Abstract
Platinum-based DNA metallointercalators are structurally different from the covalent DNA binders such as cisplatin and its derivatives but have potent in vitro activity in cancer cell lines. However, limited understanding of their molecular mechanisms of cytotoxic action greatly hinders their further development as anticancer agents. In this study, a lead platinum-based metallointercalator, [(5,6-dimethyl-1,10-phenanthroline) (1S,2S-diaminocyclohexane)platinum(II)]2+ (56MESS) was found to be 163-fold more active than cisplatin in a cisplatin-resistant cancer cell line. By using transcriptomics in a eukaryotic model organism, yeast Saccharomyces cerevisiae, we identified 93 genes that changed their expressions significantly upon exposure of 56MESS in comparison to untreated controls (p ≤ 0.05). Bioinformatic analysis of these genes demonstrated that iron and copper metabolism, sulfur-containing amino acids and stress response were involved in the cytotoxicity of 56MESS. Follow-up experiments showed that the iron and copper concentrations were much lower in 56MESS-treated cells compared to controls as measured by inductively coupled plasma optical emission spectrometry. Deletion mutants of the key genes in the iron and copper metabolism pathway and glutathione synthesis were sensitive to 56MESS. Taken together, the study demonstrated that the cytotoxic action of 56MESS is mediated by its ability to disrupt iron and copper metabolism, suppress the biosynthesis of sulfur-containing amino acids and attenuate cellular defence capacity. As these mechanisms are in clear contrast to the DNA binding mechanism for cisplatin and its derivative, 56MESS may be able to overcome cisplatin-resistant cancers. These findings have provided basis to further develop the platinum-based metallointercalators as anticancer agents.
Electronic supplementary material
The online version of this article (doi:10.1007/s12154-011-0070-x) contains supplementary material, which is available to authorized users.
Keywords: 56MESS, Platinum, Metallointercalators, Gene, Iron, Anticancer
Introduction
Platinum-based anticancer drugs, cisplatin, carboplatin and oxaliplatin, have been used world-wide to treat various cancers including testicular, ovarian and colorectal tumours in clinical settings [16, 58]. The anticancer action of these platinum drugs results from their capacity to form DNA–platinum covalent adducts, which ultimately lead to apoptosis of cancer cells [76]. However, tumour resistance and side effects such as nephrotoxicity are often consequences of their clinical applications [5, 6, 37, 66]. The development of anticancer drugs with different mechanisms of action is urgently needed to overcome drug resistance and side effects in cancer patients [40]. The mechanisms of action of anticancer drugs ultimately depend on their chemical structures and properties. Recently, platinum drugs with fundamentally different structures and binding modes to that of cisplatin have emerged [2, 40]. Multi-nuclear platinum complexes, some of which are positively charged, capable of forming flexible and non-directional interstrand adducts with DNA have shown cytotoxic activity in tumour cells [40, 52, 78].
A class of square–planar platinum(II) metallointercalators has been developed of the type [Pt(IL)(AL)]2+, where IL can be 1,10-phenanthroline (phen) or substituted 1,10-phenanthroline and AL is achiral 1,2-diaminoethance (en) or chiral 1,2-diaminocyclohexane or (1S,3R)-1,3-diamino-1,2,2-trimethylcyclopentane [8, 35, 53]. They showed activity to a variety of cancer cell lines such as human colon cancer cell line (HCT8) and human lung cancer cell line (A-427) [45, 55]. Correlation analysis of their structures with cytotoxicity revealed that methylation of 1,10-phenanthroline, the types of ancillary ligand, chirality and the bulkiness of ancillary ligands all influence the anticancer activity [8, 42, 45]. To date, [Pt(IL)(S,S-dach)]2+ complexes have been more active than the R,R forms [18, 42]. The complex [(5,6-dimethyl-1,10-phenanthroline)(1S,2S-diaminocyclohexane)platinum(II)]2+ (56MESS) is more active than the other complexes so far tested [45, 55, 79]. 56MESS is also shown to have more potent anticancer activity than cisplatin in a range of cisplatin-sensitive cancer cell lines (Table 1). 56MESS emerges as a potent complex in this class of square–planar platinum(II) metallointercalators of the type [Pt(IL)(AL)]2+.
Table 1.
IC50 values of 56MESS and cisplatin in nine cisplatin-sensitive human cancer cell lines (averages of at least three independent experiments ± SD)
| Cancer cell line | 56MESS | Cisplatin |
|---|---|---|
| 5637 (human bladder cancer) | 82 ± 29 | 350 ± 100 |
| RT-4 (urinary bladder transitional cell) | 59 ± 10 | 1,610 ± 161 |
| LCLC-103H (human lung cancer) | 46 ± 13 | 900 ± 190 |
| DAN-G (human pancreatic cancer) | 40 ± 3 | 730 ± 340 |
| MCF-7 (human breast cancer) | 28 ± 4 | 1,380 ± 290 |
| A427 (human lung cancer) | 21 ± 12 | 1,969 ± 540 |
| IGROV1(human ovarian cancer) | 3.4 ± 1 | 10.6 ± 1.1 |
| HCT8 (human colon cancer) | 2.5 ± 0.4 | 23.1 ± 2.5 |
| HT29 (human colon cancer) | 1.1 ± 0.1 | 2.8 ± 0.7 |
Identifying the molecular target and mechanisms of action of 56MESS thus becomes critical to the further development of this complex and its analogues as anticancer agents. The molecular mechanisms of action of the [Pt(IL)(AL)]2+ complexes in eukaryotic cells are not understood although a complex of this type had been tested in mice [19]. This study therefore seeks to develop and understand the cytotoxic effect of 56MESS on eukaryotes at gene expression level by the means of transcriptomics. Transcriptomics, a platform which can reveal changes of gene expression at a genome-wide scale, is a powerful holistic approach to profile gene expression for understanding molecular pathways of preclinical and clinical anticancer drugs [7, 23, 25]. The model eukaryotic organism, yeast Saccharomyces cerevisiae, plays a significant role in this approach [68] due to its easy amendability with various experimental approaches and conservation of genes and essential cellular functions with humans [3]. Application of yeast transcriptomics has already yielded insights on the genes and molecular mechanisms of cisplatin [9, 23]. We therefore employed yeast transcriptomics in this study to uncover the genes and molecular pathways underpinning the cytotoxicity of 56MESS following the assessment of 56MESS activity in a cisplatin-resistant cancer cell line L1210cisR. The results from gene expression profiling were then corroborated with inductively coupled plasma optical emission spectrometer (ICP-OES) and yeast deletion mutant screening.
Materials and methods
Cancer cell line and yeast strains
The cisplatin-resistant mouse leukaemia cancer cell line L1210cisR was kindly provided by Dr. C. Cullinane from the Peter MacCallum Cancer Institute, Melbourne, Australia. The cells were grown in complete RPMI 1640 medium (Sigma-Aldrich, Australia), supplemented with 5% fetal bovine serum (Sigma-Aldrich, Australia) without antibiotics in a humidified cabinet with 5% CO2 at 37 °C. Yeast wild-type strain BY4743 and deletion mutants used in this study were described in Table 2. Yeast was grown in synthetic medium containing D-glucose (2% wt/vol), yeast nitrogen base (0.17% wt/vol) and ammonia sulfate (0.5% wt/vol), supplemented with adenine (10 mg/L), arginine (50 mg/L), aspartic acid (80 mg/L), histidine (20 mg/L), isoleucine (50 mg/L), leucine (100 mg/L), lysine (500 mg/L), methionine (20 mg/L), phenylalanine (50 mg/L), threonine (100 mg/L), tryptophan (50 mg/L), tyrosine (50 mg/L), valine (140 mg/L) and uracil (20 mg/L). For solid media, 2% (wt/vol) agarose was added to the synthetic media. Yeast was grown at 30 °C with shaking (a liquid cell culture occupying one fifth of the flask volume).
Table 2.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4743 | MATa/a;his3D1/his3D1;leu2D0/leu2D0;lys2D0/LYS2;MET15/met15D0;ura3D0/ura3D0 | Euroscarf |
| aft1∆ | As for BY4743 with aft1::kanMX4/aft1::kanMX4 | Euroscarf |
| aft2∆ | As for BY4743 with aft2::kanMX4/ aft2::kanMX4 | Euroscarf |
| anr1∆ | As for BY4743 with anr1::kanMX4/ anr1::kanMX4 | Euroscarf |
| arn2∆ | As for BY4743 with arn2::kanMX4/ arn2::kanMX4 | Euroscarf |
| ccc2∆ | As for BY4743 with ccc2::kanMX4/ ccc2::kanMX4 | Euroscarf |
| ctr1∆ | As for BY4743 with ctr1::kanMX4/ ctr1::kanMX4 | Euroscarf |
| ftr1∆ | As for BY4743 with ftr1::kanMX4/ ftr1::kanMX4 | Euroscarf |
| gsh1∆ | As for BY4743 with gsh1::kanMX4/ gsh1::kanMX4 | Euroscarf |
Synthesis of 56MESS
56MESS was synthesised as previously described [79].
Cytotoxicity assay in cisplatin-resistant cell line L1210cisR and yeast
The cytotoxicity of 56MESS to L1210cisR cells was measured as previously described [43]. The cells (4 × 104/ml) were plated in 96-well plates containing complete RPMI 1640 medium. 56MESS was added to cell culture to final concentrations ranging from 0.0001 to 50.0 μM. After continuous exposure for 48 h, cell viabilities were determined using standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazoliumbromide (MTT) assay [20] with incubation time in DMSO being 2 h. The 56MESS concentration that inhibits 50% growth of the cells is the IC50 value for this compound. The collated data were averages derived from between two and four independent experiments tested at least in duplicate. MTT and DMSO were obtained from Sigma-Aldrich, Australia.
For 56MESS cytotoxicity in yeast, BY4743 yeast cells were grown to exponential phase (OD600 0.8) in synthetic medium. The cells were exposed to a gradient of concentrations from 0.4 to 3.5 mM of 56MESS at 30 °C for 180 min in glass flasks. Drug-treated and untreated cells were then diluted and plated onto agar plates to establish cell colonies. The number of colonies under different drug concentrations was used to establish cell viability curve. Data were averaged from at least three biological replicates.
Transcriptomic analysis using Affymetrix cDNA microarray
Yeast cells at exponential growth phase (OD600 0.8) were treated with freshly prepared 56MESS (0.45 mM) for 60 min. Three samples (50 mL each) from each of 56MESS-treated or untreated cultures were collected prior to and at 60 min after drug addition. Cell pellets were prepared from the collected samples, snap-frozen in liquid nitrogen and then stored at −80 °C. To purify the total RNA, the frozen cell pellets were mixed with TRIzolTM reagent (Invitrogen, Australia) at 4 °C and cells were then broken with a mini-bead beater. The following steps leading to final RNA preparation were carried out as described previously [1]. The integrity of the RNA was confirmed using an RNA 6000 Nano LabChips on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). RNA was then reverse-transcribed, labelled and hybridised to GeneChip® Yeast Genome 2 Arrays (Affymetrix) at Ramaciotti Centre, UNSW, Australia. The hybridised arrays were scanned using Axon GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA, USA).
Microarray data were analysed using Partek® Genomic Suite™. Data were normalised with RMA algorithms as implemented in the programme. One-way analysis of variance (ANOVA) was used to compare transcript levels between 56MESS-treated and untreated samples and differentially expressed genes were revealed according to the criteria of gene expression fold change ≥1.5-fold and false discovery rate significance level, p, at 0.05. Identification of biological pathways over-represented by these significant genes was achieved using Yeast GO Slim Mapper tool implemented in Saccharomyces Genome Database (www.yeastgemome.org). Candidate binding motifs in a given set of genes for transcriptional factors were identified using regulatory sequence analysis (RSA) tools [71, 73]. Five to eight oligonucleotide sizes in the promoter regions of genes were chosen while searching for transcription factor binding motifs.
Phenotypic screening of yeast deletion mutants
The up- and down-regulated genes resulting from 56MESS exposure were further selectively validated through phenotypic screening of their deletion mutants. Mutant cells at exponential phase (OD600 0.8) were exposed to 56MESS in 5 mL media contained in glass flasks. A culture of 100 μL for each mutant was then collected at multiple time-points and plated on agar plates. The plates were incubated at 22 °C for 2–3 days and colony counts were used to establish the viability curves. Alternatively, spot assays using 96-well plates were employed for assessing the sensitivity and resistance of mutants to drug treatment. In this assay, the mutant strains were grown to exponential phase (OD600 0.8) and then diluted in 37 °C media to produce a culture with OD600 0.01. Cells were then exposed to a gradient of drug concentrations (ranging from 14.0 mM through eight dilution steps to 0.01 mM) in 96-well plates for 20–24 h at 30 °C. The growth of yeast cells at various time-points during this period was assessed by spotting the 96-well plates onto agar plates and cell growth within the spots was assessed and photographed.
Measurement of intracellular metals in yeast using ICP-OES
The intracellular metal contents were measured using ICP-OES. The yeast wild-type BY4743 cells were grown to exponential phase (OD600 0.8) in synthetic media. Freshly prepared drug solutions were added to the cultures up to the final concentration of 0.45 mM for 56MESS. Cell samples (50 mL each) were collected prior to and at 5, 20 and 60 min after the start of drug exposure. The collected cell culture samples were then centrifuged to remove the supernatant, and the resultant cell pellets were washed with 2 μM EDTA to chelate extracellular metals. The pellets were further washed with distilled and then deionised water. The remaining water content contained in the pellets was removed by centrifugation and drying in an oven at 55 °C. The weight of each pellet was calculated as the difference between the total weight of the pellet with the tube and the weight of the empty tube (pre-weighted).
Prior to analysis by ICP-OES, the cell pellets were digested in 1 mL 65% nitric acid for 24 h at 25 °C. The samples were then vortexed and centrifuged for 10 min at 13,000 rpm in a bench-top centrifuge. The resultant supernatants were diluted at 1:100 in MilliQ water and metal contents in these samples were measured with Varian 720-ES. Data were analysed with ICP Expert II software. ICP standard element solutions for Fe, Cu Zn, Al, Mn and Cd at 0, 0.05, 0.2, 0.5 and 1.0 ppm were prepared via appropriate dilutions of standard solutions in Ultrapure water and nitric acid at a final concentration of 65% (v/v). The metal ion concentrations were measured in parts per billion (ppb and expressed as ppb/g after normalisation against yeast dry weight).
Results
Cytotoxicity of 56MESS in the cisplatin-resistant L1210cisR cancer cell line
To assess the capacity of 56MESS in overcoming a cisplatin-resistant phenotype, the cytotoxicity of 56MESS and cisplatin in cisplatin-resistant leukaemia cancer cell line L1210cisR was measured, respectively, after continuous exposure to a gradient of drug concentrations ranging from 0.0001 to 50.0 μM for 48 h. 56MESS displayed IC50 of 0.06 ± 0.003 μM, which is 163-fold more active than cisplatin having IC50 of 9.80 ±1.400 μM. This result demonstrated that 56MESS is more potent than cisplatin in the cisplatin-resistant cancer cell line L1210cisR.
Titration of 56MESS concentrations for genome-wide gene expression profiling
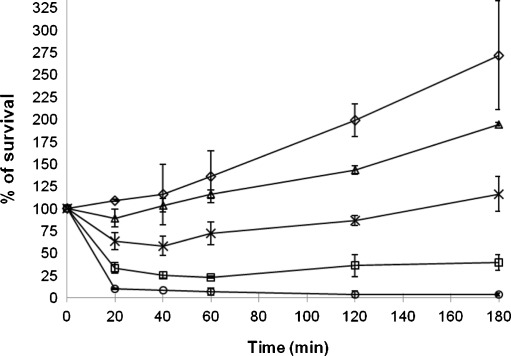
In order to apply genome-wide gene expression profiling to investigate the molecular mechanism of action of 56MESS in S. cerevisiae, we firstly examined growth inhibition to yeast cells by 56MESS at various concentrations (0.4 to 3.5 mM) over a course of 180 min (Fig. 1). A 25% growth inhibition relative to the control resulted from the treatment of 56MESS at 0.4 mM and was maintained for 180 min. 56MESS at 0.5 mM produced approximately 50% growth inhibition for the duration of the treatment. Within the same time frame, a 75% growth inhibition was achieved by 56MESS at 1.0 mM. At 3.5 mM, 56MESS led to nearly 100% cell death after exposure for 60 min. Growth inhibition of yeast by 56MESS treatment was positively correlated with drug dosage. Based on these results, the 56MESS concentration of 0.45 mM (between 0.40 and 0.50 mM), which caused significant growth inhibition, yet not substantial cell death, was chosen for genome-wide expression profiling of yeast response to 56MESS. This level of dosage was previously used in gene expression profiling studies in yeast for cellular response to drug treatments and certain environmental conditions [9, 22].
Fig. 1.
Growth inhibition of yeast cells by 56MESS. BY4743 wild-type yeast cultures at exponential growth phase (OD600 0.8) were exposed to a range of 56MESS concentrations (0.4 to 3.5 mM) for 180 min at 30 °C with agitation. Growth inhibition was expressed as the percentage of colony count at each of time-points over that at 0 time-point. Diamond, control; triangle, 0.4 mM; multiplication symbol, 0.5 mM; square, 1.0 mM; circle, 3.5 mM. The data are the averages from at least two biological replicates and error bars indicate standard deviations
Changes in gene expression and identification of major biological processes in response to 56MESS treatment
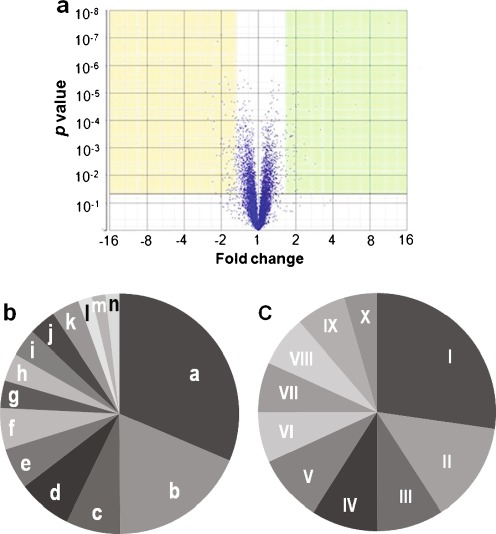
To uncover genes important to the cytotoxicity of 56MESS, cDNA microarray technology was used to measure the changes of genome-wide gene expression after treatment for 1 h with 0.45 mM 56MESS. During 56MESS challenge, 48 and 45 genes out of 5,841 genes in a yeast genome were up- and down-regulated, respectively, by at least 1.5-fold at p ≤ 0.05 compared to untreated cells (Fig. 2a). Bioinformatics analysis of these up- and down-regulated genes using Yeast GO Slim Mapper tool showed that 14 GO biological processes were represented by up-regulated genes and ten GO biological processes were represented by down-regulated genes (Fig. 2b, c). Two pathways—transport and cellular homeostasis—were highly over-represented by up-regulated genes among the 14 GO biological processes (Fig. 2b). The transport pathway was represented by 17 genes including FTR1, ARN1, ARN2, SIT1 (ARN3), ENB1 (ARN4), FRE1, FRE2, FRE3, FRE6, FIT2, FIT3, CTR1, CCC2, SIT1, PUG1, VMR1, DTR1 and RSB1. All of these genes except four (PUG1, VMR1, DTR1 and RSB1) are in the pathway of iron and copper transport across the cell membrane [24, 30, 49, 57]. For example, FTR1 gene encodes a high-affinity iron permease for the transport of iron across the plasma membrane [69] and ARN1 encodes a membrane transporter responsible for up-taking iron bounded to siderophores [30]. The cellular homeostasis pathway was represented by FTR1, ARN1, ARN2, SIT1 (ARN3), FRE3, FRE6, CCC2, TIS11, HMX1 and ENB1 (total of ten genes). All of these genes are also in the pathway of iron and copper transport and regulation of heme during iron starvation. The CTR1 is a copper transporter and CCC2 gene is Cu2+- transporting P-type ATPase [21]. Thus, the highly induced processes under 56MESS treatment are the iron and copper uptake and homeostasis.
Fig. 2.
The differentially expressed genes (a) and the major pathways (b, c) in response to 56MESS treatment. Genes whose expression changed by 1.5-fold or more at p ≤ 0.05 under 56MESS treatment were identified as differentially expressed genes using ANOVA Partek® Genomic Suite™. Each dot in (a) represents one gene. Out of 5,841 genes, 48 up-regulated genes are shown in the yellow area and 45 down-regulated genes in the green area. The major pathways represented by the up- and down-regulated genes were revealed using GO Slim Mapper and shown in b and c, respectively. Pathways represented by a to n in (b) and I to X in (c) are listed in Table S2 in “Electronic supplementary material”
The biological processes accentuated by the 45 down-regulated genes included cellular amino acid metabolic process, stress response and cellular respiration (Fig. 2c). The cellular amino acid metabolic process was represented by 12 genes of HOM3, STR3, MET13, MET17, MET28, ARG1, ARG4, ARG7, CPA2, CAR2, ILV6 and GLT1 [28, 36, 38, 50]. Among them, five genes (HOM3, STR3, MET13, MET17 and MET28) are involved in sulfur-containing amino acid (methionine and cysteine) biosynthesis, and five genes (ARG1, ARG4, ARG7, CPA2 and CAR2) in arginine biosynthesis. The gene SUL2 encoding a high-affinity sulfate permease in sulfur metabolism was also suppressed by 56MESS (see Table S1 in “Electronic supplementary material”). Furthermore, 56MESS down-regulated the transcription factor Met28p, a component of the Cbf1p–Met4p–Met28p complex which participates in the regulation of sulfur metabolism [46]. 56MESS also suppressed the expression of the following genes: PCL5, TPS2, GRX4, XBP1, MNN4 and CCP1, which were shown to be involved in stress response processes [12, 51, 56, 63]. Also down-regulated were cellular respiration genes (CYC7, SDH1, SDH2 and NDI1), implicating the involvement of mitochondria in 56MESS cytotoxicity.
Transcription regulation of the iron and copper pathway and the cytotoxicity of 56MESS
The large-scale up-regulation of iron and copper transport genes suggested that transcription factors are involved in cell response to 56MESS. The key transcription factors, Aft1p and Aft2p, were uncovered by using RSA tools (Table 3).
Table 3.
Over-represented motifs in the promoter regions of 48 genes up-regulated by 56MESS treatment
| Motifsa | Proposed transcription factors | Significance indexb |
|---|---|---|
| tgcacc | Aft1p, Aft2p | 6.55 |
| gcaccc | Aft1p, Aft2p | 9.22 |
| aaaatg | Unidentified | 0.09 |
aMotifs were obtained from analysis of promoter regions of 56MESS up-regulated genes using RSA tools [70]
bMinus log transform of the E-value, which is the product of multiplying the p-value by the number of distinct motifs. The p-value is a probability of chance occurrence of particular motifs in the promoter regions of a given list of genes. A higher significance index indicates more significant motifs
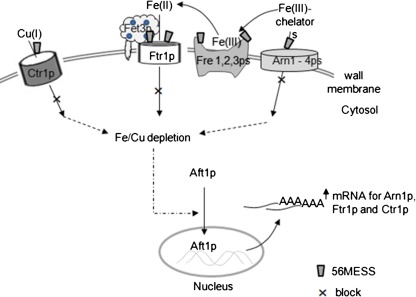
The results indicate a molecular mechanism for the up-regulation of iron and copper under 56MESS treatment (Fig. 3). In this mechanism, 56MESS targets iron and copper transporters (such as Ftr1p and Arn1p) located in the plasma membrane. We propose that the interactions of 56MESS with these transporter proteins lead to conformational changes and subsequent inhibition of iron and copper uptake, ultimately resulting in depletion of intracellular iron and copper. The depletion of intracellular iron and copper activates the translocation of Aft1p from cytoplasm into nucleus where this transcription factor binds to the motifs of the iron and copper genes as shown in Table 3. Consequently, the transcript expression of genes in the iron and copper uptake pathways was up-regulated. The key trigger for this proposed mechanism is the reduction or depletion of intracellular iron and copper, which was determined in the following experiment.
Fig. 3.
The proposed molecular mechanism for the up-regulation of iron and copper under 56MESS treatment. 56MESS interacts with iron and copper membrane transporters (Ctr1p, Ftr1p, Fre1p, Fre2p, Fre3p and Arn1–4p). Such interaction then leads to conformational changes of these transporter proteins and subsequent inhibition of iron and copper uptake, with ultimate depletion of intracellular iron and copper. The iron and copper depletion results in Aft1p translocation from cytoplasm into the nucleus where this transcription factor binds to target genes (such as ARN1 and FTR1) to trigger their expression
Determination of intracellular iron and copper concentrations
The intracellular iron and copper concentrations of yeast cells treated by 56MESS at the same concentration as used for gene expression analysis were measured with ICP-OES. Manganese, zinc, cadmium and aluminium were also included in this experiment so as to provide a more comprehensive view of metal ion profile. After 60 min of 56MESS treatment, iron and copper levels in 56MESS-treated cells were reduced to 67% and 60%, respectively, compared to those in control (Table 4). 56MESS treatment increased the retention of manganese relative to the control by 27% (Table 4) but did not affect the concentration of zinc, aluminium and cadmium (data not shown).
Table 4.
The intracellular metal ion concentrations under 56MESS treatment
| Time (min) | 56MESS-treated | Untreated | 56MESS/control (%) |
|---|---|---|---|
| Iron (ppb/g) | |||
| 0 | 83.2 ± 2.7 | 83.2 ± 2.7 | 100 |
| 5 | 79.7 ± 3.4 | 85.6 ± 2.8 | 93 |
| 20 | 58.2 ± 2.4 | 83.6 ± 5.7 | 70 |
| 60 | 63.8 ± 11.1 | 95.1 ± 19.9 | 67 |
| Copper (ppb/g) | |||
| 0 | 9.8 ± 0.3 | 9.8 ± 0.3 | 100 |
| 5 | 8.9 ± 0.1 | 10.4 ± 0.4 | 86 |
| 20 | 7.7 ± 0.3 | 11.6 ± 0.5 | 66 |
| 60 | 7.4 ± 0.5 | 12.4 ± 0.8 | 60 |
| Manganese (ppb/g) | |||
| 0 | 4.2 ± 0.2 | 4.2 ± 0.2 | 100 |
| 5 | 3.9 ± 0.1 | 3.5 ± 0.1 | 109 |
| 20 | 3.6 ± 0.2 | 3.3 ± 0.1 | 111 |
| 60 | 4.1 ± 0.2 | 3.3 ± 0.4 | 127 |
Phenotypic screening of deletion mutants of genes in iron and copper metabolism
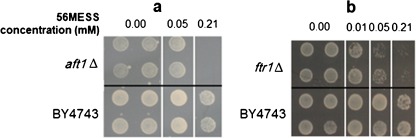
Several deletion mutants of key genes in iron and copper metabolism were screened for their growth phenotypes under 56MESS treatment in order to investigate the role of this metabolism in the cytotoxicity of this complex. The results showed that the lowest 56MESS concentration that killed 100% of aft1∆ mutant was at 0.21 mM, but it had no effect on wild-type BY4743 cells (Fig. 4a). Growth inhibition of FTR1 deletion mutant by 56MESS was evident at 0.05 mM and was more severe at 0.21 mM (Fig. 4b). The deletion mutant of CCC2 was slightly sensitive to 56MESS (data not shown). Deletion mutants of CTR1, ARN1, ARN2 and AFT2 were neither sensitive nor resistant to 56MESS. These data suggested that Ftr1p transporter, but probably not the Arn family or Ctr1p transporters, is critical to the cytotoxicity of 56MESS.
Fig. 4.
Phenotypic screening of deletion mutants of AFT1 (a) and FTR1 (b) under 56MESS treatment. The exponential phase of yeast cells was exposed to a gradient of 56MESS concentrations in 96-well plates for 22 h and then spotted to agar plates for further 48 h of incubation. The spots were the colonies of the yeast cells
Role of glutathione in 56MESS cytotoxicity
The role of glutathione in the cytotoxicity of 56MESS was first investigated by exposing 56MESS to yeast gsh1∆ mutant in which synthesis of glutathione is blocked. 56MESS at 2.5–5.0 mM caused initial growth inhibition after 20 min and 82.5% inhibition after 120 min relative to untreated cells (Fig. 5a). The glutathione deletion mutant gsh1∆ was therefore sensitive to 56MESS.
Fig. 5.
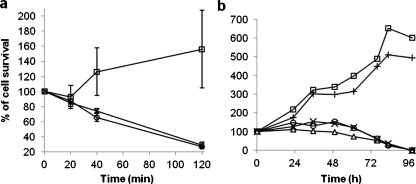
Glutathione involvement in the cytotoxicity of 56MESS. a The growth inhibition of the gsh1Δ mutant under 56MESS treatment. Square, untreated; multiplication symbol, 2.5 mM 56MESS; circle, 5.0 mM 56MESS. b Effects of supplementing glutathione into L1210 cell culture on the 56MESS’ cytotoxicity. Square, 1.2 mM GSH only; cross, control; circle, 0.045 mM 56MESS with 0.6 mM GSH; multiplication symbol, 0.045 mM 56MESS with 1.2 mM GSH; triangle, 0.045 mM 56MESS only
Further experiments were carried out to assess the effect of glutathione on the cytotoxicity of 56MESS in cancer cells by supplementing glutathione in the cell culture media before exposure to 56MESS (Fig. 5b). Addition of 1.2 mM glutathione alone to cell culture promoted 6.7% to 28.1% cell growth in comparison to the control, whereas 56MESS treatment (0.045 mM) alone inhibited cell growth by 26% at 61 h and killed 100% of cells at 96 h. The cytotoxicity that resulted from treatment with 56MESS was reduced by 10% to 51% from 23 to 76 h with the addition of glutathione (0.6 or 1.2 mM) into L1210 cell culture prior to 56MESS exposure. No reduction was observed at longer incubation periods of 83 and 96 h (Fig. 5b).
Discussion
This study shows that 56MESS is more effective in killing cisplatin-resistant L1210cisR cells than cisplatin does. This result reinforces the view that 56MESS has the potential to be a potent anticancer agent [45, 55] and highlights the possibility of 56MESS in overcoming cisplatin-resistant cancer cells. Transcriptomic data and intracellular metal content analysis with ICP-OES have delineated the details of molecular mechanisms of the cytotoxic action of the platinum metallointercalator, 56MESS.
The large-scale up-regulation of genes in the iron and copper homeostasis pathway in response to 56MESS treatment was the initial indication of the molecular mechanisms of 56MESS’ cytotoxic action. Up-regulation of these genes likely resulted from a negative feedback mechanism, where 56MESS causes a reduction of intracellular iron and copper concentrations and this subsequently activates these genes through the transcription factor Aft1p [64, 80]. This is supported by the finding that the concentrations of intracellular iron and copper were indeed low in 56MESS-treated yeast cell. Hence, disruption of iron and copper metabolism is a key biological mechanism for 56MESS cytotoxicity.
The reason for iron and copper deficiency in the cells upon 56MESS treatment could be due to the impairment of iron and copper uptake from the extracellular environment. Such impairment may have resulted from the binding of 56MESS to iron and copper transporters in the plasma membrane. Our results showed that mutants of an iron transporter Ftr1p were sensitive to 56MESS (Fig 4b), indicating that the Ftr1p transporter is a possible target of the compound. Ftr1p is a high-affinity iron transporter [3] and its inhibition can greatly reduce iron uptake, leading to a low level of intracellular iron. It may be argued that iron could still enter cells via ARN family transporters. However, under the experimental condition of this study, it is unlikely because ARN transporters only transport siderphore-chelated iron and only FeCl3 is available in the media of this study. Deletion mutants of ARN genes are neither sensitive nor resistant to 56MESS.
The ensuing cytotoxicity caused by the 56MESS-induced iron and copper depletion is explained by their essential physiological roles performed by these metal ions. Iron and copper act as cofactors in enzymes, such as superoxide dismutase [14, 60], involved in respiration, scavenging reactive oxygen species and in a wide variety of other metabolic processes [74]. Insufficient concentrations of iron and copper can therefore impede cellular functions. Indeed cells starved of iron are unable to progress from the G1 phase to the S phase in the cell cycle process [47].
The transcriptomic analysis also showed that 56MESS suppressed the biosynthesis of sulfur-containing amino acids—methionine and cysteine. Suppression of this pathway could lead to reduced production of the tri-peptide glutathione. Down-regulation of a di- or tri-peptide transporter gene PTR2 by 56MESS (see Table S1 in “Electronic supplementary material”) indicates that the uptake of glutathione could also be decreased. These findings were reinforced by the result that the gsh1∆ mutant, in which glutathione synthesis is blocked, is sensitive to 56MESS. As glutathione plays essential cellular functions including maintenance of mitochondrial genome, sporulation and detoxification of harmful compounds such as oxidants [27, 48], glutathione deficiency can weaken cellular defence systems, thus exacerbating the cytotoxic action of 56MESS.
The molecular mechanism of glutathione involvement in the cytotoxic action of 56MESS is more complex than direct glutathione binding by 56MESS. Addition of glutathione to cancer cell culture provided a certain degree of protection against 56MESS at early exposure time (from 23 to 76 h), but such protection was diminished after longer exposure (83–96 h). If direct degradation of 56MESS by glutathione occurs, then longer exposure of 56MESS to glutathione-supplemented cells would lead to further rescue effect instead of diminishing one as observed (Fig. 5b) because glutathione is stoichiometrically in excess compared to 56MESS in the experimental condition. The partial protection is likely due to the growth-promoting effect of glutathione as demonstrated by the finding that glutathione alone promotes cell growth (Fig. 5b), possibly via its antioxidant effects [54, 67]. The ability of 56MESS in overcoming the initial glutathione-mediated protection could be due to the down-regulation of glutathione production and uptake by 56MESS as discussed above. Cancer resistance to cisplatin mediated through direct glutathione binding to it is common [4, 26, 61] and therefore 56MESS may be useful in combination therapy with cisplatin to reduce glutathione concentration.
In addition to iron and copper metabolism and glutathione involvement of 56MESS cytotoxicity, the perturbation of redox homeostasis in 56MESS-treated cells was evident by the down-regulation of CCP1, GRX4 and TPS2, which are genes involved in oxidative stress response. For example, CCP1 is a mitochondrial cytochrome-c peroxidase for breaking down reactive oxygen species [11, 31]. Suppression of these genes suggests that 56MESS might have weakened cellular defence. Another finding was the increased manganese (Mn) level upon 56MESS treatment (Table 4). As Mn is a cofactor of the antioxidant superoxide dismutase [75], the elevation of the Mn may be a secondary cellular response to the 56MESS-induced disruption of the cellular antioxidant system.
The data demonstrated that the disruption of iron and copper metabolism in conjunction with suppression of sulfur-containing amino acids and weakened cellular anti-oxidant capacity are the key molecular mechanisms for 56MESS. The fact that no significant genes related to DNA repair were uncovered suggests that interacting with DNA is not the major action of 56MESS, although microprobe super resolution X-ray fluorescence elemental maps of human lung cancer cells (A549) following treatment with 56MESS did reveal platinum localisation within the nucleus [13]. Previous finding using DNase I footprinting, DNA unwinding and DNA binding affinity indicated that 56MESS–DNA interactions did not account for its cytotoxicity [42, 45]. The molecular mechanisms elucidated for 56MESS are in clear contrast to the DNA binding mechanism of anticancer drug cisplatin [15, 76]. This may explain why 56MESS is potent to cisplatin-resistant cancer cells, including L1210cisR [45, 55], where increased resistance to cisplatin can be mediated by elevated glutathione [61] and increased DNA repair [10, 76].
The iron and copper metabolism, being essential to cellular functions, is highly conserved between yeast and humans [3]. Iron and copper are required by proliferating cancer cells during carcinogenesis [32, 39, 59, 77] and high levels of iron and copper were found in certain tumour cells and tissues, including breast cancer cells [17, 33]. As iron and copper metabolism is up-regulated in neoplastic tumours [29, 39, 41, 44, 47, 59, 62, 65, 72], 56MESS and other complexes in this class should be assessed, in combination with cisplatin, in the cancer models that have elevated iron and copper metabolism. Indeed chelating cellular copper has recently been shown to selectively enhance the therapeutic efficacy of cisplatin in a mouse model of human cervical cancer [34].
In conclusion, this study has confirmed that 56MESS is highly cytotoxic in cisplatin-resistant cancer cell line. This capacity is mediated via disruption of iron and copper metabolism along with suppression of sulfur-containing amino acids and attenuation of cellular defence capacity. The high-affinity iron transporter Ftr1p may be one of the key targets of 56MESS. On the basis of these findings, 56MESS is currently being investigated for its effect, in combination with cisplatin, on cancers with elevated iron and copper metabolism.
Electronic Supplementary Materials
(PDF 30 kb)
(PDF 126 kb)
Acknowledgements
The authors thank the Australian Government for the International Science Linkage Grant and the University of Western Sydney for financial support through internal research grants (JRAW and MW). S.W. was supported by APA Award and UWS Top-Up Award from the University of Western Sydney. We also thank Ramaciotti Centre at UNSW for Affymetrix microarray, Dr. C. Cullinane from the Peter MacCallum Cancer Centre for supplying the L1210cisR cancer cell line and K. McNamara for technical assistance.
Contributor Information
Janice R. Aldrich-Wright, Phone: +61-2-46203218, FAX: +61-2-46203025, Email: j.aldrich-wright@uws.edu.au
Ming J. Wu, Phone: +61-2-46203089, FAX: +61-2-46203025, Email: m.wu@uws.edu.au
References
- 1.Alic N, Felder T, Temple MD, Gloeckner C, Higgins VJ, Briza P, Dawes IW. Genomewide transcriptional responses to a lipid hydroperoxide: adaptation occurs without induction of oxidant defenses. Free Radic Biol Med. 2004;37:23–35. doi: 10.1016/j.freeradbiomed.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Ang WH, Khalaila I, Allardyce CS, Juillerat-Jeanneret L, Dyson PJ. Rational design of platinum(IV) compounds to overcome glutathione-S-transferase mediated drug resistance. J Am Chem Soc. 2005;127:1382–1383. doi: 10.1021/ja0432618. [DOI] [PubMed] [Google Scholar]
- 3.Askwith C, Kaplan J. Iron and copper transport in yeast and its relevance to human disease. Trends Biochem Sci. 1998;23:135–138. doi: 10.1016/S0968-0004(98)01192-X. [DOI] [PubMed] [Google Scholar]
- 4.Balendiran G, Dabur R, Fraser D. The role of glutathione in cancer. Cell Biochem and Funct. 2004;22:343–352. doi: 10.1002/cbf.1149. [DOI] [PubMed] [Google Scholar]
- 5.Barabas K, Milner R, Lurie D, Adin C. Cisplatin: a review of toxicities and therapeutic applications. Vet Comp Oncol. 2008;6:1–18. doi: 10.1111/j.1476-5829.2007.00142.x. [DOI] [PubMed] [Google Scholar]
- 6.Basu A, Krishnamurthy S (2010) Cellular responses to cisplatin-induced DNA damage. J Nucleic Acids. doi:10.4061/2010/201367 [DOI] [PMC free article] [PubMed]
- 7.Boyer J, Allen WL, McLean EG, Wilson PM, McCulla A, Moore S, Longley DB, Caldas C, Johnston PG. Pharmacogenomic identification of novel determinants of response to chemotherapy in colon cancer. Cancer Res. 2006;66:2765–2777. doi: 10.1158/0008-5472.CAN-05-2693. [DOI] [PubMed] [Google Scholar]
- 8.Brodie CR, Grant Collins J, Aldrich-Wright JR (2004) DNA binding and biological activity of some platinum(II) intercalating compounds containing methyl-substituted 1,10-phenanthrolines. Dalton Trans:1145–1152 [DOI] [PubMed]
- 9.Caba E, Dickinson DA, Warnes GR, Aubrecht J. Differentiating mechanisms of toxicity using global gene expression analysis in Saccharomyces cerevisiae. Mutat Res. 2005;575:34–46. doi: 10.1016/j.mrfmmm.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Calsou P, Barret J-M, Cros S, Salles B. DNA excision–repair synthesis is enhanced in a murine leukemia L1210 cell line resistant to cisplatin. Eur J Biochem. 1993;211:403–409. doi: 10.1111/j.1432-1033.1993.tb17563.x. [DOI] [PubMed] [Google Scholar]
- 11.Charizanis C, Juhnke H, Krems B, Entian K. The mitochondrial cytochrome c peroxidase Ccp1 of Saccharomyces cerevisiae is involved in conveying an oxidative stress signal to the transcription factor Pos9 (Skn7) Mol Gen Genet. 1999;262:437–447. doi: 10.1007/s004380051103. [DOI] [PubMed] [Google Scholar]
- 12.Virgilio C, Bürckert N, Bell W, Jenö P, Boller TAW. Disruption of TPS2, the gene encoding the 100-kDa subunit of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae, causes accumulation of trehalose-6-phosphate and loss of trehalose-6-phosphate phosphatase activity. Eur J Biochem. 1993;212:315–323. doi: 10.1111/j.1432-1033.1993.tb17664.x. [DOI] [PubMed] [Google Scholar]
- 13.Dillon CT. Synchrontron radiation x-ray spectroscopy for investigations of introcellular metallointercalators: x-ray fluorescence imaging and x-ray absorption spectroscopy. In: Aldrich-Wright JR, editor. Metallointercalators, synthesis and techniques to probe their interactions with biomolecules. New York: Springer; 2011. pp. 273–298. [Google Scholar]
- 14.Djinovic K, Gatti G, Coda A, Antolini L, Pelosi G, Desideri A, Falconi M, Marmocchi F, Rotilio GMB. Crystal structure of yeast Cu, Zn superoxide dismutase. Crystallographic refinement at 2.5 A resolution. J Mol Biol. 1992;225:791–809. doi: 10.1016/0022-2836(92)90401-5. [DOI] [PubMed] [Google Scholar]
- 15.Eastman A. The mechanism of action of cisplatin: from adducts to apoptosis. In: Lippert B, editor. Cisplatin: chemistry and biochemistry of a leading anticancer drug. Zürich: VHCA; 1999. pp. 111–134. [Google Scholar]
- 16.Einhorn LH. Curing metastatic testicular cancer. Proc Natl Acad Sci USA. 2002;99:4592–4595. doi: 10.1073/pnas.072067999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farquharson MJ, Al-Ebraheem A, Falkenberg G, Leek R, Harris AL, Bradley DA. The distribution of trace elements Ca, Fe, Cu and Zn and the determination of copper oxidation state in breast tumour tissue using muSRXRF and muXANES. Phys Med Biol. 2008;53:3023–3037. doi: 10.1088/0031-9155/53/11/018. [DOI] [PubMed] [Google Scholar]
- 18.Fisher DM, Bednarski PJ, Grünert R, Turner P, Fenton RR, Aldrich-Wright JR. Chiral platinum(II) metallointercalators with potent in vitro cytotoxic activity. ChemMedChem. 2007;2:488–495. doi: 10.1002/cmdc.200600211. [DOI] [PubMed] [Google Scholar]
- 19.Fisher DM, Fenton RR, Aldrich-Wright JR. In vivo studies of a platinum(II) metallointercalator. Chem Commun. 2008;43:5613–5615. doi: 10.1039/b811723c. [DOI] [PubMed] [Google Scholar]
- 20.Freshney R. Culture of animal cells: a manual of basic technique. 5. New York: Wiley-Liss; 2005. pp. 359–373. [Google Scholar]
- 21.Fu D, Beeler TJ, Dunn TM. Sequence, mapping and disruption of CCC2, a gene that cross complements the Ca2 + 517 -sensitive phenotype of csg1 mutants and encodes a P-type ATPase belonging to the Cu2 + 518 -ATPase subfamily. Yeast. 1995;11:283–292. doi: 10.1002/yea.320110310. [DOI] [PubMed] [Google Scholar]
- 22.Gasch A, Spellman P, Kao C, Carmel-Harel O, Eisen M, Storz G, Botstein D, Brown P. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgatsou E, Alexandraki D. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3065–3073. doi: 10.1128/mcb.14.5.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson G, Muse SV. A prime of genome science. 3. Sunderland: Sinauer; 2009. pp. 191–258. [Google Scholar]
- 26.Godwin AK, Meister A, O'dwyer PJ, Huang CS, Hamilton TC, Anderson ME. High resistance to cisplatin in human ovarian cancer cell lines is associated with marked increase of glutathione synthesis. Proc Natl Acad Sci USA. 1992;89:3070–3074. doi: 10.1073/pnas.89.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant CM. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol Microbiol. 2001;39:533–541. doi: 10.1046/j.1365-2958.2001.02283.x. [DOI] [PubMed] [Google Scholar]
- 28.Hansen J, Johannesen PF. Cysteine is essential for transcriptional regulation of the sulfur assimilation genes in Saccharomyces cerevisiae. Mol Gen Genet. 2000;263:535–542. doi: 10.1007/s004380051199. [DOI] [PubMed] [Google Scholar]
- 29.Head JF, Wang F, Elliott RL. Antineoplastic drugs that interfere with iron metabolism in cancer cells. Adv Enzyme Regul. 1997;37:147–169. doi: 10.1016/S0065-2571(96)00010-6. [DOI] [PubMed] [Google Scholar]
- 30.Heymann P, Ernst JF, Winkelmann G. Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol Lett. 2000;186:221–227. doi: 10.1111/j.1574-6968.2000.tb09108.x. [DOI] [PubMed] [Google Scholar]
- 31.Ho PS, Hoffman BM, Solomon N, Kang CH, Margoliash E. Kinetics and energetics of intramolecular electron transfer in yeast cytochrome c peroxidase. Biochemistry. 1984;23:4122–4128. doi: 10.1021/bi00313a017. [DOI] [PubMed] [Google Scholar]
- 32.Hu GF. Copper stimulates proliferation of human endothelial cells under culture. J Cell Biochem. 1998;69:326–335. doi: 10.1002/(SICI)1097-4644(19980601)69:3<326::AID-JCB10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 33.Ionescu JG, Novotny J, Stejskal V, Latsch A, Blaurock-Busch E, Eisenmann-Klein M. Increased levels of transition metals in breast cancer tissue. Neuroendocrin Lett. 2006;21:36–39. [PubMed] [Google Scholar]
- 34.Ishida S, McCormick F, Smith-McCune K, Hanahan D. Enhancing tumor-specific uptake of the anticancer drug cisplatin with a copper chelator. Cancer Cell. 2010;17:574–583. doi: 10.1016/j.ccr.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaramillo D, Buck DP, Collins JG, Fenton RR, Stootman FH, Wheate NJ, Aldrich-Wright JR. Synthesis, characterisation and biological activity of chiral platinum(II) complexes. Eur J Inorg Chem. 2006;2006:839–849. doi: 10.1002/ejic.200500932. [DOI] [Google Scholar]
- 36.Jauniaux JC, Urrestarazu LA, Wiame JM. Arginine metabolism in Saccharomyces cerevisiae: subcellular localization of the enzymes. J Bacteriol. 1978;133:1096–1107. doi: 10.1128/jb.133.3.1096-1107.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson SW, Ferry KV, Hamilton TC. Recent insights into platinum drug resistance in cancer. Drug Resist Update. 1998;1:243–254. doi: 10.1016/S1368-7646(98)80005-8. [DOI] [PubMed] [Google Scholar]
- 38.Jones EW, Fink GR. Regulation of amino acid and nucleotide biosynthesis in yeast. In: Strathern JN, Jones EW, Broach JR, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. New York: CSHL; 1982. pp. 181–299. [Google Scholar]
- 39.Kabat GC, Rohan T. Does excess iron play a role in breast carcinogenesis? An unresolved hypothesis. Cancer Cause Control. 2007;18:1047–1053. doi: 10.1007/s10552-007-9058-9. [DOI] [PubMed] [Google Scholar]
- 40.Kelland LR. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 41.Kemp JD. Iron deprivation and cancer: a view beginning with studies of monoclonal antibodies against the transferrin receptor. Histol Histopathol. 1997;12:291–296. [PubMed] [Google Scholar]
- 42.Kemp S, Wheate NJ, Buck DP, Nikac M, Collins JG, Aldrich-Wright JR. The effect of ancillary ligand chirality and phenanthroline functional group substitution on the cytotoxicity of platinum(II)-based metallointercalators. J Inorg Biochem. 2007;101:1049–1058. doi: 10.1016/j.jinorgbio.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 43.Kemp S, Wheate NJ, Wang S, Collins J, Ralph S, Day A, Higgins VJ, Aldrich-Wright JR. Encapsulation of platinum(II)-based DNA intercalators within cucurbit[6,7,8]urils. J Biol Inorg Chem. 2007;12:969–979. doi: 10.1007/s00775-007-0269-z. [DOI] [PubMed] [Google Scholar]
- 44.Kicic A, Chua ACG, Baker E. Effect of iron chelators on proliferation and iron uptake in hepatoma cells. Cancer. 2001;92:3093–3110. doi: 10.1002/1097-0142(20011215)92:12<3093::AID-CNCR10107>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 45.Krause-Heuer AM, Grünert R, Kühne S, Buczkowska M, Wheate NJ, Pevelen DD, Boag LR, Fisher DM, Kasparkova J, Malina J, Bednarski PJ, Brabec V, Aldrich-Wright JR. Studies of the mechanism of action of platinum(II) complexes with potent cytotoxicity in human cancer cells. J Med Chem. 2009;52:5474–5484. doi: 10.1021/jm9007104. [DOI] [PubMed] [Google Scholar]
- 46.Kuras L, Barbey R, Thomas D. Assembly of a bZIP–bHLH transcription activation complex: formation of the yeast Cbf1–Met4–Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J. 1997;16:2441–2451. doi: 10.1093/emboj/16.9.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le NT, Richardson DR. The role of iron in cell cycle progression and the proliferation of neoplastic cells. Biochim Biophys Acta. 2002;1603:31–46. doi: 10.1016/s0304-419x(02)00068-9. [DOI] [PubMed] [Google Scholar]
- 48.Lee J, Straffon MJ, Jang T, Higgins VJ, Grant CM, Dawes IW. The essential 582 and ancillary role of glutathione in Saccharomyces cerevisiae analysed using a grande gsh1 disruptant strain. FEMS Yeast Res. 2001;1:57–65. doi: 10.1111/j.1567-1364.2001.tb00013.x. [DOI] [PubMed] [Google Scholar]
- 49.Lesuisse E, Blaiseau PL, Dancis A, Camadro J. Siderophore uptake and use by the yeast Saccharomyces cerevisiae. Microbiology. 2001;147:289–298. doi: 10.1099/00221287-147-2-289. [DOI] [PubMed] [Google Scholar]
- 50.Lim AL, Powers-Lee SG. Requirement for the carboxyl-terminal domain of Saccharomyces cerevisiae carbamoyl-phosphate synthetase. J Biol Chem. 1996;271:11400–11409. doi: 10.1074/jbc.271.19.11400. [DOI] [PubMed] [Google Scholar]
- 51.Mai B, Breeden L. Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol Cell Biol. 1997;17:6491–6501. doi: 10.1128/mcb.17.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Margiotta N, Ostuni R, Gandin V, Marzano C, Piccinonna S, Natile G. Synthesis, characterization, and cytotoxicity of dinuclear platinum-bisphosphonate complexes to be used as prodrugs in the local treatment of bone tumours. Dalton Trans. 2009;48:10904–10913. doi: 10.1039/b919721d. [DOI] [PubMed] [Google Scholar]
- 53.McFadyen WD, Wakelin LPG, Roos IAG, Leopold VA. Activity of platinum(II) intercalating agents against murine leukemia L1210. J Med Chem. 1985;28:1113–1116. doi: 10.1021/jm00146a026. [DOI] [PubMed] [Google Scholar]
- 54.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 55.Moretto J, Chauffert B, Ghiringhelli F, Aldrich-Wright JR, Bouyer F. Discrepancy between in vitro and in vivo antitumor effect of a new platinum(II) metallointercalator. Invest New Drugs. 2010;29:1164–1176. doi: 10.1007/s10637-010-9461-z. [DOI] [PubMed] [Google Scholar]
- 56.Odani T, Shimma Y, Wang XHYJ. Mannosylphosphate transfer to cell wall mannan is regulated by the transcriptional level of the MNN4 gene in Saccharomyces cerevisiae. FEBS Lett. 1997;420:186–190. doi: 10.1016/S0014-5793(97)01513-5. [DOI] [PubMed] [Google Scholar]
- 57.Protchenko O, Philpott C. Regulation of intracellular heme levels by HMX1, a homologue of heme oxygenase, in Saccharomyces cerevisiae. J Biol Chem. 2003;278:36582–36587. doi: 10.1074/jbc.M306584200. [DOI] [PubMed] [Google Scholar]
- 58.Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reizenstein P. Iron, free radicals and cancer: a review. Med Oncol Tumor Pharmacother. 1991;8:229–233. doi: 10.1007/BF02987191. [DOI] [PubMed] [Google Scholar]
- 60.Richardson J, Thomas KA, Rubin BH, Richardson DC. Crystal structure of bovine Cu, Zn superoxide dismutase at 3 A resolution: chain tracing and metal ligands. Proc Nat Acad Sci. 1975;72:1349–1353. doi: 10.1073/pnas.72.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richon VM, Schulte N, Eastman A. Multiple mechanisms of resistance to cis diamminedichloroplatinum(II) in murine leukemia L1210 Cells. Cancer Res. 1987;47:2056–2061. [PubMed] [Google Scholar]
- 62.Robbins E, Pederson T. Iron: its intracellular localization and possible role in cell division. Proc Natl Acad Sci USA. 1970;66:1244–1251. doi: 10.1073/pnas.66.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez-Manzaneque MT, Ros J, Cabiscol E, Sorribas A, Herrero E. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8180–8190. doi: 10.1128/mcb.19.12.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rutherford JC, Jaron S, Winge DR. Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J Biol Chem. 2003;278:27636–27643. doi: 10.1074/jbc.M300076200. [DOI] [PubMed] [Google Scholar]
- 65.Seligman P, Schleicher R, Siriwardana G, Domenico J, Gelfand E. Effects of agents that inhibit cellular iron incorporation on bladder cancer cell proliferation. Blood. 1993;82:1608–1617. [PubMed] [Google Scholar]
- 66.Siddik Z. Cisplatin: mode of cytotoxic action and molecular basis of 623 resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 67.Sies H. Glutathione and its role in cellular functions. Free Radic Biol Med. 1999;27:916–921. doi: 10.1016/S0891-5849(99)00177-X. [DOI] [PubMed] [Google Scholar]
- 68.Simon J, Bedalov A. Yeast as a model system for anticancer drug discovery. Nat Rev Cancer. 2004;4:1–8. doi: 10.1038/nrc1372. [DOI] [PubMed] [Google Scholar]
- 69.Stearman R, Yuan DS, Yamaguchi-Iwai Y, Klausner RD, Dancis A. A permease–oxidase complex involved in high-affinity iron uptake in yeast. Science. 1996;271:1552–1557. doi: 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- 70.Teixeira M, Monteiro P, Jain P, Tenreiro S, Fernandes A, Mira N, Alenquer M, Freitas A, Oliveira A, Sa-Correia I. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:D446–451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomas-Chollier M, Sand O, Turatsinze J, Janky R, Defrance M, Vervisch E, Brohe’e S, Helden J. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toyokuni S. Iron-induced carcinogenesis: the role of redox regulation. Free Radic Bio Med. 1996;20:553–566. doi: 10.1016/0891-5849(95)02111-6. [DOI] [PubMed] [Google Scholar]
- 73.Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ho A, Ward DM, Kaplan J. Transition metal transport in yeast. Annu Rev Microbiol. 2002;56:237–261. doi: 10.1146/annurev.micro.56.012302.160847. [DOI] [PubMed] [Google Scholar]
- 75.Loon AP, Pesold-Hunt B, Schatz G. A yeast mutant lacking mitochondrial manganese superoxide dismutase is hypersensitive to oxygen. Proc Natl Acad Sci USA. 1986;83:3820–3824. doi: 10.1073/pnas.83.11.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:308–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 77.Weinberg ED. Iron therapy and cancer. Kidney Int. 1999;55:S131–S134. doi: 10.1046/j.1523-1755.1999.055Suppl.69131.x. [DOI] [PubMed] [Google Scholar]
- 78.Wheate NJ, Collins JG. Multi-nuclear platinum drugs: a new paradigm in chemotherapy. Curr Med Chem - Anti-Cancer Agents. 2005;5:267–279. doi: 10.2174/1568011053765994. [DOI] [PubMed] [Google Scholar]
- 79.Wheate NJ, Taleb RI, Krause-Heuer AM, Cook RL, Wang S, Higgins VJ, Aldrich-Wright JR. Novel platinum(II)-based anticancer complexes and molecular hosts as their drug delivery vehicles. Dalton Trans. 2007;43:5055–5064. doi: 10.1039/b704973k. [DOI] [PubMed] [Google Scholar]
- 80.Yamaguchi-Iwai Y, Ueta R, Fukunaka A, Sasaki R. Subcellular localization Aft1 transcription factor responds to iron status in Saccharomyces cerevisiae. J Biol Chem. 2002;277:18914–18918. doi: 10.1074/jbc.M200949200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 30 kb)
(PDF 126 kb)