI] [PubMed] [Google Scholar]
- 22.Matias Rodrigues JF, Wagner A. Evolutionary plasticity and innovations in complex metabolic reaction networks. PLoS Comput Biol. 2009;5:e1000613. doi: 10.1371/journal.pcbi.1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samal A, Matias Rodrigues JF, Jost J, Martin OC, Wagner A. Genotype networks in metabolic reaction spaces. BMC Syst Biol. 2010;4:30. doi: 10.1186/1752-0509-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fong SS, Joyce AR, Palsson BØ. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res. 2005;15:1365–1372. doi: 10.1101/gr.3832305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fong SS, Marciniak JY, Palsson BØO. Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model. J Bacteriol. 2003;185:6400–6408. doi: 10.1128/JB.185.21.6400-6408.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibarra RU, Edwards JS, Palsson BO. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- 27.Goto S, Nishioka T, Kanehisa M. LIGAND: Chemical database of enzyme reactions. Nucleic Acids Res. 2000;28:380–382. doi: 10.1093/nar/28.1.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goto S, Okuno Y, Hattori M, Nishioka T, Kanehisa M. LIGAND: Database of chemical compounds and reactions in biological pathways. Nucleic Acids Res. 2002;30:402–404. doi: 10.1093/nar/30.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 30.Mengin-Lecreulx D, van Heijenoort J. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J Biol Chem. 1996;271:32–39. doi: 10.1074/jbc.271.1.32. [DOI] [PubMed] [Google Scholar]
- 31.Kawai S, Mori S, Mukai T, Hashimoto W, Murata K. Molecular characterization of Escherichia coli NAD kinase. Eur J Biochem. 2001;268:4359–4365. doi: 10.1046/j.1432-1327.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 32.Burgard AP, Nikolaev EV, Schilling CH, Maranas CD. Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res. 2004;14:301–312. doi: 10.1101/gr.1926504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maltsev N, Glass EM, Ovchinnikova G, Gu Z. Molecular mechanisms involved in robustness of yeast central metabolism against null mutations. J Biochem. 2005;137:177–187. doi: 10.1093/jb/mvi017. [DOI] [PubMed] [Google Scholar]
- 34.Samal A, et al. Low degree metabolites explain essential reactions and enhance modularity in biological networks. BMC Bioinformatics. 2006;7:118. doi: 10.1186/1471-2105-7-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitkup D, Kharchenko P, Wagner A. Influence of metabolic network structure and function on enzyme evolution. Genome Biol. 2006;7:R39. doi: 10.1186/gb-2006-7-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becker D, et al. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature. 2006;440:303–307. doi: 10.1038/nature04616. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y-M, Rock CO. Thematic review series: Glycerolipids. Acyltransferases in bacterial glycerophospholipid synthesis. J Lipid Res. 2008;49:1867–1874. doi: 10.1194/jlr.R800005-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brand LA, Strauss E. Characterization of a new pantothenate kinase isoform from Helicobacter pylori. J Biol Chem. 2005;280:20185–20188. doi: 10.1074/jbc.C500044200. [DOI] [PubMed] [Google Scholar]
- 40.Leonardi R, et al. A pantothenate kinase from Staphylococcus aureus refractory to feedback regulation by coenzyme A. J Biol Chem. 2005;280:3314–3322. doi: 10.1074/jbc.M411608200. [DOI] [PubMed] [Google Scholar]
- 41.Koonin EV, Mushegian AR, Bork P. Non-orthologous gene displacement. Trends Genet. 1996;12:334–336. [PubMed] [Google Scholar]
- 42.Galperin MY, Walker DR, Koonin EV. Analogous enzymes: Independent inventions in enzyme evolution. Genome Res. 1998;8:779–790. doi: 10.1101/gr.8.8.779. [DOI] [PubMed] [Google Scholar]
- 43.Houten SM, Waterham HR. Nonorthologous gene displacement of phosphomevalonate kinase. Mol Genet Metab. 2001;72:273–276. doi: 10.1006/mgme.2000.3133. [DOI] [PubMed] [Google Scholar]
- 44.Khersonsky O, Tawfik DS. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 45.Kim J, Kershner JP, Novikov Y, Shoemaker RK, Copley SD. Three serendipitous pathways in E. coli can bypass a block in pyridoxal-5′-phosphate synthesis. Mol Syst Biol. 2010;6:436. doi: 10.1038/msb.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raghunathan A, Reed J, Shin S, Palsson B, Daefler S. Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host-pathogen interaction. BMC Syst Biol. 2009;3:38. doi: 10.1186/1752-0509-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang X, Wallqvist A, Reifman J. Development and analysis of an in vivo-compatible metabolic network of Mycobacterium tuberculosis. BMC Syst Biol. 2010;4:160. doi: 10.1186/1752-0509-4-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oberhardt MA, Goldberg JB, Hogardt M, Papin JA. Metabolic network analysis of Pseudomonas aeruginosa during chronic cystic fibrosis lung infection. J Bacteriol. 2010;192:5534–5548. doi: 10.1128/JB.00900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yus E, et al. Impact of genome reduction on bacterial metabolism and its regulation. Science. 2009;326:1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
- 50.Förster J, Famili I, Palsson BO, Nielsen J. Large-scale evaluation of in silico gene deletions in Saccharomyces cerevisiae. OMICS. 2003;7:193–202. doi: 10.1089/153623103322246584. [DOI] [PubMed] [Google Scholar]
- 51.Becker SA, Palsson BØ. Genome-scale reconstruction of the metabolic network in Staphylococcus aureus N315: An initial draft to the two-dimensional annotation. BMC Microbiol. 2005;5:8. doi: 10.1186/1471-2180-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henry CS, et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 53.Matias Rodrigues JF, Wagner A. Genotype networks, innovation, and robustness in sulfur metabolism. BMC Syst Biol. 2011;5:39. doi: 10.1186/1752-0509-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y, et al. Blueprint for antimicrobial hit discovery targeting metabolic networks. Proc Natl Acad Sci USA. 2010;107:1082–1087. doi: 10.1073/pnas.0909181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knuth K, Niesalla H, Hueck CJ, Fuchs TM. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol Microbiol. 2004;51:1729–1744. doi: 10.1046/j.1365-2958.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- 56.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 58.Jacobs MA, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liberati NT, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Motter AE, Gulbahce N, Almaas E, Barabási AL. Predicting synthetic rescues in metabolic networks. Mol Syst Biol. 2008;4:168. doi: 10.1038/msb.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arora N, Banerjee AK, Murty US. In silico characterization of Shikimate Kinase of Shigella flexneri: A potential drug target. Interdiscip Sci. 2010;2:280–290. doi: 10.1007/s12539-010-0012-2. [DOI] [PubMed] [Google Scholar]
- 62.Dias MV, et al. Chorismate synthase: An attractive target for drug development against orphan diseases. Curr Drug Targets. 2007;8:437–444. doi: 10.2174/138945007780058924. [DOI] [PubMed] [Google Scholar]
- 63.Pillai B, et al. Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target. Proc Natl Acad Sci USA. 2006;103:8668–8673. doi: 10.1073/pnas.0602537103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pillai B, et al. Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase. Biochem Biophys Res Commun. 2007;363:547–553. doi: 10.1016/j.bbrc.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 65.Usha V, Dover LG, Roper DI, Fütterer K, Besra GS. Structure of the diaminopimelate epimerase DapF from Mycobacterium tuberculosis. Acta Crystallogr D Biol Crystallogr. 2009;65:383–387. doi: 10.1107/S0907444909002522. [DOI] [PubMed] [Google Scholar]
- 66.Brunetti L, Galeazzi R, Orena M, Bottoni A. Catalytic mechanism of l,l-diaminopimelic acid with diaminopimelate epimerase by molecular docking simulations. J Mol Graph Model. 2008;26:1082–1090. doi: 10.1016/j.jmgm.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 67.Velasco AM, Leguina JI, Lazcano A. Molecular evolution of the lysine biosynthetic pathways. J Mol Evol. 2002;55:445–459. doi: 10.1007/s00239-002-2340-2. [DOI] [PubMed] [Google Scholar]
- 68.Chan JNY, Nislow C, Emili A. Recent advances and method development for drug target identification. Trends Pharmacol Sci. 2010;31:82–88. doi: 10.1016/j.tips.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 69.Gant TW, Zhang S-D, Taylor EL. Novel genomic methods for drug discovery and mechanism-based toxicological assessment. Curr Opin Drug Discov Devel. 2009;12:72–80. [PubMed] [Google Scholar]
- 70.Sioud M. Main approaches to target discovery and validation. Methods Mol Biol. 2007;360:1–12. doi: 10.1385/1-59745-165-7:1. [DOI] [PubMed] [Google Scholar]
- 71.Oh Y-K, Palsson BO, Park SM, Schilling CH, Mahadevan R. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J Biol Chem. 2007;282:28791–28799. doi: 10.1074/jbc.M703759200. [DOI] [PubMed] [Google Scholar]
- 72.Oberhardt MA, Puchałka J, Fryer KE, Martins dos Santos VAP, Papin JA. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J Bacteriol. 2008;190:2790–2803. doi: 10.1128/JB.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jamshidi N, Palsson BØ. Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst Biol. 2007;1:26. doi: 10.1186/1752-0509-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Price ND, Reed JL, Palsson BØ. Genome-scale models of microbial cells: Evaluating the consequences of constraints. Nat Rev Microbiol. 2004;2:886–897. doi: 10.1038/nrmicro1023. [DOI] [PubMed] [Google Scholar]