Abstract
Transforming growth factor-β (TGF-β) functions as a tumor suppressor of the prostate through mechanisms that remain unresolved. Although TGF-β receptors directly activate both Smads 2 and 3, to date Smad3 has been shown to be the essential mediator of most Smad-dependent TGF-β responses, including control of gene expression, cell growth, apoptosis and tumor suppression. Using a robust lentiviral short hairpin RNA (shRNA) system to silence Smads 2 and/or 3 in the NRP-152 non-tumorigenic rat prostate basal epithelial cell line, we provide the first evidence for Smad2 as a critical mediator of TGF-β-induced apoptosis and gene expression. Parallel analyses revealed that Smad3 is the major mediator of TGF-β-induced transcriptional and apoptotic responses in the NRP-154 rat prostate carcinoma cell line. Remarkably, silencing Smad2 alone caused malignant transformation of NRP-152 cells, as assayed by s.c. tumor growth in athymic mice; whereas silencing of Smad3 alone did not induce tumors. Nevertheless, tumors induced by silencing both Smads 2 and 3 were larger than those from silencing Smad2 alone. Given previous reports that NRP-152 cells have stem cell phenotype, we speculate a critical role for Smad2 as a tumor suppressor in the basal epithelial or stem cell compartment of the prostate.
Keywords: prostate cancer, Smad, lentivirus, shRNA, nude mice
Introduction
TGF-β functions as a candidate tumor suppressor of the prostate, likely through its robust cytostatic effects (1). TGF-β signals through engaging TGF-β receptors type II (TβRII) and type I (TβRI) to form a tetrameric complex, leading to activation of TβRI. TβRI in turn phosphorylates Smad2 and Smad3 causing their homo-oligomerization and oligomerization with Smad4, which together translocate to the nucleus to regulate transcription (2). TβRI can also directly activate other intracellular signaling effectors (3).
Studies reported so far have portrayed a critical role of Smad3, but not of Smad2, in TGF-β-induce apoptosis and transcriptional control. First, Smad3 (and also Smad4) binds to Smad-binding elements (SBEs), whereas the common splice form of Smad2 does not bind DNA (2). Second, microarray analysis of Smad2 or Smad3 knockout (KO) mouse embryonic fibroblasts showed that Smad3 was essential for the regulation of immediate-early target genes of TGF-β, whereas Smad2 only transmodulated Smad3-dependent transcription (4). Finally, Smad3, but not Smad2, was shown to be required for the induction of apoptosis/growth inhibition elicited by TGF-β in various keratinocytes and epithelial cells (5).
Despite the crucial roles of Smad3 in TGF-β signaling, Smad3 mutations are detected at extremely low frequency in cancer (6), while mutations in and/or protein loss of Smad2 have been reported to be far more common (7). Although several earlier publications found that Smad2 loss/mutation occurs rarely in prostate cancer (8, 9), a recent report found that phospho-Smad2 levels are significantly reduced in prostate tumors (10), suggesting that Smad2 is inactivated during prostate carcinogenesis. Moreover, although Smad3 KO mice are viable, Smad2 KO mouse embryos are not viable, indicating an indispensable role for Smad2 in development (5). Overall, these findings implicate important roles for Smad2 in tissue homeostasis during development and tumorigenesis.
Danielpour and colleagues have previously derived two spontaneously-immortalized cell lines (NRP-152 and NRP-154) from the preneoplastic prostate of the Lobund-Wistar rat (11), and both cell lines are exquisitely sensitive to TGF-β-induced apoptosis (12). NRP-154 has a luminal epithelial phenotype and forms tumors when implanted s.c. in athymic mice, whereas NRP-152 is a non-tumorigenic basal epithelial cell line that undergoes malignant transformation following suppression of TGF-β signaling by retroviral transduction of dominant-negative (DN) TβRII (11, 13). Interestingly, NRP-152 has stem cell-like properties, and possesses the unique ability to form morphologically normal prostate ductal structures under the renal capsule of athymic mice when recombined with normal urogenital sinus mesenchyme (14, 15).
In the current study we explored the roles of endogenous Smad2 and Smad3 in TGF-β-mediated apoptosis, gene expression and tumor suppression of NRP-152 cells, and compared those responses in NRP-154 cells. Here we report the first evidence that Smad2 can function as a tumor suppressor and critical mediator of TGF-β responses in prostate epithelial cells.
Materials and Methods
Reagents
Sources were: α-Caspase-3 active [AF-835], recombinant human TGF-β1 (R&D systems, Minneapolis, MN); α-β-actin (Sigma, St. Louis, MO); α-Smad3 [sc-8332],α-Smad4 [sc-7966] (Santa Cruz, Santa Cruz, CA); α-Smad2 and Matrigel (BD Biosciences, San Jose, CA).
Cell Culture
NRP-152, NRP-154 cell lines were maintained in GM2.1 medium, and treated with TGF-β1 in GM3 medium as before (16).
Plasmids
3TP-Lux and SBE4-Lux were generous gifts from Dr. Joan Massague and Dr. Bert Vogelstein, respectively. AP-1-Lux, NF-κB-Lux minimal response elements are cis-acting PathDetect™ constructs from Stratagene (La Jolla, CA). Plasmids involved in lentivirus production and Smad silencing were described earlier (16).
Lentivirus-mediated Stable Gene Silencing
The production of shRNA-expression lentiviruses were previously described (16). For transduction and establishment of stable gene silencing, multiplicity of infection=50 of lentiviruses harboring shRNA-LacZ, shRNA-Smad2 and/or shRNA-Smad3 were used for NRP-152 cells (to derive NRP-152-shRNA cells). NRP-154-tTR-shRNA cells were derived as described previously (16).
Western Blot
Cell lysates were prepared and analyzed by Western blot as described previously (16).
Luciferase Reporter Assay
Cells were transfected, treated, and assayed similarly as before (16). For each well, 0.2 μg of individual luciferase reporter vector, 20 ng of CMV-renilla, and 0.8 μg of pcDNA3 were used.
Cell number assay
Control or Smad-ablated NRP-152 and NRP-154 cells, after various treatments, were washed with PBS and detached by Trypsin-EDTA solution. Cell number was enumerated using a Coulter Counter.
Flow Cytometry
For cell cycle analysis, cells (1.5 × 106) were detached from plates, washed once in PBS, and then fixed in 90% methanol. Following treatment with 0.1 mg/ml of RNase A, cells were incubated with 50 μg/ml of propidium iodide and then analyzed with an EPICS-XL MCL flow cytometer. Sub-G1 cells, which have less than 2n DNA content, are considered apoptotic.
In Vivo Tumorigenicity Studies
Six- to seven-week-old intact male athymic nude (NCr:NU) mice were injected s.c. on opposing flanks with 0.1 ml of cell suspension (2 × 106 cells) in DMEM/F12:Matrigel (1:1, v/v). Tumor volume (length × width2 × 0.5) was assessed weekly with the use of fine calipers. Excised tumors were fixed in formalin, embedded in paraffin and H&E stained.
Results and Discussion
Lentivirus-mediated Smad silencing
We used NRP-152 (basal) and NRP-154 (luminal) rat prostate epithelial cell lines to define the role of Smads 2 and 3 in TGF-β signaling in prostate epithelial lines derived from the pre-neoplastic prostate of Lobund-Wistar rats (11). Both cells lines are exquisitely sensitive to growth suppression and apoptosis by TGF-β (12), the NRP-154 cell line forms differentiated tumors when implanted s.c. in athymic mice (11) and the NRP-152 non-tumorigenic cell line undergoes malignant transformation upon disruption of TGF-β signaling by retroviral transduction of DN-TβRII (13). We investigated the roles of Smads 2 and 3 in mediating key TGF-β responses associated with apoptosis and gene expression in both NRP-152 and NRP-154 cells and tumor suppression in NRP-152 cells by stably silencing Smads 2 and/or 3 using a lentivirus-mediated silencing strategy involving stable expression of small hairpin (sh)-Smad2 and sh-Smad3 (Fig. 1A), as described before (17). This strategy enabled us to achieve >95% knockdown of Smad2 and Smad3 in both NRP-152 (Fig. 1A) and NRP-154 cells (16).
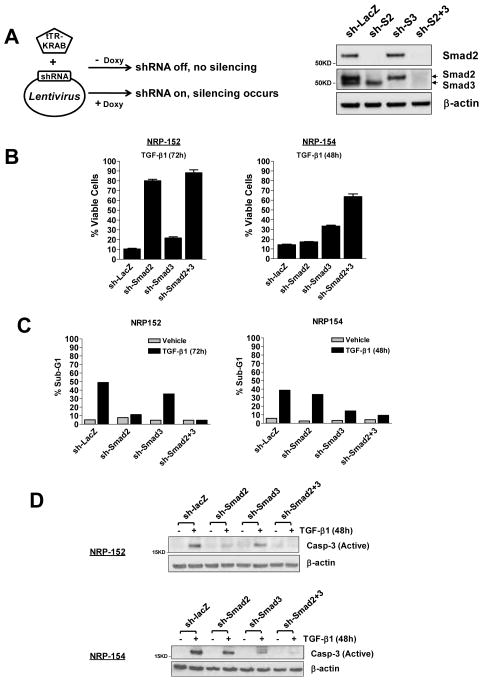
Fig. 1. Smad2 is essential for TGF-β-induced apoptosis of NRP-152 basal epithelial cells, while Smad3 is the major mediator of TGF-β-induced apoptosis in NRP-154 luminal epithelial cells.
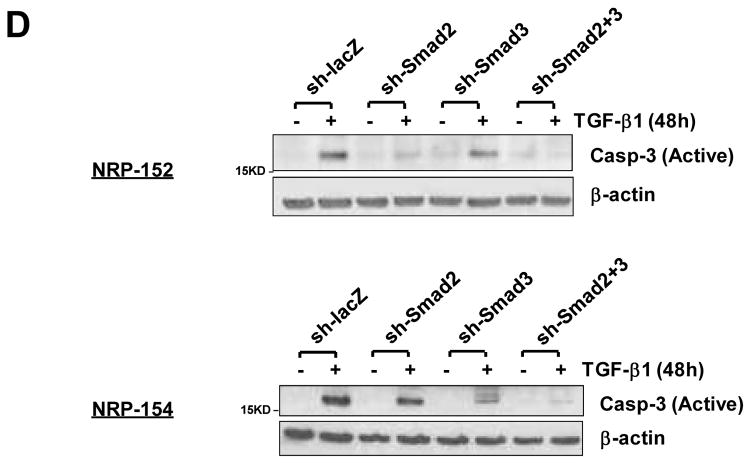
(A) Smads 2 and 3 were effectively silenced individually or in combination, as exemplified at the protein levels in NRP-152 cells. Note that Smad3 antibody recognizes both Smad2 and Smad3, as indicated by the arrows. (B) NRP-152-shRNA cells or NRP-154-tTR-shRNA cells incubated with Doxy were treated with 10 ng/ml TGF-β1 or vehicle for 72h or 48h respectively, and the attached cells were counted using a Coulter counter. % Viable cells represents the percentage of viable cells after TGF-β treatment, and was calculated from three independent experiments. (C, D) Cells were plated and treated as in (B). Apoptosis was measured using FACS analysis of sub-G1 population (C, indicated as % Sub-G1 and plotted in a bar graph format), or an antibody detecting the active form of caspase-3 (D).
The relative dependence on Smad2 and Smad3 for apoptotic responses initiated by TGF-β
We first explored whether silencing either or both of the TGF-β receptor Smads would inhibit TGF-β-induced apoptosis in NRP-152 and NRP-154 cells. NRP-152- and NRP-154-shRNA cells were treated with vehicle or TGF-β for 72 h or 48 h, respectively. The percentage of viable cells following TGF-β treatment was determined in the above silenced cells. We observed that sh-Smad2 substantially blocked TGF-β-induced death of NRP-152 cells, in contrast to a minimal effect on viability of NRP-154 cells (Fig. 1B). In the latter cell line, sh-Smad3 significantly reduced cell death induced by TGF-β, while Smad2 silencing was effective in suppressing such cell death only when co-silenced with Smad3 (Fig. 1B). Similar patterns were observed with other indicators of apoptosis such as sub-G1 population (Fig. 1C) and caspase-3 activation (Fig. 1D). The basis for such differential dependence on R-Smad for TGF-β-mediated apoptosis in these two cell lines, according to our preliminary data, could be related to their distinct differentiation states. NRP-154 cells are tumorigenic luminal epithelial cells, and the observation that Smad3 is their major apoptosis mediator for TGF-β is reminiscent of previous studies in keratinocytes and several cancer cell lines (5). In addition, a report by Kim et al. revealed opposite effects of Smad2 and Smad3 in TGF-β’s cytostatic responses in an array of tumor cells (18). Instead, we observed a cooperative effect of these R-Smads in both NRP-154 and NRP-152 cells (Fig. 1). More interestingly, silencing Smad2 alone in NRP-152 cells, which have stem cell-like properties (15), robustly inhibited apoptosis by TGF-β (Fig. 1). This finding by itself is quite novel, and could represent a more general mechanism used by TGF-β to induce apoptosis in undifferentiated, stem cell-like, pluripotent or basal cells.
Smad2 is critical to transcriptional activation by TGF-β in NRP-152 cells
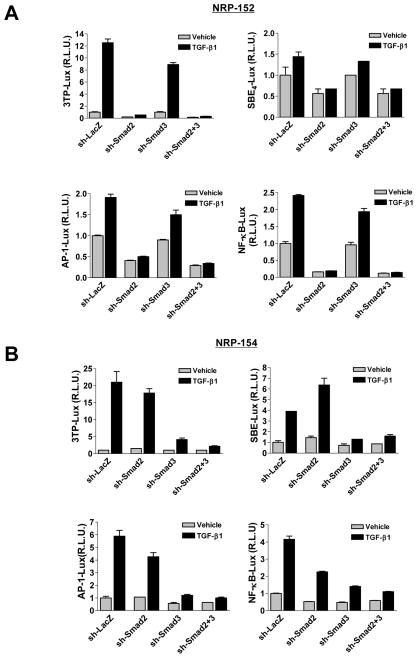
We next examined whether Smad2 was also required for transcriptional responses by TGF-β in NRP-152 cells. To this end, we used the PAI promoter construct (3TP-Lux) and several basic response-element reporters (SBE4-luciferase, AP-1-luciferase, and NF-κB-luciferase). Interestingly, silencing Smad2 abrogated most of the TGF-β-induced promoter activities in NRP-152 cells (Fig. 2A), unlike NRP-154 cells in which silencing Smad3 alone suppressed most of those responses (Fig. 2B). In NRP-152 cells, silencing Smad2 alone substantially suppressed the basal activity of all four promoters, consistent with our previous report that NRP-152 cells produce active autocrine TGF-β (15). Collectively, our data indicate that Smad2 in NRP-152 cells and Smad3 in NRP-154 cells are the major mediators of transcriptional responses byTGF-β. The mechanism by which Smad2 or Smad3 mediate TGF-β-induced transcriptional activation of the AP-1 and NF-κB response elements is likely to be indirect as they lack SBEs.
Fig. 2. The dependence on Smad2 and Smad3 for TGF-β-mediated transcriptional responses.
(A and B) NRP-152-shRNA (A) or NRP-154- tTR-shRNA (B) cells were plated in GM2.1 medium or GM2.1 medium + Doxy (0.1 μg/ml) respectively, kept for two days, and then transfected with various luciferease reporter constructs (3TP-Lux, SBE4-Lux, AP-1-Lux, or NF-κB-Lux) and CMV-driven renilla vector. TGF-β1 (10 ng/ml) or vehicle was added 24 h after transfection, and cells were incubated for an additional 24 h before assay. Firefly luciferase values were normalized to renilla luciferase, and the value of the first bar in every graph is arbitrarily set as 1. R.L.U., relative luciferase unit.
Smad2 functions as a tumor suppressor in NRP-152 cells
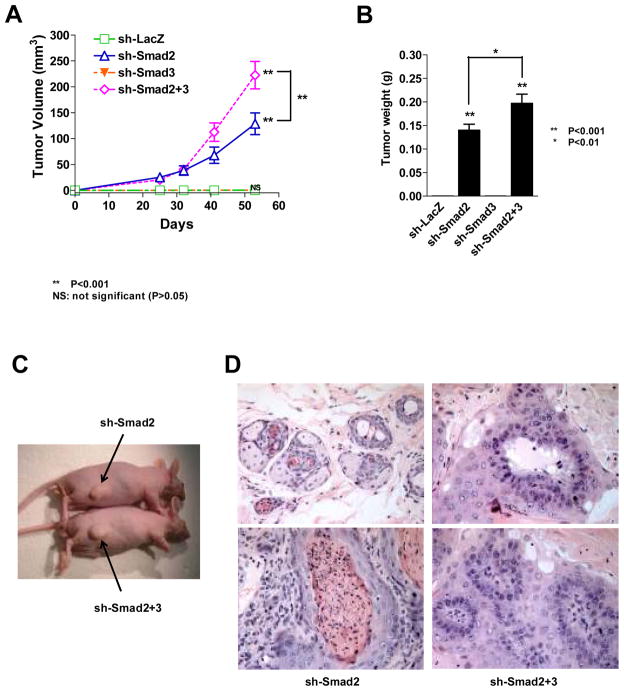
Previous work demonstrated that retroviral transduction of DN-TβRII, which blocks TGF-β-induced cell death, caused malignant transformation of NPR-152 prostate basal epithelial cells (13). Since Smad2 is critical for TGF-β-mediated cellular responses in NRP-152 cells, and TGF-β functions as a tumor suppressor of those cells (13), we hypothesized that Smad2 is a tumor suppressor in NRP-152 cells. To test this hypothesis, NRP-152-sh-LacZ, -sh-Smad2, -sh-Smad3, or -sh-Smad2+3 cell pools were individually inoculated s.c. above both hind flanks of male athymic mice, using 10 mice/group. Mice injected with sh-Smad2 or sh-Smad2+3 cells formed palpable lesions by 4 weeks, whereas no lesions were detectable even up to 4 months in mice receiving the sh-LacZ or sh-Smad3 cells. By Day 41, 15 of 18 injection sites (83%) in sh-Smad2 group developed tumors (Table 1). Although silencing Smad3 alone did not cause tumors, it enhanced tumor incidence to 94% and significantly increased tumor size when concomitantly silenced with Smad2 (Table 1, Fig. 3A). By Day 53 both sh-Smad2 and sh-Smad2+3 groups progressed to 100% tumor incidence (Table 1) and had larger tumor burden (Fig. 3C). The kinetics of tumor volume (Fig. 3A) as well as the tumor weight (Fig. 3B) demonstrated rapid growth of both sh-Smad2 and sh-Smad2+3 tumors, with greater growth rate in the latter group (P<0.001, Fig. 3A). These results support that Smad2 but not Smad3 functions as a tumor suppressor in NRP-152 cells, although loss of Smad3 enhanced tumor growth and incidence in the Smad2 deficient cells.
Table 1. Incidence of tumor formation by NRP-152-shRNA cells in nude mice.
Injection of individual NRP-152 cell derivatives into nude male mice was described in the Materials and Methods. Tumor incidence was represented by the number of palpable lesions (>2×2 mm2 in area) out of total number of injection sites at Day 41 (D41) and Day 53 (D53) after injection, and the percentages were indicated in the parentheses.
| Cell type injected | Tumor incidence (D41) | Tumor incidence (D53) |
|---|---|---|
| NRP-152-sh-LacZ | 0/20 (0%) | 0/20 (0%) |
| NRP-152-sh-Smad2 | 15/18 (83%) | 18/18 (100%) |
| NRP-152-sh-Smad3 | 0/20 (0%) | 0/20 (0%) |
| NRP-152-sh-Smad2+3 | 16/17 (94%) | 17/17 (100%) |
Fig. 3. Loss of Smad2 in prostate basal epithelial cells promotes tumorigenicity in nude mice.
(A) Kinetics of tumor growth of individual NRP-152-shRNA cell pools injected s.c. into male nude mice. Tumor size was measure and tumor volume was calculated as described in Materials and Methods. Statistical analysis comparing tumor growth of sh-Smad2, sh-Smad3, sh-Smad2+3 relative to the control (sh-LacZ) group at Day 53 is indicated at the end of the three curves. In addition, the difference in tumor growth between sh-Smad2 and sh-Smad2+3 groups was also analyzed statistically. Note that sh-Smad3 tumor growth curve overlaps with that of the control. (B) Tumor weight was measured at Day 53, when mice were sacrificed. The statistical significance of tumor weight relative to the control is indicated on top of the bars, and the difference between sh-Smad2 and sh-Smad2+3 tumor weights is also indicated. **P<0.001, *P<0.01; NS: not significant (P>0.05). (C) Representative mice bearing tumor from sh-Smad2 or sh-Smad2+3 xenografts at Day 53. (D) Histological features of representative tumors from sh-Smad2 (left) and sh-Smad2+3 (right) groups. The pictures are of 40x magnification. P-value is calculated using ONE-WAY ANOVA method.
Histologically, tumors in sh-Smad2 and sh-Smad2+3 groups had similar features, containing cells with large pleiomorphic nuclei and prominent nucleoli (Fig. 3D). Clusters of adenocarcinoma cells were frequently observed, often with necrotic centers, and there were significant areas of squamous carcinoma (Fig. 3D). Overall, the sh-Smad2 +3 tumors were less desmoplastic and more glandular than sh-Smad2 tumors. In general, tumors in both groups have features typical of those generated from NRP-152 cells expressing DN-TβRII (13)
The above results suggest that Smad2 functions as a tumor suppressor of prostate epithelial cells. To our knowledge, this is the first report demonstrating critical roles of Smad2 in various TGF-β responses including apoptosis, and the first report that directly links loss of Smad2 to tumorigenesis. A previous study using a Smad3 C-terminal deletion construct, which reduces the phosphorylation of both Smad2 and Smad3, speculated a role for both of these Smads in tumor suppression of breast epithelial cells (19). Another report suggesting a tumor suppressive function of Smad2 was shown in a conditional KO mouse model in which KO of Smad2 enhanced the onset, incidence and stage of chemically-induced skin tumors (20). It is likely that other reports overlooked the relative importance of Smad2 in the cytostatic responses of TGF-β owing to limitations in the types of cell lines used (i.e., tumorigenic, differentiated cells).
The derivation of NRP-152 cells from a pre-neoplastic prostate, their non-tumorigenic phenotype (11), their stem cell-like features (15) and their ability to reconstitute a functional prostate epithelium in vivo (14) make them an ideal model to study early stages of tumorigenesis. Given these properties, we speculate that Smad2 is a critical part of the tumor suppressor pathway of TGF-β in the prostate, and this function of Smad2 may be particularly important in the basal or stem cell compartment of this gland. Consistent with the refractory nature of silencing Smad3 in NRP-152 cells, total Smad3 and TGF-β-induced phospho-Smad3 levels in this line are substantially less than in NRP-154 cells, while those of Smad2 are similar in both lines (supplementary Fig. 1A). Interestingly, Smad3 is selectively induced in NRP-152 cells undergoing transdifferentiation towards a luminal phenotype (Supplementary Fig. 1B, C), and we observed relatively higher Smad2 to Smad3 ratios in basal versus luminal rat prostate epithelial cell lines (Supplementary Fig. 1B). We thus speculate that relative higher Smad2 to Smad3 ratio could be a general property of prostate basal versus luminal epithelial cells.
The possibility that Smad2 has a selective role in stem cell function is consistent with the fact that knockout of Smad2 but not Smad3 causes embryonic death (5), and with the appearance of many more inactivating mutations in Smad2 versus in Smad3 in a variety of tumors (7). We thus speculate that the cytostatic or tumor suppressor function of Smad2 may be lost during differentiation of normal tissues or during prostatic carcinogenesis. If so, identifying the molecular basis for such loss/inactivation and finding ways to restore the cytostatic function of Smad2 holds great promise for the therapeutic intervention of prostate cancer.
Supplementary Material
Acknowledgments
We thank Earl Lawrence for help with pathohistology. This research was supported by NCI grants R01CA092102 and R01CA102074 (to D. Danielpour) and the Flow Cytometry, Athymic Mouse, and Histology Core Facilities of the Case Comprehensive Cancer Center (P30 CA43703). Reema Wahdan was supported bypredoctoral fellowship from Case Comprehensive Cancer Center Research Oncology Training grant 5T32CA059366-15.
References
- 1.Danielpour D. Functions and regulation of transforming growth factor-beta (TGF-beta) in the prostate. Eur J Cancer. 2005;41:846–57. doi: 10.1016/j.ejca.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 3.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 4.Yang YC, Piek E, Zavadil J, et al. Hierarchical model of gene regulation by transforming growth factor beta. Proc Natl Acad Sci U S A. 2003;100:10269–74. doi: 10.1073/pnas.1834070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown KA, Pietenpol JA, Moses HL. A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling. J Cell Biochem. 2007;101:9–33. doi: 10.1002/jcb.21255. [DOI] [PubMed] [Google Scholar]
- 6.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 7.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17:41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Latil A, Pesche S, Valeri A, Fournier G, Cussenot O, Lidereau R. Expression and mutational analysis of the MADR2/Smad2 gene in human prostate cancer. Prostate. 1999;40:225–31. doi: 10.1002/(sici)1097-0045(19990901)40:4<225::aid-pros3>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 9.Yin Z, Babaian RJ, Troncoso P, et al. Limiting the location of putative human prostate cancer tumor suppressor genes on chromosome 18q. Oncogene. 2001;20:2273–80. doi: 10.1038/sj.onc.1204310. [DOI] [PubMed] [Google Scholar]
- 10.Perttu MC, Martikainen PM, Huhtala HS, et al. Altered levels of Smad2 and Smad4 are associated with human prostate carcinogenesis. Prostate Cancer Prostatic Dis. 2006;9:185–9. doi: 10.1038/sj.pcan.4500871. [DOI] [PubMed] [Google Scholar]
- 11.Danielpour D, Kadomatsu K, Anzano MA, Smith JM, Sporn MB. Development and characterization of nontumorigenic and tumorigenic epithelial cell lines from rat dorsal-lateral prostate. Cancer Res. 1994;54:3413–21. [PubMed] [Google Scholar]
- 12.Hsing AY, Kadomatsu K, Bonham MJ, Danielpour D. Regulation of apoptosis induced by transforming growth factor-beta1 in nontumorigenic rat prostatic epithelial cell lines. Cancer Res. 1996;56:5146–9. [PubMed] [Google Scholar]
- 13.Tang B, de Castro K, Barnes HE, et al. Loss of responsiveness to transforming growth factor beta induces malignant transformation of nontumorigenic rat prostate epithelial cells. Cancer Res. 1999;59:4834–42. [PubMed] [Google Scholar]
- 14.Hayward SW, Haughney PC, Lopes ES, Danielpour D, Cunha GR. The rat prostatic epithelial cell line NRP-152 can differentiate in vivo in response to its stromal environment. Prostate. 1999;39:205–12. doi: 10.1002/(sici)1097-0045(19990515)39:3<205::aid-pros9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 15.Danielpour D. Transdifferentiation of NRP-152 rat prostatic basal epithelial cells toward a luminal phenotype: regulation by glucocorticoid, insulin-like growth factor-I and transforming growth factor-beta. J Cell Sci. 1999;112 (Pt 2):169–79. doi: 10.1242/jcs.112.2.169. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, Song K, Krebs TL, Jackson MW, Danielpour D. Rb/E2F4 and Smad2/3 link survivin to TGF-beta-induced apoptosis and tumor progression. Oncogene. 2008;27:5326–38. doi: 10.1038/onc.2008.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–61. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SG, Kim HA, Jong HS, et al. The endogenous ratio of Smad2 and Smad3 influences the cytostatic function of Smad3. Mol Biol Cell. 2005;16:4672–83. doi: 10.1091/mbc.E05-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian F, DaCosta Byfield S, Parks WT, et al. Reduction in Smad2/3 signaling enhances tumorigenesis but suppresses metastasis of breast cancer cell lines. Cancer Res. 2003;63:8284–92. [PubMed] [Google Scholar]
- 20.Hoot KE, Lighthall J, Han G, et al. Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J Clin Invest. 2008;118:2722–32. doi: 10.1172/JCI33713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.