SUMMARY
In the differentiating alphaproteobacterium Caulobacter crescentus, organelle synthesis at cell poles is critical to forming different progeny after cell division. Coordination of polar organelle synthesis, including pili and holdfast, and flagellum ejection, is mediated in part by the scaffolding protein PodJ. At the time of cell division, PodJ undergoes regulated processing to a short form that persists at the flagellar pole of swarmer cells. This study analyzes how PodJ’s role in structural and signaling protein localization impacts organelle synthesis. A PodJ mutant with an internal deletion exhibits reduced sensitivity to pili-tropic phage ΦCbK, resulting from reduced pilA gene expression, which can be linked to altered signaling protein localization. The phage sensitivity defect of a ΔpodJ mutant can be partially suppressed by ectopic pilA expression. Induction of PodJ processing, by manipulation of podJ itself or controlled perP expression, resulted in decreased pilus biogenesis and, when coupled with a podJ mutation that reduced pilA expression, led to complete loss of phage sensitivity. As a whole, the results show that PodJ’s scaffolding role for structural and signaling proteins both contribute to flagellar pole organelle development.
INTRODUCTION
While many bacteria replicate by binary fission, a large number of bacteria replicate using complex life cycles, often employing differentiated cell types at different stages of the cell cycle. Cell division can be used to produce different progeny cell types by segregation of cellular structures and protein complexes such that when cell division occurs, each daughter cell receives different organelles and proteomes. This style of cellular differentiation is used in the developmental cycle of Caulobacter crescentus (for a review of C. crescentus development, see (Curtis & Brun, 2010)). The cell cycle of C. crescentus begins with a stalked cell, a cell with a thin extension of the cell envelope at one cell pole (the stalk) tipped with an adhesive polysaccharide organelle called the holdfast. The stalked cell elongates, becoming a predivisional cell. During this time, a flagellum is formed at the pole opposite the stalk. Just prior to cell division, flagellar rotation is activated, followed by extrusion of pili from that same pole after cell division. Cell division yields a daughter stalked cell that can reenter the replication cycle, and a motile swarmer cell that cannot replicate. After an obligatory period in the non-replicative phase, the swarmer cell differentiates, ejecting its flagellum, losing its pili, and synthesizing the holdfast and stalk at the previous flagellar cell pole.
Understanding the mechanisms of polar localization in C. crescentus is critical to understanding how development is coordinated not only in this organism, but many related bacteria as well. While many alphaproteobacteria do not display morphologically distinct cell types as seen for C. crescentus, it is becoming increasingly evident that the underlying mechanisms for differential cell division are conserved. For example, another alphaproteobacterium, Agrobacterium tumefaciens, produces two different daughter cells, one that is stationary (analogous to the C. crescentus stalked cell) and one that is motile (like the C. crescentus swarmer cell) (Kahng & Shapiro, 2001), and synthesizes a unipolar polysaccharide adhesin similar to the holdfast (Tomlinson & Fuqua, 2009). While C. crescentus exhibits zonal polar growth to synthesize a stalk (Brown et al., 2011), A. tumefaciens exhibits zonal polar growth that results in the bulk of cell elongation emanating from a single pole (Brown et al., 2012). Additionally, key C. crescentus developmental proteins such as CtrA, CcrM, DivK and PleC are conserved throughout the alphaproteobacteria (Bellefontaine et al., 2002, Hallez et al., 2007, Lam et al., 2003, Robertson et al., 2000, Brassinga et al., 2002, Brilli et al., 2010).
One key protein involved in polar organelle development in C. crescentus is PodJ (Wang et al., 1993). PodJ is a large protein (974 amino acids), with a single transmembrane domain causing the C-terminal third of the protein to reside in the periplasm (Crymes et al., 1999, Hinz et al., 2003, Viollier et al., 2002b). PodJ is produced in stalked cells and localizes to the pole opposite the stalk. Given that PodJ has no demonstrable enzymatic activity, and that it contains a number of protein-protein interaction domains (Figure 1A, (Lawler et al., 2006)), including three predicted coiled-coil domains in the cytoplasmic portion and three predicted tetratricopeptide repeat domains (as well as a putative peptidoglycan binding domain) in the periplasmic portion, it is likely that PodJ serves a scaffolding role. In fact, it has been shown that PodJ is necessary for localization of the developmentally important histidine kinase PleC to the flagellar pole (Hinz et al., 2003, Viollier et al., 2002b), as well as pilus assembly and secretion proteins CpaE and CpaC (Viollier et al., 2002a), holdfast attachment proteins HfaDAB (Hardy et al., 2010), and the cyclic-di-GMP effector protein PopA (Duerig et al., 2009). PodJ is largely conserved throughout the alphaproteobacteria, and homologs almost always contain the C-terminal TPR repeats, while conservation of the coiled-coil and peptidoglycan binding domains are variable.
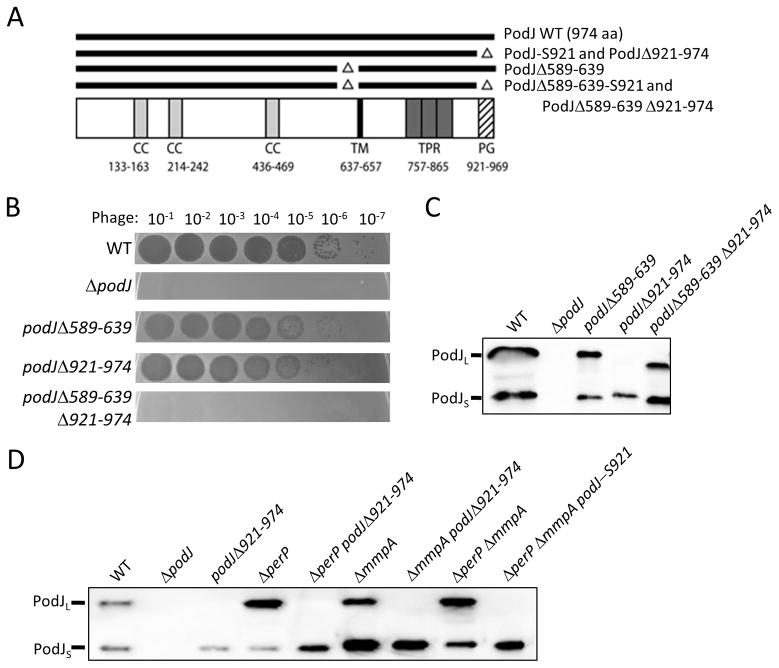
Figure 1.
Two deletions in podJ abolish phage sensivity. A: Schematic of PodJ, with predicted coiled coil (CC), transmembrane (TM), tetratricopeptide repeat (TPR) and peptidoglycan binding (PG) domains indicated. The numbers above the predicted domain indicate the amino acid region for each domain. Important mutants used in this study are depicted as lines above the schematic. B: Phage sensitivity assays of various podJ mutants. Cell cultures were OD600 normalized then mixed with soft agar and plated on agarose plates. After hardening, serial dilutions of bacteriophage ΦCbK were spotted onto the plates and allowed to dry prior to incubation. C: Western analysis of various podJ mutants. Western analysis was performed using whole cell lysates from OD600 normalized cell cultures and anti-sera raised against the cytoplasmic portion of PodJ to show PodJL and PodJS. D: Western analysis of the fast processing podJΔ921-974 allele in combination with protease mutants. Western analysis was performed using whole cell lysates from OD600 normalized cell cultures and anti-sera raised against the cytoplasmic portion of PodJ to show PodJL and PodJS. ΔperPΔmmpA podJ-S921 used a stop codon at the amino acid 921 codon to remove the C-terminal 53 amino acids of PodJ and results in the synthesis of the same protein as the podJΔ921-974 mutant.
PodJ is synthesized and localized in stalked cells as a full-length protein (long form, PodJL). Coincident with cell division, PodJ is processed to a short form (PodJS) by removal of the periplasmic portion of the protein (Viollier et al., 2002b, Hinz et al., 2003). PodJS persists throughout the swarmer cell phase. Upon swarmer cell differentiation, PodJS is cleared from the cell prior to re-synthesis of PodJ in the stalked cell. Major regulation of PodJL to PodJS proteolysis is performed at the time of cell division by the periplasmic protease PerP as perP mutants accumulate PodJL (Chen et al., 2006). PerP is positively regulated by CtrA. CtrA is a two component transcriptional regulator that regulates 26% of all cell cycle-regulated genes (Laub et al., 2000). CtrA is subject to complex regulation (see (Curtis & Brun, 2010) for an in-depth discussion). In the predivisional cell, two histidine kinases are localized at opposite ends of the cell. DivJ is localized at the stalked pole, while PleC is localized at the flagellar pole. Both histidine kinases act on the single domain response regulator, DivK, but in opposite fashion. DivJ phosphorylates DivK, whereas PleC desphosphorylates it. DivK diffuses between the cell poles, becoming bipolarly localized and in the process alternates phosphorylation states and allows the cell poles to communicate. Cytokinesis isolates DivJ and PleC activities from one another, leading to rapid dephosphorylation of DivK in the nascent swarmer cell. Non-phosphorylated DivK promotes activation of CtrA (Tsokos et al., 2011). Thus compartmentalization of the predivisional cell leads to increased amounts of activated CtrA, which leads to expression of PerP in the swarmer cell compartment and initiation of PodJ processing. This explains why PodJ processing is coincident with cell division. However, in a perP mutant, some processing of PodJL to PodJS persists, indicating redundancy in the processing system, and making it difficult to determine the biological purpose of PodJ processing. Clearing of PodJS during swarmer cell differentiation is thought to be regulated by the membrane metalloprotease MmpA as mutants in this protease accumulate PodJS, and PodJS is not removed during the swarmer to stalked cell transition (Chen et al., 2005).
podJ mutants, in addition to having delocalized PleC, do not produce holdfast or pili, do not eject the flagellum efficiently, and display reduced motility in low percentage agar plates (although they are observed to swim by microscopy) (Hinz et al., 2003, Viollier et al., 2002b, Crymes et al., 1999). Because PodJ is tied to the localization of so many proteins and involved in so many processes, the mechanistic reasons behind the phenotypes of podJ mutants have been difficult to determine and the larger role that PodJ plays in polar development is not well understood. The work presented here uses different mutations in podJ to isolate the deficiencies that contribute to the pilus and holdfast synthesis defect. In addition, various roles for PodJ in scaffolding and signaling are determined, demonstrating that PodJ is integrated into polar developmental pathways in multiple ways and highlighting its importance in coordinating polar development processes into a cohesive and efficient whole.
RESULTS
The podJΔ589-639Δ921-74 mutant has altered phage resistance
A ΔpodJ mutant is deficient in pili synthesis (Hinz et al., 2003, Viollier et al., 2002b). Expression of the pilus subunit gene pilA and localization of the pili assembly factor CpaE are impaired in a ΔpodJ mutant (Lawler et al., 2006, Viollier et al., 2002b). However, reduced pilA expression is also found in mutants lacking PleC (Viollier et al., 2002a), whose localization depends on PodJ (Lawler et al., 2006, Hinz et al., 2003, Viollier et al., 2002b), it remained unclear whether the defects of a ΔpodJ mutant are due to the scaffolding role of PodJ, to a potential signaling role, or both. To begin identifying the reasons for the pilus synthesis defect in ΔpodJ, mutations were sought within podJ that had effects on pilus biogenesis. Sequence analysis led to the identification of a fifty amino acid region (from amino acids 589 to 639, directly preceding the transmembrane domain) that had 12 lysine or arginine residues, indicating an unusual charge density and a feature of the protein worthy of investigation. A mutant was constructed such that the coding region for these amino acids was deleted (Figure 1A). Additionally, previous analysis had shown that deletion of the region encoding the putative peptidoglycan binding domain (amino acids 921 to 974, see Fig. 1A) caused a reduction in the ability of that mutant to support the reproduction of bacteriophage ΦCbK (Lawler et al., 2006), which infects C. crescentus via the pilus (Chen et al., 2006, Hinz et al., 2003, Skerker & Shapiro, 2000). The two individual mutations (podJΔ589-639 and podJΔ921-974) were also combined into the same strain to create podJΔ589-639Δ921-974. The mutant strains, as well as control strains wild-type and ΔpodJ, were then assessed for ΦCbK phage resistance as an indicator of pilus production, by plating cells in soft agar and applying serial dilutions of phage to the agar surface (Figure 1B). After incubation, areas of infection had no cell growth (as seen in wild-type), while mutants resistant to the phage had confluent cell growth (as seen with ΔpodJ). Both the podJΔ589-639 and podJΔ921-974 mutants had an approximately 10-fold reduction in phage sensitivity compared to wild-type, particularly noticeable in the 10−5 phage dilution spot. Interestingly, the double mutant was resistant to the phage and was virtually identical to the ΔpodJ control.
To determine if the lack of phage sensitivity in podJΔ589-639Δ921-974 was due to altered PodJ stability, western blot analysis was performed on the strains using antibody raised against the N-terminal portion of PodJ. This antibody detects both PodJL and PodJS. As seen in the western blot (Figure 1C), both PodJL and PodJS are detectable in wild-type whole cell lysates, while no protein is detectable in ΔpodJ. Both PodJL and PodJS were detected in podJΔ589-639, but with a slight downshift in size due to the internal deletion. Only PodJS was detectable in podJΔ921-974, consistent with previous reports that deletion of the putative peptidoglycan binding domain leads to rapid processing of the protein ((Lawler et al., 2006), Supplemental Figures S2–3). In fact, the Δ921-974 mutation induces processing independent of the normal processing proteases, as PodJΔ921-974 is still rapidly processed in the ΔperP, ΔmmpA, and ΔperP ΔmmpA mutant backgrounds (Figure 1D).
It is thought that the fast processing of the PodJΔ921-974 mutant is what gives rise to the reduction in phage sensitivity and presumed reduction in pilus biogenesis. Processing is essentially the removal of the PodJ periplasmic domain. The phenotype of a transposon insertion in the periplasmic domain coding region of podJ implicated that domain in pilus biogenesis (Viollier et al., 2002b). Additionally, when stop codons were placed in podJ that resulted in increasingly longer truncations of the protein from the C terminus, stop codons which removed large portions of the periplasmic domain had no discernable effect on holdfast synthesis or swarming motility, but did cause an increase in resistance to infection by ΦCbK (Lawler et al., 2006). It should also be noted that many of the stop codon mutations that caused truncations in the PodJ periplasmic domain led to immediate PodJL processing, and that the degree of proteolytic processing was often correlated with the degree of disruption of ΦCbK infection (Lawler et al., 2006). Surprisingly, even though the podJΔ589-639Δ921-974 mutant contains the Δ921-974 mutation, which normally causes rapid processing, it has a strong PodJL signal (Figure 1C). This result indicates that the Δ589-639 mutation slows down the fast-processing phenotype of the Δ921-974 mutation. If the rapid processing of PodJΔ921-974 increases phage resistance, then the podJΔ589-639Δ921-974 mutant should be less resistant than podJΔ921-974 because the Δ589-639 mutation suppresses the fast processing of PodJΔ921-974; however, the opposite result was obtained. podJΔ589-639Δ921-974 is more phage resistant than podJΔ921-974 and is, in fact, nearly as phage resistance as the podJ null mutant. The Δ589-639 and Δ921-974 mutations are additive in terms of phage resistance, but antagonistic in terms of PodJ processing.
Induction of PodJ processing causes reduced pilus biogenesis in wild-type and podJΔ589-639
To try to resolve the puzzling discrepancy between the pili synthesis and PodJ processing phenotypes of the podJΔ589-631 Δ921-974 mutant, we first investigated the basis of its slow processing phenotype. Since the deleted 589-631 amino acids lie in the cytoplasmic domain of PodJ, we reasoned that this deletion was unlikely to impact processing of the periplasmic domain for a structural reason but was more likely to reduce processing for a regulatory reason. We therefore tested the effect of the ΔpodJ and of the podJΔ589-631 mutation on expression of the processing protease gene perP and found that it was significantly reduced in both mutants (Figure 2A), suggesting that the slower PodJ processing in the podJΔ598-631 mutant is due to lower PerP level. To test this hypothesis, perP was introduced ectopically at the xyl locus under the control of xylose and the effect of perP induction was analyzed in wild-type and in the podJΔ598-631 mutant. Western blot analysis showed that following 4 hours of perP induction, no PodJL was detectable in a wild-type background, and there was a significant reduction of PodJL in the podJΔ589-639 mutant (Figure 2B). Induction of perP expression in a wild-type background increased phage resistance by roughly two orders of magnitude, while in podJΔ589-639 phage sensitivity was virtually abolished despite the fact that a significant level of PodJL was still present (Figure 2C). The same alterations in phage sensitivity were obtained when the analysis was performed with strains carrying the PpilA-lacZ construct (data not shown) used later to assess the expression of pilA in these conditions. These results indicate that the podJΔ589-631 mutation reduces PodJ processing by impacting the expression of PerP, although this is not the only reason. These results agree with the previous study of Chen et al. (Chen et al., 2006) where induction of PerP synthesis led to a loss of wild-type PodJL detection and correlated loss of pilus biogenesis.
Figure 2.
perP expression in podJ mutants and analysis of the effect of ectopic expression of perP on PodJ processing and pilus biogenesis. A: Gene expression of perP was measured using β-galactosidase assays of exponentially growing cultures in wild-type, ΔpodJ, and podJΔ589-639 strains. B: Overnight wild-type and podJΔ589-639 with and without Pxyl-perP cultures were diluted to OD600 = ~0.2 and allowed to grow for 4 hours under non-inducing (0.3% glucose; (−)) or inducing (0.3% xylose; (+)) conditions. Cultures were normalized to the same OD600 and whole cell lysates were prepared from cell cultures and subjected to Western blot analysis using PodJ anti-sera raised against the cytoplasmic portion of the protein. C: Cultures from B with the Pxyl-perP construct were used to inoculate molten soft agar with antibiotic and appropriate sugar and poured onto plates containing antibiotic and appropriate sugar. The agar was allowed to harden and 10-fold dilutions of phage ΦCbK were spotted onto the surface and allowed to dry prior to incubation. D: β-galactosidase assays were performed on cell cultures with the Pxyl-perP construct as previously described. E: Whole cell lysates with the Pxyl-perP construct were separated on Tricine SDS-PAGE gels and subjected to Western blot analysis using anti-PilA anti-sera. Three separate blots were performed using independent cell cultures. F: Band intensities of PilA Western blots were measured using ImageJ software. Each density was normalized to a percentage of the wild-type control (without Pxyl-perP) on each blot and the results from the three blots were averaged.
Ectopic expression of the periplasmic protease PerP could lead to reduced pilus biogenesis not through any effect on PodJ, but by altering gene expression indirectly, or by degrading the pilus biogenesis machinery directly. This hypothesis was tested by monitoring the expression and stability of various pilus components in response to ectopic PerP expression. pilA gene expression was unperturbed by perP overexpression in the wild-type or the podJΔ589-639 background (Figure 2D), and minor effects on PilA protein levels (Figure 2E–F). Induction of perP expression also had no effect on the stability or abundance of pilus biogenesis protein CpaE (data not shown).
Induction of PodJL processing, either by introduction of the podJΔ921-974 mutation or by induction of PerP, results in an increase in phage resistance in both the wild-type (elimination of PodJL, see Figure 1B–C, Figure 2B–C) and in the slow processing podJΔ589-631 background (decrease in the PodJL/PodJS ratio, see Figure 1B–C, Figure 2B–C). Surprisingly, induction of PodJL processing has a more severe effect in the podJΔ589-631 background than in the wild-type background, causing near complete phage resistance despite the fact that the PodJL form can still be detected, albeit at a reduced level. These results suggest that the podJΔ589-631 deletion impacts signaling. In the following sections, we analyze the effect of the podJ mutations on gene expression and protein localization in order to decipher the basis for these phenotypes.
podJΔ589-639 has predominantly mis-localized cpaE but correctly positioned pili
Thus far, pilus biogenesis in the various strains has been inferred from phage sensitivity assays. Pilus biogenesis in the mutants was investigated more directly by a number of means. First, localization of the pilus biogenesis protein CpaE was detected by GFP-labeling. CpaE has been previously shown to localize to the flagellar pole in predivisional cells by this method (Viollier et al., 2002b). Its localization was also shown to be dependent on PodJ as no fluorescent focus could be detected in ΔpodJ cells. A gfp-cpaE gene was introduced in each strain by allelic replacement and visualized by fluorescence microscopy (Figure 3A). As previously reported, CpaE was predominantly located at the flagellar pole in wild-type predivisional cells (61 ± 11% of all predivisional cells, Figure 3A, Table 1). In contrast to previously reported results, a fluorescent CpaE focus was detectable in ΔpodJ cells. It should be noted that, like others, we had previously been unable to detect CpaE foci in ΔpodJ until recent upgrades to our microscope’s sensitivity. Interestingly, the foci in ΔpodJ were almost exclusively found at the stalked pole with 26 ± 6% at the stalked pole compared to 3 ± 3% at the flagellar pole (Table 1). Foci in podJΔ921-974 were predominantly found at the flagellar pole (27 ± 6%) but with an elevated incidence of bipolar localization (12 ± 4%) compared to other strains, indicating that the lack of a periplasmic domain does not significantly impact CpaE localization. CpaE was mostly located at the stalked pole in podJΔ589-639 (17 ± 11% stalked versus 5 ± 2% flagellar) and in podJΔ589-639Δ921-974 (13 ± 7% stalked versus 4 ± 1% flagellar); these mutants also exhibited a strong decrease in the number of cells with a CpaE focus as compared to wild-type. The reduction in detectable CpaE foci in the mutants does not appear to be the result of decreased protein synthesis as CpaE protein levels appear the same in all strains (Figure 3B). The results indicate that the PodJ589-639 region plays an important role in CpaE localization, yet the podJΔ589-639 mutant is still sensitive to phage whereas the podJ deletion mutant is resistant.
Figure 3.
Molecular analysis of pilus production. A: Localization of pili and pilus components. CpaE was visualized by GFP labeling; gfp-cpaE replaced cpaE in the chromosome. Numbers are average and standard deviation of predivisional cells with only flagellar pole CpaE localization (% Flag). Pili were visualized by adsorption of fluorescently labeled ΦCbK to cells (Fluo ΦCbK) or by immunofluorescence microscopy using antisera raised against Pilin followed by detection using FITC-conjugated secondary antibody (PilA (IMF)). Numbers are the average and standard deviation of percentage of swarmer cells labeled (% Label). B: Western analysis of gfp-cpaE strains. Western analysis was performed using whole cell lysates from OD600 normalized cell cultures and an anti-GFP monoclonal antibody.
Table 1.
Localization of developmental proteins in podJ mutants
| Strain | % Flagellar pole only* | % Stalked pole only* | % Bipolar* | % No signal detected | Total cells counted |
|---|---|---|---|---|---|
| CpaE: | |||||
| Wild-type | 61 ± 11 | 0 ± 0 | 1 ± 1 | 38 ± 11 | 1333 |
| ΔpodJ | 3 ± 3 | 26 ± 6 | 4 ± 2 | 67 ± 8 | 1228 |
| podJΔ589-639 | 5 ± 2 | 17 ± 11 | 4 ± 1 | 73 ± 14 | 1502 |
| podJΔ921-974 | 27 ± 6 | 7 ± 1 | 12 ± 4 | 55 ± 6 | 1306 |
| podJΔ589-639 Δ921-974 | 4 ± 1 | 13 ± 7 | 3 ± 2 | 81 ± 8 | 1333 |
| PleC: | |||||
| Wild-type | 44 ± 5 | 0 ± 0 | 0 ± 0 | 56 ± 5 | 2357 |
| ΔpodJ | 3 ± 1 | 9 ± 2 | 2 ± 1 | 86 ± 4 | 1203 |
| podJΔ589-639 | 16 ± 6 | 2 ± 2 | 1 ± 1 | 81 ± 9 | 900 |
| podJΔ921-974 | 36 ± 6 | 0 ± 0 | 1 ± 0 | 63 ± 6 | 853 |
| podJΔ589-639 Δ921-974 | 3 ± 1 | 5 ± 1 | 1 ± 1 | 91 ± 2 | 799 |
| DivL: | |||||
| Wild-type | 40 ± 9 | 3 ± 1 | 10 ± 6 | 47 ± 16 | 912 |
| ΔpodJ | 3 ± 1 | 30 ± 3 | 24 ± 4 | 43 ± 2 | 894 |
| podJΔ589-639 | 14 ± 0 | 13 ± 0 | 28 ± 0 | 45 ± 1 | 822 |
| podJΔ921-974 | 17 ± 1 | 11 ± 1 | 24 ± 0 | 48 ± 3 | 1190 |
| podJΔ589-639 Δ921-974 | 6 ± 3 | 23 ± 3 | 23 ± 4 | 48 ± 4 | 1241 |
| DivK: | |||||
| Wild type | 0 ± 0 | 12 ± 9 | 56 ± 4 | 32 ± 11 | 887 |
| ΔpodJ | 0 ± 0 | 39 ± 14 | 20 ± 3 | 42 ± 17 | 1299 |
| podJΔ589-639 | 0 ± 0 | 33 ± 14 | 24 ± 21 | 43 ± 21 | 901 |
| podJΔ921-974 | 0 ± 0 | 18 ± 9 | 39 ± 14 | 43 ± 21 | 899 |
| podJΔ589-639 Δ921-974 | 0 ± 0 | 40 ± 11 | 23 ± 12 | 38 ± 12 | 818 |
| CckA: | |||||
| Wild-type | 0 ± 0 | 32 ± 9 | 49 ± 1 | 19 ± 9 | 898 |
| ΔpodJ | 0 ± 0 | 56 ± 11 | 22 ± 5 | 23 ± 7 | 818 |
| podJΔ589-639 | 0 ± 0 | 49 ± 10 | 26 ± 10 | 26 ± 12 | 672 |
| podJΔ921-974 | 0 ± 0 | 42 ± 13 | 31 ± 19 | 27 ± 11 | 739 |
| podJΔ589-639 Δ921-974 | 0 ± 0 | 49 ± 5 | 26 ± 9 | 25 ± 5 | 841 |
Percentages are derived from number of cells exhibiting indicated fluorescent localization out of total predivisional cells counted (including unlabeled cells). Numbers are averages and standard deviations based on three independent trials.
The fact that CpaE mislocalizes to the stalked pole in portions of the populations for podJΔ589-639 and podJΔ921-974, and that these strains are sensitive to phage, brings up the intriguing possibility that these strains produce pili at the wrong pole. To address this possibility, cells of all strains were exposed to fluorescently labeled ΦCbK for 15 minutes prior to visualization by fluorescence microscopy. This technique has been used successfully to visualize phage binding to swarmer cells (Skerker & Shapiro, 2000, Lawler et al., 2006, Hinz et al., 2003). While wild-type swarmer cells were efficiently labeled (Figure 3A), no other strain demonstrated labeling above that of ΔpodJ, even strains that were sensitive to the phage. These results indicate that fluorescent phage labeling is not a sensitive method for the assessment of pili synthesis.
We used immunofluorescence microscopy with an anti-pilin antibody as an alternate method of detecting pili localization (Figure 3A). Pili were readily detected in phage sensitive wild-type cells, but not in the phage-resistant mutants ΔpodJ or podJΔ589-639Δ921-974. Pili were also detected cells of phage-sensitive mutants podJΔ589-639 and podJΔ921-974, indicating that these strains produce pili. These results are consistent with the phage sensitivity plaque assay results. We did not detect pili at the base of stalks (the position of mislocalized CpaE and most likely position of ectopic pili formation). These results suggest that pili are only formed at the flagellar pole, and that strains that have mislocalized CpaE only produce pili in the subpopulation of cells with correctly localized CpaE. Whether correct CpaE localization is the sole determining factor of pili formation is unknown.
ΔpodJ and podJΔ589-639Δ921-974 pilus deficiencies can be suppressed by ectopic pilA expression
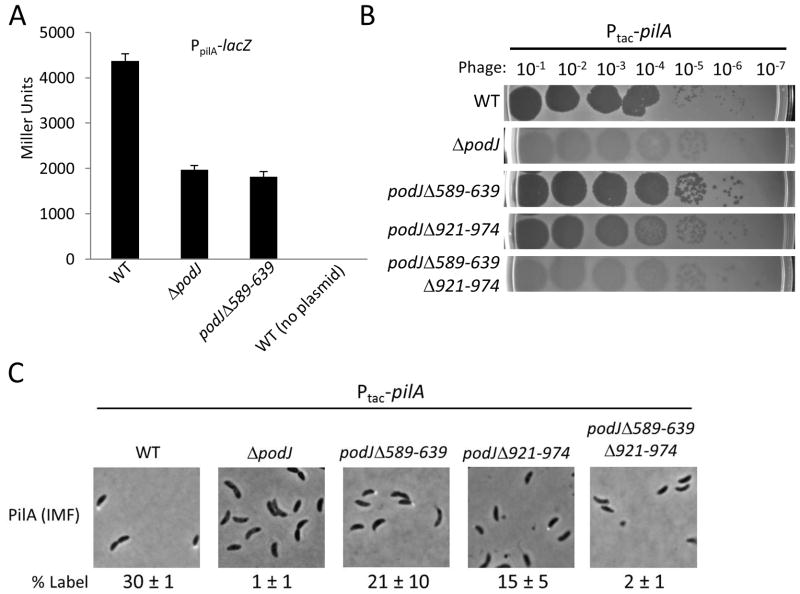
Expression of pilA is controlled by the master regulator of C. crescentus development: CtrA (Skerker & Shapiro, 2000, Gora et al., 2010). In a previous section, we showed that the transcription of perP, another gene activated by CtrA (Chen et al., 2006), is reduced in ΔpodJ and podJΔ589-639. We therefore tested if pilA transcription was altered in these mutants. β-galactosidase assays using a PpilA-lacZ fusion showed that pilA gene expression is reduced greater than two-fold in both ΔpodJ and podJΔ589-639 (Figure 4A) compared to wild-type.
Figure 4.
Effect of podJ mutations on pilA gene expression and suppression of pilus biogenesis defects by constitutive pilA expression. A: Gene expression of pilA was measured using β-galactosidase assays of exponentially growing cultures in wild-type, ΔpodJ, and podJΔ589-639 strains. B: Phage sensitivity assays were performed with strains containing constitutively expressed pilA (Ptac-pilA). Cell cultures were OD600 normalized then mixed with soft agar containing antibiotic and poured onto agarose plates with antibiotic. After hardening, serial dilutions of bacteriophage ΦCbK were spotted onto the plates and allowed to dry prior to incubation. C: Immunofluorescence microscopy was performed on strains carrying Ptac-pilA constructs using antisera raised against Pilin followed by detection using FITC-conjugated secondary antibody. Numbers are the average and standard deviation of percentage of swarmer cells labeled (% Label).
The above results suggest that the reduced phage sensitivity of podJ mutants could be partly due to reduced pilA expression. We therefore tested whether constitutively expressing pilA could suppress the phage sensitivity defect of podJ mutants. Driving pilA expression from the constitutive Ptac promoter completely suppressed the phage sensitivity defect of the podJΔ589-639 mutant (Figure 4B); it is as sensitive to phage as wild-type in this assay, as compared to Figure 1B where podJΔ589-639 has reduced sensitivity compared to wild-type in that assay. Constitutive pilA expression also partially rescued the defect of the podJΔ921-974 mutant (Figure 4B), and even slightly suppressed the phage sensitivity defect in the ΔpodJ and podJΔ589-639Δ921-974 mutants. Plaques could be seen at phage dilutions similar to wild-type, however they were very cloudy from cell growth, suggesting that increased pilA gene expression could only partly suppress the phage sensitivity defect of these strains.
Since CpaE is misslocalized in the podJ mutants, we used immunofluorescence of pilin to determine if suppression of their phage sensitivity defect was due to properly localized pili synthesis (Figure 4C). Localization of pili could be detected on swarmer cells in wild-type, podJΔ589-639 and podJΔ921-974 as before. No pili localization could be detected in ΔpodJ and podJΔ589-639Δ921-974. No stalked pole fluorescence could be observed, indicating that even with PilA overexpression, pili only form at the flagellar pole. Given the suppression of the phage sensitivity defect of the ΔpodJ and podJΔ589-639Δ921-974 mutants, it was surprising that immunofluorescence was unable to detect pili in the suppressed strains. It is possible that the pilus filaments formed by Ptac-pilA suppression in these strains are too small to detect but still allow some phage infection, or are very long and easily sheared from the cell during immunofluorescence preparation, or there are relatively few pili-producing cells in the population to be labeled in the assay, which would be consistent with the low level of suppression.
The ΔpodJ and podJΔ589-639 mutations alter PleC and DivK localization
Results described in the previous section indicate that two CtrA-regulated genes, pilA and perP, have the same approximate two-fold reduction in expression in the ΔpodJ and podJΔ589-639 mutants, suggesting that CtrA activity is partially compromised in this strain. Although theΔ921-974 mutation bypasses PerP and MmpA for processing, the reduction of perP expression could be contributing to the lack of fast processing in podJΔ589-639Δ921-974. We further investigated the reason for the reduced perP and pilA gene expression in Δ589-639 containing strains. One plausible explanation for a potential reduction in CtrA activity in podJΔ589-639 is that this region may be necessary for proper PleC activity. PodJ is required for PleC localization to the flagellar pole (Viollier et al., 2002b, Lawler et al., 2006). It is possible that PodJ is necessary not just for PleC localization, but proper PleC activity as well. Reduction in PleC activity would lead to increased DivK phosphorylation, which would then lead to decreased CtrA activation. To investigate this hypothesis, a pleC-gfp construct on a replicating plasmid was placed in each strain, and resultant strains were analyzed by fluorescence microscopy (Figure 5A, Table 1). As previously reported, PleC is mostly localized to the flagellar pole in wild-type cells (44 ± 5%), while in ΔpodJ there is a severe reduction in localized PleC, with the majority of localized protein found at the stalked pole (9 ± 2% stalked pole compared to 3 ± 1% flagellar pole). PleC localization in podJΔ921-974 is close to wild-type level (36 ± 6% flagellar pole) indicating processing has little effect on PleC localization. Both the podJΔ589-631 and podJΔ589-639Δ921-974 mutations cause a strong decrease in the number of cells with polarly localized PleC, although PleC is still mostly localized to the flagellar pole in podJΔ589-639 in the few cells with polar PleC (16 ± 6% flagellar pole, 2 ± 2% stalked pole). This reduction in localized PleC may lead to partial reduction in CtrA activity that could cause a reduction in pilA and perP gene expression.
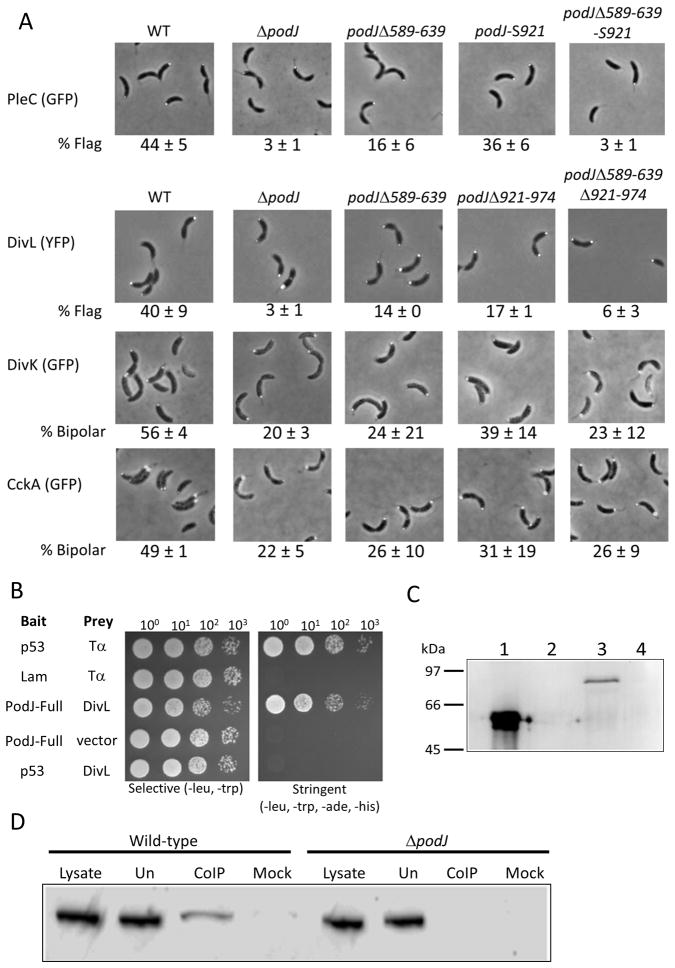
Figure 5.
PodJ mutations affect signaling protein localization. A: Localization of signaling proteins in PodJ strains. PleC was visualized using plasmid encoded pleC-gfp. podJ-S921 and podJΔ589-639-S921 strains used a stop codon at the amino acid 921 codon to remove the C-terminal 53 amino acids of PodJ and results in the synthesis of the same protein as the podJΔ921-974 mutant. Numbers are average and standard deviation of predivisional cells with only flagellar pole PleC localization (% Flag). DivL was visualized using chromosomally-encoded divL-yfp under control of the native promoter. Numbers are average and standard deviation of predivisional cells with flagellar pole only DivL localization (% Flag). DivK was visualized using plasmid encoded divK-gfp. Numbers are average and standard deviation of predivisional cells with bipolar DivK localization (% Bipolar). CckA was visualized using plasmid encoded cckA-gfp. Numbers are average and standard deviation of predivisional cells with bipolar CckA localization (% Bipolar). B: Yeast two-hybrid results of PodJ and DivL interactions. Growth on the selective plate requires presence of both bait and prey vectors while growth on the stringent plate requires interaction between the bait and prey protein fusions. p53 and Tα serve as a positive control for interaction while Lam and Tα are a negative control for interaction. C: Far-western analysis of PodJ-DivL interaction. Lane 1 – purified PodJ cytoplasmic domain. Lane 2 – E. coli whole cell lysate with plasmid expression FtsZ-intein. Lane 3 – purified DivL-His. Lane 4 – E. coli whole cell lysate with empty vector. The blot was probed with purified PodJ cytoplasmic domain followed by incubation with anti-PodJ antibody, goat-anti-rabbit secondary antibody and finally visualization. D: In vivo co-immunoprecipitation. Lysate from strains expressing DivL-YFP were incubated with anti-PodJ antibody. Antibody complexes were collected using Protein A agarose beads and collected co-immunoprecipitate was probed using anti-GFP antibody which also recognizes YFP. Lysate – whole cell lysate of strain. Un – lysate unbound to Protein A agarose beads. Co-IP – co-immunoprecipitate eluted from the beads. Mock – co-immunoprecipitate eluted from beads from a control procedure where anti-PodJ antibody had been omitted.
PodJ recruits DivL to the flagellar pole through protein-protein interaction
The pattern of DivL localization (Sciochetti et al., 2005) is consistent with the possibility that it is recruited to the incipient flagellar pole by PodJ. The localization of DivL in various podJ mutants was analyzed using a divL-yfp allele as the sole functional copy of the gene and under control of the native promotor. In wild-type, DivL localizes predominantly to the flagellar pole (40 ± 9%, Figure 5A, Table 1) with little stalked pole localization (3 ± 1%) and some bipolar localization (10 ± 6%). The ΔpodJ mutant loses flagellar pole localization of DivL (3 ± 1%) and localizes it mostly at the stalked pole (30 ± 3%), however it also has significant bipolar localization (24 ± 4%), indicating that PodJ may not be the sole flagellar pole determinant for DivL localization. It had been previously shown that DivL is delocalized in a divJ mutant (Sciochetti et al., 2005), suggesting a DivJ-dependent DivL localization factor. The podJΔ589-639Δ921-974 mutant resembles ΔpodJ, with virtually no flagellar pole localization of DivL (6 ± 3%) but with nearly equal amounts of stalked and bipolar DivL localization (24 ± 3% and 24 ± 4% respectively). podJΔ589-639 has some flagellar pole (14 ± 0%) and stalked pole (13 ± 0%) localization of DivL, but it is predominantly bipolarly localized in this strain (28 ± 0%). Interestingly, similar results were obtained for DivL localization in podJΔ921-974 (17 ± 1%, flagellar pole, 11 ± 1% stalked pole and 24 ± 0% bipolar). This result suggests that the periplasmic domain of PodJ may play a part in DivL localization.
A direct interaction between PodJ and DivL was investigated by a variety of means. First, a yeast two-hybrid screen was performed for proteins that interact with full length PodJ using a genomic prey library. From this screen, PodJ-Bait was shown to interact robustly with a DivL-Prey library plasmid that contained the C-terminal portion of DivL from amino acids 470 to the end of DivL (Fig. 5B); this portion of DivL contains part of the PAS domain as well as full histidine phosphotransfer and ATPase domains. To verify this interaction, the full length PodJ was subcloned into the prey plasmid and the DivL fragment was subcloned into the bait plasmid, co-transformed into yeast or transformed with the appropriate negative control plasmid for bait or prey. An interaction between PodJ-Prey and DivL-Bait was observed while neither plasmid alone allowed yeast to grow on stringent media when present with the negative control plasmids (DivL-Bait and PodJ-Prey, data not shown). A Far Western blotting experiment in which the cytoplasmic portion of PodJ was used to probe a Western blot containing DivL confirmed that the two proteins interact (Fig. 5C, lane 3). PodJ did not interact with FtsZ-intein or with proteins from a whole cell lysate of E. coli containing the empty pET22b vector (Figure 5C, lanes 2 and 4). In addition to these in vitro methods, PodJ and DivL were also shown to interact using co-immunoprecipitation from C. crescentus (Figure 5D). We conclude that PodJ recruits DivL to the incipient flagellar pole of predivisional cells by direct protein-protein interaction.
Compromised PleC activity leads to decreased pilA and perP gene expression in the ΔpodJ mutants
PleC affects CtrA activation by dephosphorylating the response regulator DivK, which is phosphorylated by DivJ. Phosphorylated DivK inhibits CtrA activation by binding to the tyrosine kinase DivL, which prevents DivL from binding to the hybrid histidine kinase CckA (Tsokos et al., 2011). Lack of DivK phosphorylation allows DivL to bind to CckA and promote its kinase activity, which causes phosphorylation of the histidine phosphotransfer protein ChpT, which then phosphorylates CtrA. Phosphorylated DivK, which binds to DivL at the flagellar pole of predivisional cells, can be visualized using DivK-GFP (Jacobs et al., 2001, Lam et al., 2003). In a pleC mutant, DivK becomes hyperphosphorylated and is more bipolarly localized than in wild-type (Jacobs et al., 2001). In podJ strains where PleC activity is compromised (ΔpodJ, podJΔ589-639, podJΔ589-639Δ921-974) DivK should also become hyperphosphorylated and more bipolarly localized. However, as DivL is the flagellar pole DivK anchor and DivL flagellar pole localization is perturbed in these strains, it is possible that less DivK flagellar pole localization will be observed. A divK-gfp construct on a replicating plasmid was introduced in each strain, and DivK localization was visualized by fluorescence microscopy (Figure 5A, Table 1). DivK was less abundant at the flagellar pole in ΔpodJ, podJΔ589-639 and podJΔ589-639Δ921-974. Wild-type and podJΔ921-974 showed 56 ± 4% and 39 ± 14% bipolar localization respectively, while ΔpodJ had 20 ± 3% bipolar localization, podJΔ589-639 24 ± 21% bipolar localization and podJΔ589-639Δ921-974 23 ± 12% bipolar localization. These results are consistent with less flagellar pole DivL localization.
DivL and DivK affect activation of CtrA through CckA. Non-phosphorylated DivK cannot bind to DivL, which allows DivL to bind to CckA and increase its kinase activity, leading to CtrA activation (Tsokos et al., 2011, Iniesta et al., 2010). DivL binding to CckA causes CckA to form a focus at the flagellar pole of predivisional cells, which can be detected by fluorescence microscopy. Therefore, localization of CckA to the flagellar pole is an indication of its activity. Many tagged constructs of CckA form constitutive stalk pole foci, so determination of CckA activity can be simplified to a comparison of stalked pole versus bipolar localization (Angelastro et al., 2010, Iniesta et al., 2010). CckA localization was evaluated in the podJ mutants using a GFP-labeled construct on a replicating plasmid (Figure 5A, Table 1). We scored localization of CckA in predivisional cells to facilitate the identification of the flagellar pole. We found that 49 ± 1% of wild-type predivisional cells exhibited bipolar CckA localization (Table 1). CckA localization in podJΔ921-974 was highest of the mutant strains, with 31 ± 19% bipolar localization, compared to ΔpodJ, podJΔ589-639 and podJΔ589-639Δ921-974 with 22 ± 5%, 26 ± 10% and 26 ± 9% bipolar localization in predivisional cells respectively. Conversely, the latter three strains strains showed an increased incidence of stalked pole-localized or non-localized CckA, indicating that flagellar pole CckA localization is at least partially compromised, likely from reduced DivL localization. The loss of DivK and CckA flagellar pole localization due to reduced DivL localization may prevent proper regulation of CckA activity by DivL/DivK and thus lead to inefficient CtrA activation.
The potential reduction in CtrA activation in ΔpodJ could result from decreased PleC activity leading to hyperphosphorylation of DivK, or it could result from delocalization of DivL preventing proper activation of CckA. If it is the former, then preventing DivK phosphorylation should swing the DivK~P balance back towards wild-type and suppress the signaling defects of a ΔpodJ mutant. If it is the latter, then DivK is already being appropriately dephosphorylated but DivL cannot interact with CckA, and further dephosphorylation of DivK will have no effect. To distinguish between these hypotheses, the gene encoding the DivK kinase, divJ, was deleted by allelic replacement with a spec cassette in wild-type and ΔpodJ backgrounds. Deletion of divJ results in decrease of DivK phosphorylation (Wheeler & Shapiro, 1999, Matroule et al., 2004). The resulting mutants were tested for phage sensitivity and PilA transcription/translation. Wild-type and ΔdivJ showed similar levels of phage sensitivity (Figure 6A), while ΔpodJ was phage resistant as expected. Similar to the Ptac-pilA suppression of ΔpodJ, ΔpodJΔdivJ::spec had wild-type levels of plaque formation, but the plaques were cloudy due to cell growth, indicating partial suppression. Strains carrying ΔdivJ tend to have larger plaques and zones of clearing likely due to the growth defect caused by the ΔdivJ mutation. The ΔpodJΔdivJ::spec mutant also had wild-type level of pilA gene expression (Figure 6B) and PilA protein (Figure 6C–D). These results indicate that compromised PleC activity, rather than compromised interaction between DivL and CckA, leads to decreased pilA and perP gene expression in the ΔpodJ strain.
Figure 6.
Pilus biogenesis defect of ΔpodJ suppressed by divJ deletion. A: Phage sensitivity assays of various podJ and divJ mutants. Cell cultures were OD600 normalized then mixed with soft agar and plated on agarose plates. After hardening, serial dilutions of bacteriophage ΦCbK were spotted onto the plates and allowed to dry prior to incubation. B: pilA gene expression in various podJ and divJ mutants was measured using β-galactosidase assays of exponentially growing cultures. C: Whole cell lysates of various podJ and divJ mutants were separated on Tricine SDS-PAGE gels and subjected to Western blot analysis using anti-PilA anti-sera. D: Band intensities of the PilA Western blot were measured using ImageJ software. Each density was normalized to a percentage of the wild-type control.
Effect of podJ mutations on holdfast synthesis
The work described above has focused on the consequences of PodJ processing on pili synthesis. However, PodJ is also required for holdfast synthesis (Hinz et al., 2003, Viollier et al., 2002b). Similar to pilus synthesis proteins, localization of the holdfast attachment protein HfaB (Hardy et al., 2010) and the holdfast export protein HfsD (J. Javens, unpublished) depend on PodJ. While it had been previously shown that a mutant with the fast-processing podJ921 allele still produces holdfast and has a minor reduction in phage sensitivity (Lawler et al., 2006), it was shown here that the phage sensitivity phenotype of podJ921 was exaggerated when combined with the podJΔ589-639 mutation (Figure 1B). To determine if holdfast synthesis has a similar pattern of regulation as pilus synthesis, holdfast was detected using fluorescein-conjugated wheat germ agglutinin (lectin) for various alleles of podJ (Figure 7A). Holdfast was detected in wild-type, podJΔ589-639, and podJΔ921-974 strains, but not in ΔpodJ or podJΔ589-639-S921 strains. Localization of mCherry-labeled HfaB was analyzed by fluorescence microscopy. podJ mutants that produced holdfast had detectable HfaB foci at the tips of stalks, whereas strains that did not produce holdfast had either diffuse fluorescent signal or foci located at random points along the cell (Figure 7A). The loss of HfaB polar localization indicates that the organization of the holdfast machinery is disrupted. These results are similar to the phage sensitivity assay results in that the Δ589-639 and Δ921-974 mutations individually do not result in strong holdfast-deficient phenotypes, but they abolish holdfast synthesis and localization of holdfast components when combined.
Figure 7.
Analysis of holdfast synthesis in podJ mutants. A: Cell cultures were treated with FITC-conjugated wheat germ agglutinin lectin and visualized by fluorescence microscopy. podJ-S921 used a stop codon at the amino acid 921 codon to remove the C-terminal 53 amino acids of PodJ and results in the synthesis of the same protein as the podJΔ921-974 mutant. The holdfast attachment protein HfaB was labeled by a mCherry fusion. hfaB-mCherry is in single-copy on the chromosome under control of the native hfa promoter. B: Various podJ and divJ mutant cell cultures were treated with FITC-conjugated wheat germ agglutinin lectin and visualized by fluorescence microscopy.
We tested the ability of a ΔdivJ::spec mutation to suppress the holdfast deficiency of a ΔpodJ mutant like it does for pilus synthesis. Stalked cells of wild-type and ΔdivJ::spec strains were robustly labeled by lectin staining (>70%), while no labeling of ΔpodJ cells was observed (Figure 7B). The ΔpodJΔdivJ::spec strain had 10% labeled stalked cells, indicating that the ΔdivJ::spec mutation can suppress the holdfast production defect in ΔpodJ, but the suppression is not very strong. These results suggest that loss of holdfast production in ΔpodJ likely stems more from loss of PodJ scaffolding than altered signaling.
PodJS is the result of PerP and MmpA proteolysis
Based on the importance PodJ processing has for pilus biogenesis, the mechanism of PodJ proteolysis was investigated further. PodJL is proteolytically processed to PodJS coincident with cell division, and processing appears to involve the action of the periplasmic protease PerP. PodJS persists in the swarmer cell until differentiation. It is thought that clearing of PodJS during swarmer cell differentiation is initiated by the membrane metalloprotease MmpA by cleaving off the transmembrane domain and allowing PodJ to be released from the membrane for further degradation. It was previously hypothesized that PodJS is the result of PerP proteolysis because there is reduced amount of PodJS and increased amount of PodJL in a perP mutant (Chen et al., 2006).
To further the analysis of PodJ proteolysis, the site where PerP cleaves PodJ was identified. The PerP protein lacking the secretion signal (PerPΔLS) and the periplasmic domain of PodJ (PodJPERI) were purified and used in in vitro proteolysis assays, as previously described (Chen et al., 2006). Reaction mixtures were separated by SDS-PAGE, transferred onto PVDF membrane, and visualized with Ponceau S staining (Figure 8B). Consistent with earlier results, digestion of PodJPERI produced two fragments, PodJPERI-C (~26 kDa) and PodJPERI-N (~6 kDa). N-terminal analysis of the larger peptide, PodJPERI-C, revealed a sequence that matched exactly amino acids 703-717 of PodJ: VALTTGKVVPAEVEA. Thus, PerP appears to cut between amino acids 702 and 703 of PodJ (Figure 8A).
Figure 8.
Analysis of PodJ processing. A: Schematic of PodJ, with predicted coiled coil (CC), transmembrane (TM), tetratricopeptide repeat (TPR) and peptidoglycan binding (PG) domains indicated. The numbers above the predicted domain indicate the amino acid region for each domain. Amino acids 635-711 are shown below the schematic. MmpA and PerP cut sites are depicted. B: In vitro processing of PodJ periplasmic domain (PodJPERI) by PerP lacking the secretion signal (PerPΔLS). Lane 1 – PerPΔLS only. Lane 2 – PodJPERI only. Lane 3 – PodJPERI + PerPΔLS. N-terminal (PodJPERI-N) and C-terminal (PodJPERI-C) processing fragments are indicated. C: The amino acid sequence of PodJ from amino acids 589 to 754 is shown. The underlined region indicates the transmembrane domain. The last peptide detected by mass spectrometry from immunopurified PodJS for each strain is shaded gray. For podJΔ589-639, the deleted region is indicated by strikethrough. D: Previous and alternative models of PodJ proteolysis.
The position of the site of PerP cleavage only 60 amino acids from the transmembrane domain makes it extremely difficult to distinguish the product of PerP cleavage from that of MmpA for a ~94 kDa protein. To determine if PodJS present in swarmer cells is the product of PerP or of PerP + MmpA, we used mass spectrometry to determine the terminal amino acid of immunopurified PodJS (Figure 8A, C). The terminal amino acid of the most C-terminal peptide detected was found to be four amino acids into the predicted transmembrane domain and was not a trypsin cut site, indicating that the protein ended a few amino acids into the transmembrane domain (Figure 8A, C). This was puzzling, since PerP resides in the periplasm and is unlikely to cut PodJ at the detected cleavage site. To simplify the subsequent descriptions, we will use PodJS+TM and PodJS-TM to designate the product of PerP processing that still contains the transmembrane domain and periplasmic domain up to the PerP cleavage site and the product of MmpA processing that no longer contains the transmembrane domain, respectively.
The above results suggest that MmpA generates PodJS-TM in late predivisional cells rather than degrade it during swarmer cell differentiation. To verify these results, mass spectrometry analysis was performed on immunopurified PodJS from a ΔmmpA mutant. While the terminal amino acid could not be identified due to the fact that the most C-terminal peptide detected ended in a trypsin cut site, additional protein sequence beyond the transmembrane domain was found in the ΔmmpA mutant (Figure 8A–B, supplementary material), indicating that PodJS immunopurified from wild-type cells is the result of MmpA proteolysis (Figure 8D).
An alternative explanation for the above results is that promiscuous MmpA activity during lysis of wild-type cells and extensive treatment required for immunoprecipitation generates the shortened form lacking the transmembrane domain despite the presence of protease inhibitors in the lysis buffer. If MmpA cleavage of the PodJ transmembrane domain only occurs during swarmer cell differentiation, PodJ should be predominantly membrane-associated in swarmer cells, whereas if PerP and MmpA both act at cell division, it should be predominantly soluble. In fractionation experiments, PodJ from wild-type swarmer cells was found predominantly in the soluble protein fraction, indicating a lack of the transmembrane domain, while PodJ from ΔmmpA swarmer cells was principally found in the membrane fraction and required detergent to be solubilized (Supplementary Figure S-4). These results are consistent with PerP and MmpA cleavage of PodJ occurring at cell division, although promiscuous MmpA activity during the lysis step of these experiments cannot be completely ruled out.
Results of this section suggest an alternative model of PodJ proteolysis (Figure 8D). It was previously thought that PerP generated PodJS+TM in late predivisional cells and that MmpA initiated clearing of PodJS+TM by cleaving off the transmembrane domain during the swarmer to stalked cell transition. Instead, our results suggest a model where both PerP and MmpA function in late predivisional cells to generate PodJS-TM, which lacks a transmembrane domain, and that clearing of this PodJS-TM during swarmer cell differentiation is regulated not solely by release from the membrane but by release from the membrane followed by some unknown mechanism.
Finally, we analyzed the short form of PodJ generated in the podJΔ589-631 mutant and found that the protein extended beyond the PerP cleavage site (Fig. 8C). This result is consistent with the compromized PerP activity found in this mutant as described in earlier sections.
To determine if mutations neighboring the determined cut sites could alter proteolysis by MmpA, site directed mutations were made in podJ such that the chemistry of the amino acids immediately before (A642R) and after (L643R) the MmpA cut-site were altered. Additionally, site-directed mutations were made that altered the sequence immediately preceding the transmembrane domain, changing the protein sequence from KKSKAR to AVAVAV starting at amino acid 629, thereby removing four positively charged amino acids. However, Western blot analysis demonstrated these mutants do not have dramatically altered processing compared to wild-type, nor did they have any detectable alteration in phage sensitivity, swarming in low percentage agar, or PodJ localization phenotypes (data not shown). The results suggest that individual mutations altering chemistry surrounding cut sites may be ineffective at preventing PodJ proteolysis. The lack of altered phenotypes in mutants with changes to chemistry surrounding the MmpA cut site suggests that there are redundant proteases for PodJ proteolysis, or that MmpA can use alternative cut sites. The latter case has been observed before in the autoproteolysis of the Hap autotransporter in Haemophilus influenzae (Hendrixson et al., 1997, Kenjale et al., 2009).
DISCUSSION
The studies presented here shed further light on the function of PodJ. PodJ serves a scaffolding role for pilus structural proteins, such that when processing of PodJ is induced, pilus biogenesis decreases with reduced flagellar pole CpaE localization, but without altering pilA gene expression, PilA protein levels, CpaE protein levels, or PleC localization. This scaffolding role was already suspected (Viollier et al., 2002b, Lawler et al., 2006). We show in this work that PodJ also serves a role in ensuring proper intracellular signaling. Without PodJ, PleC, DivK DivL, and CckA all show decreased localization at the flagellar pole, which leads to a decreased activity in CtrA-activated promoters. It is likely that PodJ’s impact on signaling is a different facet of its scaffolding ability. It was possible that PodJ localized signaling proteins but this localization had no impact on signaling activity. For example, polar localization by PodJ could have served mostly to ensure proper protein segregation after cell division. However, the data presented here indicate that delocalization of signaling proteins from the flagellar pole leads to alterations in signaling, therefore PodJ is necessary for proper signaling by localizing signaling proteins to the same subcellular site. These two facets of PodJ, the scaffolding of structural proteins and the maintenance of proper signaling, can be genetically separated. The podJΔ589-639 deletion impacts the signaling aspect, and although CpaE is delocalized, pilus synthesis still occurs at the correct pole. The defect in phage sensitivity caused by the podJΔ589-639 mutation can be suppressed by bypassing the signaling block (i.e. ectopic expression of pilA), indicating that the block in pilus synthesis is mostly from altered signaling. The podJΔ921-974 deletion impacts the scaffolding aspect by inducing rapid processing and causing altered pilus protein localization, while leaving the signaling aspect unharmed. DivL localization is shifted towards bipolar localization in this strain, but DivK and CckA localization are relatively unaltered. Each deletion on its own results in mild phenotypes, but when combined the resultant mutant phenocopies ΔpodJ. This result indicates that the phage resistance phenotype of ΔpodJ is the result of deficiencies in structural protein localization and altered intracellular signaling. This hypothesis is supported by the fact that ectopic pilA expression only partially suppressed the phage sensitivity defect because it could only cover the altered signaling aspect. Based on this research, when analyzing ΔpodJ phenotypes in the future both structural protein localization and signaling must be considered.
The research presented here also indicates that regulated processing of PodJ after cell division serves to shut off pilus biogenesis. PodJL is produced in the predivisional cell and is needed for assembly of the pilus base unit (Hinz et al., 2003, Viollier et al., 2002b). Upon cell division, the pilus filament PilA protein is expressed while PodJ is processed (though the presence or absence of the transmembrane domain likely has little impact on pilus biogenesis). This serves to activate pilus filament formation from base units already assembled and prevent additional base assembly. If this hypothesis is correct, why does the podJΔ921-974 mutant not display a larger reduction in phage sensitivity? It is clear that removing the periplasmic domain by deletion phenocopies ΔpodJ for phage resistance (Lawler et al., 2006). Even though the PodJΔ921-974 mutant is processed faster than wild-type, the periplasmic portion of the protein must exist for at least a limited amount of time and in fact can be detected by overloading and overexposing western blots (data not shown). Ectopic expression of perP causes a much greater decrease in phage sensitivity than the podJΔ921-974 mutant (approximately 100-fold), indicating that higher PerP expression is more efficient at degrading the periplasmic portion of PodJ than the destabilizing effect of the Δ921-974 mutation. Therefore, the small amount of time the long form is present in the Δ921-974 mutant appears to be enough to initiate some pilus synthesis.
If the cell cycle dependent processing of PodJ serves to synchronize pilus synthesis, what purpose does that accomplish for the cell? One possibility may be due to the role of pili in surface attachment. C. crescentus uses the coordinated action of the flagellum, the pili, and the holdfast to attach to surfaces (Levi & Jenal, 2006, Bodenmiller et al., 2004, Li et al., 2012). Both the flagellum and pili are needed for reversible initial attachment, and the pili are required for the transition from the reversible to irreversible phase of adhesion through their role in the surface contact stimulation of holdfast synthesis (Li et al., 2012). In addition, retraction of the pili is thought to orient the cell such that the pole that produces the holdfast is in close proximity/contact with the surface, thereby optimizing efficient holdfast attachment. Biogenesis of new pili at the same pole at the wrong time could add a counterproductive force, pushing the cell away from the surface while other pili are retracting to bring the cell in closer. Blocking new pilus synthesis would prevent such counterproductive activity.
While increasing PodJL processing by mutation of podJ or induction of PerP increased phage resistance in every case, it was surprising that podJΔ589-639Δ921-974 and podJΔ589-639 with induced PerP were completely phage resistant, like mutants deleting the whole periplasmic domain (Lawler et al., 2006), despite the presence of a significant level of PodJL. One possible explanation for the apparent contradiction is that even though PodJL is detectable in podJΔ589-639Δ921-974, it is not available to perform its role in pilus biogenesis. In both wild-type and podJΔ921-974 there is no delay between induction of processing and actual proteolysis. It is possible that the Δ589-639 mutation causes a delay between the binding of PerP (or other processing proteases) to the periplasmic domain and the processing of this domain. The PodJ periplasmic domain bound by proteases might be unavailable for its scaffolding function in pilus biogenesis proteins because it is in an unfavorable conformation, or occluded by the proteolysis enzymes.
How might the Δ589-639 mutation cause a delay in PodJL processing? Since it appears that Δ589-639 causes reduced activity from CtrA-dependent promoters, one possibility is that there is some CtrA-regulated factor necessary for efficient PodJ processing that is not properly expressed, perhaps the cryptic proteolysis mechanism that operates in the absence of PerP. Alternatively, another effect of the Δ589-639 deletion is displaced signaling proteins from the flagellar pole. PleC has a periplasmic domain (Wang et al., 1993), and while there has been no study performed that would demonstrate PodJ and PleC interacting through their periplasmic portions, it is possible that the PodJ periplasmic domain needs to be in the context of the PleC periplasmic domain for efficient processing to occur. This may explain why the in vitro processing of PodJPERI by PerP took several hours. Or it may be that PodJ interacts with itself and that self-assembly is the context needed for efficient processing. The Δ589-639 mutation may disrupt PodJ self-assembly like it disrupts assembly of PodJ with signaling proteins. Regardless, the Δ589-639 mutation appears to do what the ΔperP mutation could not, which is slow down the rapid processing of podJΔ921-974. Elucidating the mechanism by which processing is slowed when it should be rapid could finally yield a non-processing podJ allele that can help further understand the function of PodJ processing.
Degradation of PodJ is similar to the regulated intramembrane proteolytic processing of RseA in E. coli (Alba & Gross, 2004, Raivio & Silhavy, 2001). RseA is an anti-sigma factor that has a single transmembrane domain, a periplasmic portion, and a cytoplasmic portion that binds σE, inhibiting its activity (Missiakas et al., 1997, De Las Penas et al., 1997, Ades et al., 1999, Campbell et al., 2003). Envelope stress activates the periplasmic protease DegS, which removes a portion of the periplasmic C-terminal domain of RseA (Ades et al., 1999, Alba et al., 2001, Walsh et al., 2003, Wilken et al., 2004), making it a suitable substrate for degradation by the membrane metalloprotease YaeL (Alba et al., 2002, Kanehara et al., 2001). Intramembrane proteolysis results in release of the cytoplasmic portion of RseA from the membrane, leading to complete degradation of the remaining protein by ClpXP and release of σE (Flynn et al., 2003).
Previous work using SDS-PAGE to determine the size of the short form of PodJ led to the hypothesis that PodJL is processed by PerP at the time of cell division to yield PodJS+TM, whose transmembrane domain is subsequently removed by MmpA during swarmer cell differentiation (Figure 8D)(Chen et al., 2006, Chen et al., 2005). Work presented used the improved resolution of mass spectrometry to determine the end point of the short form of PodJ. Our results suggest that PerP and MmpA both function at the time of cell division and lead to the formation of PodJS lacking a transmembrane domain, while an unknown protease regulates degradation of this PodJS during the swarmerto stalked cell transition. The resulting new model of PodJ processing is in closer agreement with the RseA model. YaeL-mediated proteolysis is not a regulated step; the only factor limiting YaeL activity is presentation of a suitable substrate from DegS activity (Bohn et al., 2004, Kanehara et al., 2003). MmpA working with PerP to create PodJS lacking a transmembrane domain suggests that MmpA-mediated degradation is not regulated either, which is supported by the fact that MmpA is constitutively expressed (Chen et al., 2005). Although data presented here further strengthens ties between the RseA and PodJ proteolysis models, there is still a notable difference. In RseA, once the proteolysis cascade is initiated it does not stop until RseA is completely degraded. For PodJ, the proteolysis pauses at PodJS-TM until swarmer cell differentiation. This indicates there is an additional regulatory step not present in the RseA pathway. The nature of this regulatory step is unknown.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
Strains used in this study are listed in Table S-1. All C. crescentus strains were grown in PYE medium (Poindexter, 1964) at 30°C. Escherichia coli strains were grown in Luria-Bertani media at 37°C. Where appropriate, antibiotics were used at the following concentrations: kanamycin 10 μg/ml (liquid) and 5 μg/ml (plates), tetracycline 0.5 μg/ml (liquid) and 2 μg/ml (plates), ampicillin 100μg/ml (liquid and plates), spectinomycin 25 μg/ml (liquid) and 100 μg/ml (plates). E. coli antibiotic concentrations were: kanamycin (50 μg/ml), tetracycline (12 μg/ml), ampicillin or carbenicillin (100 μg/ml).
Electro-Ten compentent cells (Stratagene/Agilent Technologies, Santa Clara, CA) were used for transformation of Matchmaker prey library ligations and amplified in TB (Sambrook et al., 1989). BL21λDE3 was used for protein expression. Yeast strains were grown in either YPDA (20 g/L Difco Peptone, 10 g/L Yeast extract, 2% glucose, .003% adenine hemisulfate) or SD (6.7 g/L Difco yeast nitrogen base without amino acids, 2% glucose) medium. SD medium was supplemented with 10X amino acid dropout solution (0.15 g/L His; 0.2 g/L Ade, Arg, Met, Trp, Ura; 0.3 g/L Ile, Lys, Tyr; 0.5 g/L Phe; 1 g/L Leu; 1.5 g/L Val; 2 g/L Thr), missing Leu, Trp, Ade, and/or His as appropriate and 2% Bacto Difco agar for plates.
Strain construction
Primers used in cloning are listed in Table S-2. To create a clean deletion of the codons encoding PodJ amino acids 921-974, an upstream fragment was generated by PCR using oligonucleotides FDSPGEco and RDSPGXba2 (596 bp) with a EcoRI site at the 5′ end and an XbaI site at the 3′ end, and a downstream fragment was generated with podJXbaU3 and podJbamd2 (671 bp) with an XbaI site at the 5′ end and a BamHI site at the 3′ end. The fragments were digested with the appropriate enzymes and cloned into pNPTS138 (M.R.K. Alley, unpublished) to create NPTS138podJΔ921, which was checked by sequencing using the M13 forward and reverse oligonucleotides. pNPTS138podJΔ921 was then introduced into CB15 and ΔmmpA by conjugation. The in-frame deletion was created by two-step selection, first for kanamycin resistance and sucrose sensitivity and then for sucrose resistance and kanamycin sensitivity (Stephens et al., 1996). The final construct was amplified from the C. crescentus chromosome.
To create perP deletion mutants, a fragment upstream of perP was generated by PCR using oligonucleotides FdperPup3 and RdperPup3 (469 bp) with a HindIII site at the 5′ end and an XbaI site at the 3′ end, and a fragment downstream of perP was generated with FdperPdn2 and RdperPdn3 (465 bp) with an XbaI site at the 5′ end and an EcoRI site at the 3′ end. The fragments were digested with the appropriate enzymes and cloned into pNPTS138 to create pNPTS138ΔperP, which was sequenced with M13 forward and reverse oligonucleotides. One T to C mutation was found early in the plasmid sequence, but was not incorporated after integration into the C. crescentus chromosome. pNPTS138ΔperP was introduced into CB15, ΔmmpA and podJΔ921-974 by conjugation. The final construct was amplified from the C. crescentus chromosome and sequenced. To create ΔmmpA ΔperP podJ-S921, plasmid pBGST18-podJ921 (Lawler et al., 2006) into ΔmmpA ΔperP by electroporation, followed by kanamycin selection.
To create an in-frame clean deletion of the codons encoding PodJ amino acids 589-639, a 435 bp upstream fragment was generated by PCR using oligonucleotides d589Hind and d589XbaDn with a HindIII site at the 5′ end and an XbaI site at the 3′ end, and a 467 bp downstream fragment was generated with d639XbaUp and 591mutEcodn with an XbaI site at the 5′ end and an EcoRI site at the 3′ end. The products were digested with the appropriate enzymes and cloned into pNPTS138 to create pNPTS138podJΔ589, and sequenced with M13 forward and reverse oligonucleotides. pNPTS138podJΔl589 was then introduced into CB15 by conjugation using E. coli strain S-17. The podJ locus was amplified from the C. crescentus chromosome and sequenced to confirm the deletion. pNPTS138podJΔ589 was electroporated into CB15 and podJΔ921-974. Two-step selection and deletion verification were performed as before. To create podJΔ589-639-S921 and ΔmmpA ΔperP podJ-S921, plasmid pBGST18-podJ921 (Lawler et al., 2006) was conjugated into recipient strains followed by kanamycin selection.
To create site-directed mutants in podJ, an approximately 1 kb fragment spanning the middle third of podJ was generated by PCR using oligonucleotides, with a HindIII site at the 5′ end and an EcoRI site at the 3′ end. The fragment was digested with the appropriate enzymes and cloned into pSKII+ (Stratagene) and sequenced. The Quikchange mutagenesis kit (Stratagene) was then used to alter protein sequence from KKSKAR to AVAVAV starting at amino acid 629, as well as A642R, and L643R mutations create various point mutations. To mutate the basic region preceding the PodJ transmembrane domain, sense oligonucleotide PodJKKAV and antisense oligonucleotide PodJKKAVanti were used. To create the A642R mutation, sense oligonucleotide PodJTTR and antisense oligonucleotide PodJTTRanti were used. To create the L643R mutation, sense oligonucleotide PodJRVV and antisense oligonucleotide PodJRVVvanti were used. The constructs were sequenced, and those with the appropriate mutations were subcloned out into pNPTS138 and introduced into C. crescentus. The final constructs were amplified from the C. crescentus genome and sequenced to verify the mutations.
A GFP-CpaE strain was constructed based upon the YFP-CpaE strain described in (Viollier et al., 2002a), using different restriction sites and using eGFP instead of YFP. A ~700 bp upstream fragment made with primers cpaEupF and cpaEupR, terminating with the cpaE start codon followed by a BamHI site, was cloned into pNPTS138 using HindIII and BamHI. eGFP from pXGFPC-2 (Thanbichler et al., 2007) was PCR amplified using primers cpaEgfpnewF and cpaEgfpnewR such that the start and stop codons were omitted, bounded by BamHI on the 5′ end and EcoRI on the 3′ end. The eGFP fragment was cloned into the previous plasmid using BamHI and EcoRI. Finally, a ~600 bp fragment beginning with the second cpaE codon, bounded by an EcoRI site on the 5′ terminus and NheI on the 3′ end, was amplified using primers cpaEdownF and cpaEdownR. The downstream fragment was cloned into the previous plasmid, creating plasmid pPDC1. The plasmid construct was verified by sequencing, and electroporated into various strains. The presence of the correct fusion was verified by PCR and Western blot analysis. The construct was electroporated into various strains using previously established conditions followed by two-step selection, and the correct allelic replacement was verified by PCR.
To generate strains tracking perP expression, plasmid pJC327 (PperP-lacZ, (Chen et al., 2006)) was electroporated into recipient strains using previously established conditions followed by tetracycline selection. To generate strains for tracking pilA expression, plasmid pJS70 (PpilA-lacZ, (Skerker & Shapiro, 2000)) was electroporated into recipient strains using previously established conditions, followed by tetracycline selection. Strains expressing pilA constitutively were constructed by electroporating plasmid pJS96 (Ptac-pilA, (Skerker & Shapiro, 2000)) into recipient strains using previously established electroporation conditions followed by spectinomycin selection. To generate strains tracking PleC localization, plasmid pPleC-GFP (pleC-gfp, (Wheeler & Shapiro, 1999)) was transferred into recipient strains by conjugal mating, followed by tetracycline selection. Strains containing divK-gfp were constructed by mating an S17 E. coli strain with plasmid pMR20divK-EGFP (Jacobs et al., 2001) with recipient strains, followed by tetracycline selection. Strains containing cckA-gfp were constructed by mating an S17 E. coli strain with plasmid pMR20cckAgfp (Jacobs et al., 1999) with recipient strains, followed by tetracycline selection. Strains with perP expressed ectopically at the xyl locus under xylose control were constructed by electroporating pJC317 (Pxyl-perP, (Chen et al., 2006)) into recipient strains, followed by kanamycin selection. To generate strains tracking HfaB localization, plasmid pCHYChfaB (hfaB-mCherry, (Hardy et al., 2010)) was transferred into recipient strains by conjugal mating. pCHYChfaB integrates into the chromosome, creating a merodiploid of hfaB. The hfaB-mCherry allele created by integration is under control of the native hfa promoter.
A ΦCr30 lysate of NA1000 ΔdivJ::spec (Pierce et al., 2006) was made and ΔdivJ::spec was transduced into wild-type to create ΔdivJ::spec, and into ΔpodJ to create ΔpodJΔdivJ::spec. A ΦCr30 lysate of NA1000 divL-yfp (Reisinger et al., 2007) was made and divL-yfp was transduced into wild-type, ΔpodJ, podJΔ589-639, podJΔ921-974 and podJΔ589-638Δ921-974. ΦCr30 transducing phage lysates were prepared as described previously (Ely & Johnson, 1977a, Ely & Johnson, 1977b). All transductions were done using a modification of West et al. (West et al., 2002).
For screening the Caulobacter yeast two-hybrid library, a PodJ-Bait plasmid was generated. First the full length podJ gene was amplified in two pieces; a 1368 bp 5′ region of podJ using primers 2HPodJERUp and 2HpodJSpDn and another 1590 bp 3′ region of podJ using primers Y2HPodJSpUp and Y2HPodJBHDn. The 5′ PCR product was digested with EcoRI/SphI, the 3′ PCR product was digested with SphI/BamHI with and the pBBR1MCS2 plasmid was digested with EcoRI/BamHI; a three part ligation was performed, resulting in pBBR1MCS2-podJ-full. The 2.9 kb EcoRI/BamHI fragment was cut out of pBBR1MCS2-podJ-full and ligated into pGBKT7, resulting in pGBKpodJfull yeast 2-hybrid binding domain/bait vector. For verifying protein-protein interactions pulled up from the yeast two-hybrid C. crescentus library screen, the same 2.9 kb EcoRI/BamHI fragment of podJ was cloned into the pGADT7 yeast two-hybrid activation domain/prey vector, resulting in plasmid pGADpodJfull, which generates a fusion of PodJ to the GAL4 activation domain. The plasmid containing divL from the library screen (pGADDivL) was digested with EcoRI and XhoI. The ~1.9 kb fragment was then cloned into pGBKT7 (digested with EcoRI and SalI; SalI and XhoI have compatible overhangs). The resulting plasmid (pGBKDivL) was sequenced to confirm the 5′ fusion site was unchanged.
SDS-PAGE, Tricine SDS-PAGE, and Western blotting
Cell lysates were prepared from exponentially growing cells (OD600 = 0.35 – 0.7). The equivalent of 1.0 ml of culture at OD600 = 0.7 was centrifuged at 16,000 × g for 2 min at room temperature. Supernatant was removed and cell pellets were resuspended in 50 μl 10 mM Tris pH 8.0. 50 μl 2X SDS-PAGE loading buffer was added and the lysate was boiled for 2 min. Lysates were loaded onto 7.5% SDS-PAGE gels. For analysis of PilA protein levels, cell lysates were separated on 10% total acrylamide/3% bis-acrylamide tricine SDS-PAGE gels according to the instructions of Schagger and von Jagow (Schagger & von Jagow, 1987).
Separated proteins were transferred to nitrocellulose and subjected to Western blot analysis using standard laboratory practices. Western blots for PodJ used rabbit antisera raised against the cytoplasmic portion of PodJ at 1:10,000 dilution (Hinz et al., 2003). Western blots for GFP used mouse monoclonal JL-8 GFP antibodies (Clontech) at 1:1,000 dilution. Blots for PilA used purified PilA antibodies at 1:5,000 dilution (Skerker & Shapiro, 2000) (Viollier et al., 2002a). Detection was done using Supersignal West Pico chemiluminescence substrate (Pierce) and either anti-mouse or anti-rabbit HRP-conjugated secondary antibodies (Bio-Rad) at 1:10,000 dilution. PilA Western blots were performed in triplicate and densitometry of the PilA bands was performed using ImageJ software (Abramoff et al., 2004) according to software instructions, normalizing the amount of PilA protein in each sample as a percentage of the wild-type control on each blot.
ΦCbK phage assays
Exponential growing cultures of strains to be assayed for phage sensitivity were normalized to OD600 = 0.5. Two hundred microliters of normalized cell culture were mixed with 2.5 ml molten PYE 0.3% agar and poured on to the surface of warmed PYE plates containing appropriate antibiotics and, for Pxyl-perP strains, sugars. Ten-fold dilutions of a 1 X l010 pfu/ml ΦCbK stock was made in PYE broth and 10 μl drops were placed on the surface of the hardened soft agar and allowed to air dry prior to plate inversion. Plates were incubated at room temperature (~23°C) for 2 days.
β-galactosidase assays
β-galactosidase assays were performed according to Miller (Miller, 1972). Briefly, 200 μl of OD600 = 0.4 – 0.6 cells were added to 600 μl Z buffer. 50 μl chloroform was added to each sample, vortexed, and incubated at 30°C for 5 min. 200 μl 4 mg/ml ONPG was added and the samples were incubated at 30°C. When significant yellow color had developed, the incubation time was noted and 400 μl 1 M Na2CO3 was added. A420 was measured and Miller units for each reaction were calculated using the following formula: (A420)1000/(OD)(t)(v) where OD = OD600 of the culture, t = time in min, and v = volume of the culture in ml.
Fluorescence and immunofluorescence microscopy
Agarose pads were made using PYE broth, 1.0% electrophoresis-grade agarose. Cells were spotted on a pad and immediately viewed. A Nikon Eclipse E800 with 100× Plan Apo oil immersion objective was used for both fluorescent and phase contrast microscopy. Imaging of was performed using a Nikon FITC-HyQ filter cube for GFP and 83000 triple filter cube for mCherry (Chroma Technologies) with a Photometrics Cascade 1K cooled charged-coupled device camera. Screen captures of both phase contrast and GFP emission were generated using Metamorph software (Molecular Devices) and overlaid. To enumerate GFP-CpaE, PleC-GFP, DivK-GFP and CckA-GFP cells, predivisional cells in each sample preparation in which stalked poles could be differentiated from flagellar poles were evaluated as to whether a fluorescent focus was present at a given pole, and each sample condition was repeated for a total of 3 times using different source cultures. To enumerate ΦCbK labeled swarmer cells, stalkless cells were evaluated as to whether a fluorescent focus was present or not, and each sample condition was repeated a total of 3 times using different cell cultures. Fluorescent wheat germ agglutinin (lectin) binding assays were performed as previously described (Janakiraman & Brun, 1999). To enumerate fluorescent wheat germ agglutinin labeled cells, stalked cells in each sample preparation were evaluated as to whether a fluorescent focus was present at the end of the stalk.
Immunofluorescence microscopy of PilA was performed on unpermeabilized cells with a modification to the protocol previously described for permeabilized cells (Quardokus et al., 2001). Briefly, cells were subcultured from overnight cultures and grown for 4 hours to an OD600= 0.5, 1 μl of PilA antibody was added directly to 1.7 ml microcentrifuge tubes and then 100 μl of cells added to the tube and incubated 30 minutes. Cells were spun at 7000 × g for 8 min at 4oC and washed three times with phosphate-buffered saline (PBS), pH 7.4 and then fixed in phosphate buffered 2.5% formaldehyde (Ted Pella, Inc., Redding, CA) for 30 min followed by another set of washes. The protocol was then followed as published starting with the blocking step. Goat anti-rabbit FITC (Jackson Immunoresearch, Inc., West Grove, PA) antibody was used at a 1:100 for detecting PilA. Epifluorescence microscopy was performed on a Nikon Eclipse 800 light microscope equipped with an FITC-HyQ cube for PilA samples. Images were captured using a Photometrics Cascade 1K camera and Metamorph Imaging Software package.
C. crescentus genomic Matchmaker prey library construction
C. crescentus strain CB15 genomic DNA was partially digested with HinP1I. The digested DNA was ligated into plasmid pGADT7 (Clontech), which was ClaI digested and calf intenstinal alkaline phosphatase treated (CIAP). The ligation was transformed into ElectroTen-Blue (Stratagene, La Jolla, CA) competent cells and plated to determine the number of colonies per transformation. To achieve ~95% coverage of the 4 MB Caulobacter genome (5 fold coverage), 20,000 colonies per transformation multiplied by 6 reading frames would mean that at last 120, 000 transformants are needed. Based on this calculation, 12 transformations were done to get 150,000 transformants to achieve >95% coverage of the C. crescentus genome in 6 reading frames. The library was pooled and 6 transformations were amplified in 3 liters of TB containing 200 μg/ml ampicillin for 13.5 hours at 37C. Plasmid DNA (870 ng/μl ) was isolated from the culture using the Qiagen Tip-500 plasmid purification system, 200 μl aliquots were frozen at −80°C.
Prey library screen
Plasmid pGBK-PodJ-Full was transformed into yeast strain AH109 using the transformation protocol recommended in the Matchmaker v.3 manual (BD/Clontech), generating strain YB3577. The same protocol was used to transform YB3577 with the genomic library in pGADT7. Briefly, strain YB3577 was grown in 1.0 L SD (-Trp) liquid media to an OD600 of 0.4. Cells were collected at 1000 × g at 25°C, washed with sterile dH2O, and resuspended in 8 mL TE/LiAc (10 mM Tris-HCl, 1 mM EDTA, pH 7.5, 100 mM lithium acetate). To the cells the following was added: 0.35 mg library DNA, 4 mg salmon sperm DNA, and 60 mL PEG/LiAc (40% polyethylene glycol 3350, 10 mM Tris-HCl, 1 mM EDTA, pH 7.5, 100 mM lithium acetate), and the mixture was incubated at 30°C with shaking for 1 hour. Then 7 mL 100% dimethyl sulfoxide (DMSO) was added, and the mix was incubated at 42°C for 15 minutes. After chilling the cells on ice for 2 minutes, they were collected at 1000 × g at 25°C and resuspended in 10 mL TE (10 mM Tris-HCl, 1 mM EDTA). Cells were plated (200 μL per plate) on SD (-Leu Trp) agar plates containing 0.02 mg/mL X-α-gal (Glycosynth Warrington Cheshire, England) and incubated at 30°C for 3–4 days. This yielded ~1000 colonies per plate among which 21 dark blue colonies were spotted and were restreaked onto fresh selective (SD (Leu –Trp)) and stringent (SD (-Leu –Trp –Ade –His)) plates.
Isolation of plasmid from yeast
To isolate plasmid from yeast, colonies were inoculated in 7 mL YPDA medium and grown overnight at 30°C. Cells were collected and rinsed in 1 mL SE buffer (0.9 M sorbitol, 0.1 M EDTA, pH 8.0), then resuspended in 400 μL SEB (SE with 14 mM BME). To this was added 20μL (~100 U) lyticase (Sigma Chemicals) and suspension was incubated at 37°C for 30 minutes. Cells were then collected and resuspended in 400 μL TE (10 mM Tris-HCl, 1 mM EDTA, pH 7.5), then 90 μL of ETS (0.3 M EDTA, 0.4 M Tris-HCl pH 7.0, 2.2% SDS) was added and mixture was incubated at 70°C for 30 minutes. After this incubation, 80 μL 5 M KAc was added to lyse the cells, and the mixture was incubated on ice for 2 hours. Unlysed cells and debris was pelleted, and nucleic acid was precipitated from the supernatant with ethanol, pelleted, and resuspended in TE. 25 μg RNAse was then added and the remaining plasmid DNA was precipitated with isopropanol, washed with ethanol, pelleted, and resuspended in TE. This plasmid suspension contains both pGAD- and pGBK-derived plasmids, which was transformed into DH5α competent cells. Colonies were isolated on either LB containing ampicillin (for pGAD) or kanamycin (for pGBK). Individual plasmids were isolated and the inserts identified by direct sequencing of the plasmid.
Protein overexpression and purification for Far Western blot analysis
To purify the cytoplasmic portion of PodJ, YB3044 BL21 (λDE3) containing pET22b-PodJ was grown in LB medium at 37°C. The culture was induced at an OD600 of 0.5–0.6 with 0.5 mM IPTG and grown at 30°C for 2 hours. The cells were harvested, resuspended in equilibration buffer (50 mM Tris, 100 mM KCl, 0.05% Dextran Sulfate, pH8.3) and lysed by sonication with a Misonix Sonicator 3000 in the presence of protease inhibitor cocktail tablets without EDTA (Roche Indianapolis, IN). After centrifugation to remove cellular debris, the supernatant containing soluble, recombinant PodJ-His was purified over a Nickel-charged His-Bind resin column as described by the manufacturer (Novagen). The fusion protein was eluted with 20mM Tris pH7.9, 500mM NaCl and 1M imidazole. The final concentration of PodJ-His was 380μg/ml using BCA Protein Assay Kit (Pierce).
To purify cytoplasmic DivL (missing the N-terminal transmembrane domain), BL21 (λDE3) containing pET21b-DivL (YB4711) was grown in LB medium at 37°C. The culture was induced at an OD600 of 0.6 with 0.2 mM IPTG and grown at 16°C for 22 hours. The cells were harvested, and resuspended in equilibration buffer (50 mM Tris, 500 mM NaCl, 5 mM EDTA, pH8.0). The cells were lysed and DivL-His was purified as described above for PodJ. The final concentration was of DivL was 990μg/ml.
Far-Western blot analysis
For Far-Western blot analysis, samples were run on a 10% SDS-PAGE and transferred to a nitrocellulose membrane (Whatman Schleicher and Schuell BA85). After Ponceau-S staining to confirm the transfer, the blot was blocked with 5% non-fat dry milk with TTBS buffer (20 mM Tris pH 7.6, 137 mM NaCl, 0.05% Tween20) for 1 hour at room temperature. The blot was incubated with 2.28μg/ml PodJ-His (Bait protein) in 5ml 5% non-fat dry milk with TTBS buffer at 4°C for 14 hours. The blot was washed with TTBS buffer three times, 5 minutes, 5 minutes and 15 minutes. The blot was incubated with affinity-purified anti-PodJ antibody against the cytoplasmic portion of PodJ (Hinz et al., 2003) at a 1:750 dilution, at 4°C for 14 hours. After washing as above, the blot was treated with secondary antibody Goat Anti-Rabbit HRP-Conjugate 10μg/ml (Pierce) at a dilution of 1:10000, at room temperature for 1.5 hours. The blot was washed with TTBS buffer three times, 10 minutes, 10 minutes and 15 minutes. To detect the PodJ-DivL interaction, the blot was incubated with Super Signal West Dura Extended Duration substrate (Thermo Scientific/Pierce) for 10 minutes, and analyzed using a Kodak Image Station 440CF.
A serial dilution of purified DivL-His from 3.96μg down to 0.099μg was run on a 10% SDS-PAGE and transferred to nitrocellulose to determine the specificity of the interaction between PodJ and DivL. On a separate blot, For 38 ng of PodJ was loaded as a positive control for the PodJ antibody, 2.475μg purified DivL-His was loaded to demonstrate PodJ-DivL interaction, as negative controls, two strains were used YB3562 BL21λDE3 containing pET22b without insert and YB1848 BL21λDE3 containing pTYB4-FtsZ to over-express FtsZ-intein. The YB3562 and YB1848 were grown in LB medium at 37°C until OD600 =0.6 and induced with 0.5 mM IPTG at 30°C for 2.5 hours. The final OD600 was 1.77 for FtsZ-intein (~ 99kD) and 1.37 for pET22b. For sample preparation, 1 ml of culture was centrifuged for 2 minutes. The supernatant removed and the pellet resuspended in 50μl ddH2O. 50μl 2X SDS sample buffer was added and boiled for at least 6 minutes. Samples were loaded on the 10% SDS-PAGE gel. Co-immunoprecipitation and Western blot analysis
Affinity purified antibodies were prepared as described in (Din et al., 1998) and concentrated 4:1 using an Amicon Ultra 15, 10,000 MWCO centrifugal filter (Millipore Ireland ltd) and concentrated 4:1 again using a Microcon centrifugal filters ultracel YM-10 10000 MWCO (Millipore Ireland ltd), aliquoted and frozen at −80°C until used for co-immunoprecipitation experiments. Co-immunoprecipitation was performed on C. crescentus cells using a modification of the procedure described by Radhakrishnan (Radhakrishnan et al., 2008). Briefly, Mid-log phase cells (100 ml) were harvested by centrifugation at 6200 × g for 10 minutes. Cells were washed once in buffer 1 (50 mM sodium phosphate (Na3PO4) pH 7.4, 150 mM sodium chloride (NaCl), 1 mM EDTA, and then centrifuged at 6200 × g for 10 minutes. Cells were lysed at room temperature for 40 minutes in buffer 2 (50 mM Na3PO4 pH 7.4, 150 mM NaCl, 1 mM EDTA, 10 mM magnesium chloride (MgCl2), 0.5% n-dodecyl-β-D-maltoside (DDM), 18,000 units Ready-lyseTM lysozyme (Epicentre Technologies, Madison, WI), 30 units DNase I (Roche Applied Science, Indianapolis, IN) and 1X protease inhibitor cocktail (Complete™ EDTA-free, (Roche Applied Science, Indianapolis, IN)). Cellular debris was removed by centrifugation at 9200 × g for 5 min at 4°C. Supernatant was pre-cleared with 60 μl Protein A-agarose (Roche Applied Science, Indianapolis, IN) for 30 minutes at 4°C with shaking, centrifuged 5 minutes, and collected (about 900 μl). To 450 μl supernatant, affinity purified anti-PodJ antibodies (1:100 dilution) were added and incubated for 90 min at 4°C. Antibody-protein complexes or the negative control (supernatant incubated without antibody) was added to 50 μl Protein A-agarose beads and incubated for 1 hr at 4°C. Beads were centrifuged 10 minutes at 4°C, washed three times with 1 ml buffer 3 (Na3PO4 pH 7.4, 150 mM NaCl, 1 mM EDTA, 1.05% NP40), then washed twice with 1 ml 10% buffer 3. The beads were resuspended in 40 μl 2X SDS loading buffer and boiled for 10 min. The beads were removed by centrifuging 1 min and the solubilized proteins run on a 10% SDS-PAGE. Co-immunoprecipitated proteins were identified by immunoblotting using monoclonal antibodies to GFP (JL-8, 1:625, Clontech, Mountain View, CA) to detect DivL-YFP. Blots were washed in 1X phosphate buffered saline (PBS) with 0.05% Tween 20 three times and incubated with horseradish peroxidase conjugated goat anti-mouse (1:10,000) for 1 hour at room temperature, then washed again, followed by incubation with Super Signal West Dura Substrate (Thermo Scientific, Waltham, MA) for 10 minutes detecting the chemiluminescent signal using a 4000 MM Pro image station (Carestream Health, Rochester, NY).
Sample preparation for liquid chromatography-tandem mass spectrometry (LC-MS/MS)
For analysis of PodJS produced in C. crescentus, 40 ml of either wild-type strain CB15, the ΔmmpA mutant, or the podJΔ589-639 mutant was grown in PYE to an OD600 of 0.7. The sample was then lysed and immunoprecipitated as described (Chen et al., 2006). Cultures were separated into 20 ml aliquots, and each aliquot was processed as follows: cells were pelleted and resuspended in 1 ml SDS buffer (10 mM Tris-HCl pH 8, 1% SDS, 1 mM EDTA), and divided into two microcentrifuge tubes and boiled for 2.5 min. The boiled lysates were added to 9 ml RIPA buffer (50 mM tris pH 8, 150 mM NaCl, 1 mM EDTA, 1% NP40 v/v, 0.5% deoxycholic acid w/v, 0.1% SDS w/v), along with one mini EDTA free protease inhibitor tablet (Roche). 60 μl washed Protein A agarose (Roche) were then added, and the mixture was incubated on ice with shaking for 15 min to preclear nonspecific proteins. The soluble portion was transferred to two microfuge tubes and spun for 2 min at 16,000 × g. Supernatants were transferred to a 15 ml conical tube, and 60 μl crude antiserum raised against the cytoplasmic portion of PodJ was added (Hinz et al., 2003), and rocked overnight at 4°C. 60 μl washed Protein A agarose (Roche) was then added and rocked at room temperature for 1 hour. The Protein A agarose beads were pelleted by centrifugation in a single microcentrifuge tube, by spinning aliquots, removing the supernatants and repeating. Beads were boiled in 60 μl SDS loading buffer (1X buffer was made by mixing 2X buffer (0.125M Tris-HCl pH 6.8, 20% glycerol, 4% SDS, 1.43 M 2-mercaptoethanol, 0.005% Bromophenol blue) 1:1 with 10 mM Tris pH 8.0) 4.5 min, and centrifuged to pellet beads. The supernatant was loaded on a 7.5% 1.5 mm SDS-PAGE gel and stained with Coomassie brilliant blue dye; the bands corresponding to PodJL and PodJS were excised with a clean razor blade. Extracted gel bands were analyzed in the Indiana University Proteomics Facility by in-gel trypsin digestion followed by LC-MS/MS analysis.
In-gel trypsin digestion
Dithiothreitol (DTT) and iodoacetamide (IAM) were obtained from Bio-Rad (Hercules, CA). HPLC grade water and acetonitrile were purchased from EMD Chemicals (Darmstadt, Germany). Proteomics grade trypsin and formic acid were obtained from Sigma-Aldrich (St. Louis, MO). Ammonium bicarbonate was purchased from Mallinckrodt Baker Inc. (Paris, KY).Proteins were digested in the gel bands by a standard procedure described previously (Rosenfeld et al., 1992). Briefly, each excised gel spot was cut into small pieces (less than 1 mm in each dimension), placed in a 0.6-ml microcentrifuge tube, and covered with 75 μl of 25 mM ammonium bicarbonate in 50% acetonitrile and vortexed for 10 min at room temperature to destain the proteins. The solution was removed from the tube and discarded. This destaining step was repeated twice. Gel pieces were vacuum-centrifuged for 25 min at 30°C to dryness. Disulfide bonds were reduced by the addition of 50 μl of 10 mM DTT with incubation at 60°C for 45 to 60 min. After removing and discarding the solution, 50 μl of 55 mM IAM was added and the sample incubated in the dark at room temperature for 45 to 75 min to alkylate reduced cysteines. After removing and discarding the solution, 100 μl of 25 mM ammonium bicarbonate, followed by two 75 μl aliquots of 25 mM ammonium bicarbonate in 50% acetonitrile, were added and the sample vortexed for ten min. The solution was removed and discarded each time after vortexing. Gels were again vacuum-centrifuged for 20 min at 30°C to dryness. Seventy-five microliters of 12.5 μg/ml trypsin solution in 25 mM ammonium bicarbonate was added to dried gel pieces. The gel pieces were rehydrated at 4°C for 5 to 10 min, then incubated at 37oC for 18 to 21 hours (overnight). Following digestion, the liquid was removed from gel pieces and transferred to a new 0.6-mL microfuge tube. Two 50-μl aliquots of 5% formic acid in 50% acetonitrile were added to the gel pieces and vortexed for 10 min, then removed and added to the tube containing the digest solution. Trypsin digests (approximately 120 μl) were vacuum-centrifuged at 30°C to dryness (3 to 5 hours), reconstituted in 10 μl of water, and transferred to autosampler vials for LC-MS/MS analysis.
LC/MS-MS
Six microliters of protein digest was loaded onto a 15 mm × 100 μm i.d. trapping column packed with 5 μm, 200 A Magic C18AQ packing material (Microm BioResourses Inc., Auburn, CA). Peptides were eluted through a 150 mm × 75 μm i.d. analytical column with the same packing material, except 100 A pore size, using a 30-minute gradient from 97% to 60% solvent A, 97:3:0.1 water/acetonitrile/formic acid (Solvent B is 0.1% formic acid in acetonitrile.) at 250 nl/min using an Eksigent nanoLC-2D system (Eksigent Technologies, Dublin, CA). From the end of the column, ions were electrosprayed directly into a ThermoFinnigan (San Jose, CA) LCQ Deca XP ion-trap mass spectrometer which recorded mass spectra and data-dependent tandem mass spectra of the peptide ions. Tandem mass spectra were searched against the known amino acid sequence for PodJ from C. crescentus (Nierman et al., 2001) without enzyme specificity (a “no enzyme” search) using a licensed copy of Mascot (Perkins et al., 1999) for peptide identification.
In vitro proteolysis assays
For in vitro proteolysis assays, periplasmic domains of PerP and PodJ (PerPΔLS and PodJPERI, respectively) were purified and mixed, as previously described (Chen et al., 2006). After incubation at 37°C for 18 hours, samples were resolved using 12% SDS-PAGE, transferred onto PVDF membrane, and stained with 0.5% Ponceau S in 1% acetic acid. The membrane was allowed to air dry; then the desired protein band was cut out and subjected to Edman sequencing, which was performed by the Protein and Nucleic Acid Facility at Stanford University.
Supplementary Material
Acknowledgments
We thank P. Viollier for providing PodJ antibody, L. Shapiro for providing PilA antibody, and G. Hardy for help in strain construction. We are grateful to Danielle O’Donnol for constructing the C. crescentus yeast 2-hybrid prey library, to David Larson for constructing pBBR1MCS2-podJ-full, to Yang Wang for ftsZ-intein, and to Deanne Pierce for DivL-His. We thank members of our laboratory for critical reading of the manuscript. This work was supported by grants GM51986 and GM077648 from the National Institutes of Health to YVB. P.D.C. was supported by a postdoctoral National Institutes of Health National Research Service Award F32GM084618 from the National Institute of General Medical Sciences. M.L.L was supported by a National Institutes of Health Predoctoral Fellowship (GM07757). J.C.C. is supported by Award Number SC2GM082318 from the National Institute of General Medical Sciences. A portion of the work was conducted while J.C.C. was a postdoctoral fellow in Lucy Shapiro’s group, a position funded in part by NIH grants GM32506 and F32 G067472.
References
- Abramoff M, Magelhaes P, Ram S. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Ades SE, Connolly LE, Alba BM, Gross CA. The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 2002;16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba BM, Zhong HJ, Pelayo JC, Gross CA. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide sigma (E) activity. Mol Microbiol. 2001;40:1323–1333. doi: 10.1046/j.1365-2958.2001.02475.x. [DOI] [PubMed] [Google Scholar]
- Angelastro PS, Sliusarenko O, Jacobs-Wagner C. Polar localization of the CckA histidine kinase and cell cycle periodicity of the essential master regulator CtrA in Caulobacter crescentus. Journal of bacteriology. 2010;192:539–552. doi: 10.1128/JB.00985-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefontaine AF, Pierreux CE, Mertens P, Vandenhaute J, Letesson JJ, De Bolle X. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol Microbiol. 2002;43:945–960. doi: 10.1046/j.1365-2958.2002.02777.x. [DOI] [PubMed] [Google Scholar]
- Bodenmiller D, Toh E, Brun YV. Development of surface adhesion in Caulobacter crescentus. J Bacteriol. 2004;186:1438–1447. doi: 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn C, Collier J, Bouloc P. Dispensable PDZ domain of Escherichia coli YaeL essential protease. Mol Microbiol. 2004;52:427–435. doi: 10.1111/j.1365-2958.2004.03985.x. [DOI] [PubMed] [Google Scholar]
- Brassinga AK, Siam R, McSween W, Winkler H, Wood D, Marczynski GT. Conserved response regulator CtrA and IHF binding sites in the alpha-proteobacteria Caulobacter crescentus and Rickettsia prowazekii chromosomal replication origins. J Bacteriol. 2002;184:5789–5799. doi: 10.1128/JB.184.20.5789-5799.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol. 2010;4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. Polar growth in the Alphaproteobacterial order Rhizobiales. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Kysela DT, Brun YV. Polarity and the diversity of growth mechanisms in bacteria. Semin Cell Dev Biol. 2011;22:790–798. doi: 10.1016/j.semcdb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- Chen JC, Hottes AK, McAdams HH, McGrath PT, Viollier PH, Shapiro L. Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. EMBO J. 2006;25:377–386. doi: 10.1038/sj.emboj.7600935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- Crymes WB, Jr, Zhang D, Ely B. Regulation of podJ expression during the Caulobacter crescentus cell cycle. J Bacteriol. 1999;181:3967–3973. doi: 10.1128/jb.181.13.3967-3973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Penas A, Connolly L, Gross CA. SigmaE is an essential sigma factor in Escherichia coli. J Bacteriol. 1997;179:6862–6864. doi: 10.1128/jb.179.21.6862-6864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din N, Quardokus EM, Sackett MJ, Brun YV. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol Microbiol. 1998;27:1051–1063. doi: 10.1046/j.1365-2958.1998.00752.x. [DOI] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B, Johnson RC. Generalized transduction in Caulobacter crescentus. Genetics. 1977a;87:391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B, Johnson RC. Generalized Transduction in Caulobacter crescentus. Genetics. 1977b;87:391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Gora KG, Tsokos CG, Chen YE, Srinivasan BS, Perchuk BS, Laub MT. A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol Cell. 2010;39:455–467. doi: 10.1016/j.molcel.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallez R, Mignolet J, Van Mullem V, Wery M, Vandenhaute J, Letesson JJ, Jacobs-Wagner C, De Bolle X. The asymmetric distribution of the essential histidine kinase PdhS indicates a differentiation event in Brucella abortus. EMBO J. 2007;26:1444–1455. doi: 10.1038/sj.emboj.7601577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy GG, Allen RC, Toh E, Long M, Brown PJ, Cole-Tobian JL, Brun YV. A localized multimeric anchor attaches the Caulobacter holdfast to the cell pole. Mol Microbiol. 2010;76:409–427. doi: 10.1111/j.1365-2958.2010.07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson DR, de la Morena ML, Stathopoulos C, St Geme JW., 3rd Structural determinants of processing and secretion of the Haemophilus influenzae Hap protein. Molecular microbiology. 1997;26:505–518. doi: 10.1046/j.1365-2958.1997.5921965.x. [DOI] [PubMed] [Google Scholar]
- Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- Iniesta AA, Hillson NJ, Shapiro L. Cell pole-specific activation of a critical bacterial cell cycle kinase. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7012–7017. doi: 10.1073/pnas.1001767107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C, I, Domian J, Maddock JR, Shapiro L. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- Jacobs C, Hung D, Shapiro L. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc Natl Acad Sci U S A. 2001;98:4095–4100. doi: 10.1073/pnas.051609998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janakiraman RS, Brun YV. Cell cycle control of a holdfast attachment gene in Caulobacter crescentus. J Bacteriol. 1999;181:1118–1125. doi: 10.1128/jb.181.4.1118-1125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahng LS, Shapiro L. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J Bacteriol. 2001;183:3065–3075. doi: 10.1128/JB.183.10.3065-3075.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Akiyama Y, Ito K. Characterization of the yaeL gene product and its S2P-protease motifs in Escherichia coli. Gene. 2001;281:71–79. doi: 10.1016/s0378-1119(01)00823-x. [DOI] [PubMed] [Google Scholar]
- Kanehara K, Ito K, Akiyama Y. YaeL proteolysis of RseA is controlled by the PDZ domain of YaeL and a Gln-rich region of RseA. EMBO J. 2003;22:6389–6398. doi: 10.1093/emboj/cdg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenjale R, Meng G, Fink DL, Juehne T, Ohashi T, Erickson HP, Waksman G, St Geme JW., 3rd Structural determinants of autoproteolysis of the Haemophilus influenzae Hap autotransporter. Infect Immun. 2009;77:4704–4713. doi: 10.1128/IAI.00598-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Matroule JY, Jacobs-Wagner C. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev Cell. 2003;5:149–159. doi: 10.1016/s1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- Lawler ML, Larson DE, Hinz AJ, Klein D, Brun YV. Dissection of functional domains of the polar localization factor PodJ in Caulobacter crescentus. Mol Microbiol. 2006;59:301–316. doi: 10.1111/j.1365-2958.2005.04935.x. [DOI] [PubMed] [Google Scholar]
- Levi A, Jenal U. Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J Bacteriol. 2006;188:5315–5318. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Molecular microbiology. 2012;83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell. 2004;118:579–590. doi: 10.1016/j.cell.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Miller J. A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 1972. Experiments in Molecular Genetics. [Google Scholar]
- Missiakas D, Mayer MP, Lemaire M, Georgopoulos C, Raina S. Modulation of the Escherichia coli sigmaE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol Microbiol. 1997;24:355–371. doi: 10.1046/j.1365-2958.1997.3601713.x. [DOI] [PubMed] [Google Scholar]
- Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen J, Heidelberg JF, Alley MR, Ohta N, Maddock JR, Potocka I, Nelson WC, Newton A, Stephens C, Phadke ND, Ely B, DeBoy RT, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Kolonay JF, Smit J, Craven MB, Khouri H, Shetty J, Berry K, Utterback T, Tran K, Wolf A, Vamathevan J, Ermolaeva M, White O, Salzberg SL, Venter JC, Shapiro L, Fraser CM. Complete genome sequence of Caulobacter crescentus. Proceedings of the National Academy of Science, U S A. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Pierce DL, O’Donnol DS, Allen RC, Javens JW, Quardokus EM, Brun YV. Mutations in DivL and CckA rescue a divJ null mutant of Caulobacter crescentus by reducing the activity of CtrA. J Bacteriol. 2006;188:2473–2482. doi: 10.1128/JB.188.7.2473-2482.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter JS. Biological properties and classification of the Caulobacter group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quardokus EM, Din N, Brun YV. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol Microbiol. 2001;39:949–959. doi: 10.1046/j.1365-2958.2001.02287.x. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio TL, Silhavy TJ. Periplasmic stress and ECF sigma factors. Annu Rev Microbiol. 2001;55:591–624. doi: 10.1146/annurev.micro.55.1.591. [DOI] [PubMed] [Google Scholar]
- Reisinger SJ, Huntwork S, Viollier PH, Ryan KR. DivL performs critical cell cycle functions in Caulobacter crescentus independent of kinase activity. J Bacteriol. 2007;189:8308–8320. doi: 10.1128/JB.00868-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson GT, Reisenauer A, Wright R, Jensen RB, Jensen A, Shapiro L, Roop RM., 2nd The Brucella abortus CcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J Bacteriol. 2000;182:3482–3489. doi: 10.1128/jb.182.12.3482-3489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory; New York: 1989. [Google Scholar]
- Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sciochetti SA, Ohta N, Newton A. The role of polar localization in the function of an essential Caulobacter crescentus tyrosine kinase. Mol Microbiol. 2005;56:1467–1480. doi: 10.1111/j.1365-2958.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci U S A. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Iniesta AA, Shapiro L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 2007;35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson AD, Fuqua C. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr Opin Microbiol. 2009;12:708–714. doi: 10.1016/j.mib.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokos CG, Perchuk BS, Laub MT. A Dynamic Complex of Signaling Proteins Uses Polar Localization to Regulate Cell-Fate Asymmetry in Caulobacter crescentus. Developmental cell. 2011;20:329–341. doi: 10.1016/j.devcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J. 2002a;21:4420–4428. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci U S A. 2002b;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- Wang SP, Sharma PL, Schoenlein PV, Ely B. A histidine protein kinase is involved in polar organelle development in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1993;90:630–634. doi: 10.1073/pnas.90.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West L, Yang D, Stephens C. Use of the Caulobacter crescentus genome sequence to develop a method for systematic genetic mapping. Journal of bacteriology. 2002;184:2155–2166. doi: 10.1128/JB.184.8.2155-2166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T. Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease. Cell. 2004;117:483–494. doi: 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.