Summary
Coenzyme A (CoA) is an essential cofactor for all forms of life. The biochemistry underpinning the assembly of CoA in Escherichia coli and other enterobacteria is well understood, except for the events leading to maturation of the L-aspartate-α-decarboxylase (PanD) enzyme that converts pantothenate to β-alanine. PanD is synthesized as pro-PanD, which undergoes an auto-proteolytic cleavage at residue Ser25 to yield the catalytic pyruvoyl moiety of the enzyme. Since 1990, it has been known that Escherichia coli yhhK strains are pantothenate auxotrophs, but the role of YhhK in pantothenate biosynthesis remained an enigma. Here we show that Salmonella enterica yhhK strains are also pantothenate auxotrophs. In vivo and in vitro evidence show that YhhK interacts directly with PanD, and that such interactions accelerate pro-PanD maturation. We also show that S. enterica yhhK strains accumulate pro-PanD,and that not all pro-PanD proteins require YhhK for maturation. For example, the Corynebacterium glutamicum panD+ gene corrected the pantothenate auxtrophy of a S. enterica yhhK strain, supporting in vitro evidence obtained by others that some pro-PanD proteins autocleave at faster rates. We propose the name PanM for YhhK to reflect its role as a trigger of pro-PanD maturation by stabilizing pro-PanD in an autocleavage-prone conformation.
Keywords: coenzyme A biosynthesis, enzyme maturation, aspartate decarboxylase, YhhK, GCN5 acetyltransferases, pyruvoyl enzymes
Introduction
Coenzyme A (CoA) is essential to all forms of life, with its role in metabolism spanning anabolic and catabolic processes (Abiko, 1975). Pantothenate is a precursor of CoA that animals must acquire through their diets (Smith & Song, 1996). In contrast, most bacteria synthesize this precursor de novo (Leonardi & Jackowski, 2007) via the condensation of pantoate and β-alanine (Fig. 1). Most bacteria, including S. enterica and E. coli, synthesize β-alanine from aspartate, a reaction catalyzed by aspartate α-decarboxylase (PanD; EC 4.1.1.11) (Cronan et al., 1982, Genschel, 2004). PanD belongs to a class of decarboxylases and reductases whose activities require a pyruvoyl moiety (van Poelje & Snell, 1990). PanD is synthesized as an inactive 14-kDa protein (pro-PanD) that undergoes proteolytic cleavage between residues Gly45 and Ser25 to generate active PanD (Ramjee et al., 1997). This cleavage is a self-catalyzed backbone rearrangement that yields a 2.8-kDa β-subunit and 11-kDa α-subunit modified with an N-terminal pyruvoyl moiety (Albert et al., 1998, Ramjee et al., 1997). In vitro, the rate of pro-PanD cleavage is slower than other pyruvoyl enzymes, leading to the proposal that another factor may be necessary for maturation in vivo (Ramjee et al., 1997).
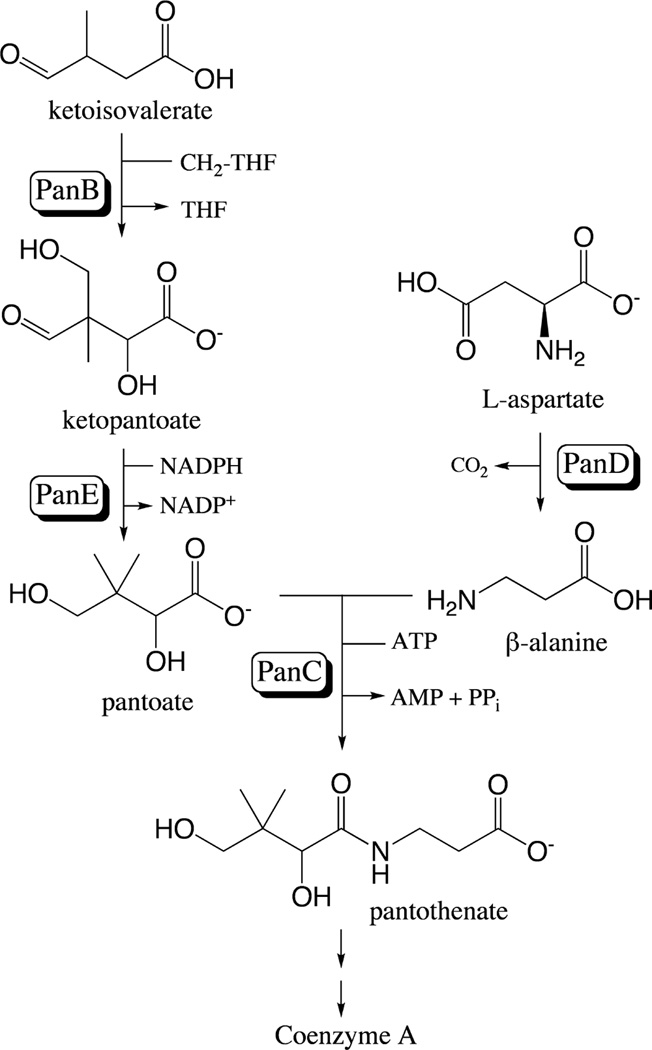
Figure 1. Pantothenate biosynthesis in S. enterica and E. coli.
The enzymes catalyzing each reaction with genes encoding them in parentheses are as follows: ketopantoate hydroxymethyltransferase (panB; EC 2.1.2.11), ketopantoate reductase (panE; EC 1.1.1.169), aspartate decarboxylase (panD; EC 4.1.1.11), and pantothenate synthetase (panC; EC 6.3.2.1). CH2-THF, methylene-tetrahydrofolate; THF, tetrahydrofolate. Modified from (Jones et al., 1993).
The uncharacterized YhhK protein has been known for over two decades to play a role in pantothenate synthesis since inactivation of yhhK results in a pantothenate auxotrophy (Adams et al., 1990, Koyanagi et al., 2004). This result was intriguing because the yhhK gene is located in the liv locus encoding Leu/Ile/Val transporters (14 min on E. coli chromosome), rather than in the panBCD locus (3 min). Surprisingly, the function of YhhK has not been established. Despite its location in the liv operon, YhhK is not thought to play a role in amino acid transport (Adams et al., 1990).
Since each step of the pantothenate biosynthetic pathway can be reconstituted in vitro, it is unlikely that YhhK affects the synthesis of pantothenate directly (Miyatake et al., 1979, Teller et al., 1976, Williamson & Brown, 1979, Zheng & Blanchard, 2000). Thus we hypothesized that YhhK might play an ancillary role in pantothenate biosynthesis. Here we show that YhhK triggers the conversion of inactive pro-PanD into active PanD. We propose a change in the name of the YhhK protein to PanM to reflect its role as a maturation factor that triggers pro-PanD cleavage.
Results
YhhK (hereafter PanM) is necessary for β-alanine synthesis in S. enterica
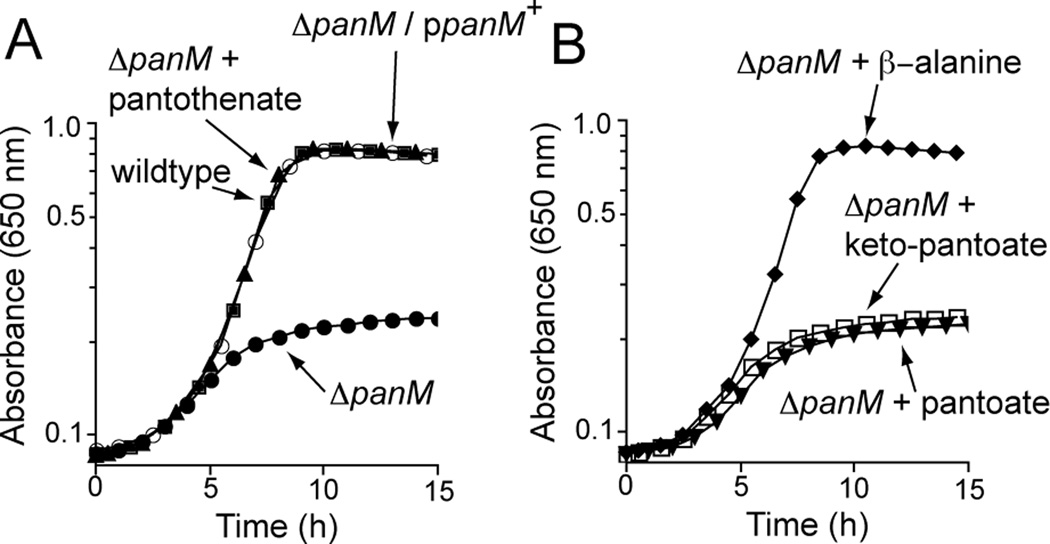
As in E. coli (Adams et al., 1990), the deletion of panM in S. enterica (Fig. 2A) resulted in a pantothenate auxotrophy that was corrected upon expression of panM+ in trans. The latter indicated that the absence of PanM was solely responsible for the phenotype. To determine which step of pantothenate synthesis was blocked in a panM strain, we performed feeding experiments using pathway intermediates. Only the addition of β-alanine restored growth of the panM mutant (JE12555) in minimal medium devoid of pantothenate, indicating that synthesis of β-alanine was impaired in cells lacking PanM (Fig. 2B). However, it was unclear from this information whether the absence of PanM had a direct or indirect effect on β-alanine synthesis.
Figure 2. Growth of panM supplemented with pantothenate precursors.
A. S. enterica wildtype (DM10310; squares) and panM (JE12555; closed circles) on minimal glycerol medium. Growth of a S. enterica panM strain in minimal medium supplemented with pantothenate (triangles), or complemented with panM+ in trans from plasmid pPAN65 (open circles). B. Growth of a S. enterica panM strain in minimal glycerol medium supplemented with intermediates of the pantothenate biosynthesis pathway: keto-pantoate (open squares), pantoate (triangles) and β-alanine (diamonds). SEM for all data was <0.04 absorbance units.
Deletion of panM does not affect panD expression
We investigated whether PanM was needed for panD expression. To do this, we used a strain in which a transposition-deficient MudI17134 (lacZ+ kan+) element (Castilho et al., 1984) was inserted into the panD gene, placing lacZ+ under the control of the panD promoter (PpanD, strain DM380, Table S1). Expression of the PpanD-lacZ+ reporter was analyzed as a function of PanM in panD::MudJ panM+ (JE13233) and panD::MudJ panM (JE13234) strains (Table S1). Levels of β-galactosidase activity were measured in cells grown on minimal glycerol medium. Under the conditions used, the level of PpanD-lacZ+ expression was not significantly different in strain JE13233 (743 ± 2 Miller units) and JE13234 (758 ± 3 Miller units), indicating that PanM did not affect panD expression.
PanM interacts directly with PanD
An alternative hypothesis was that PanM somehow controlled PanD activity, either directly or indirectly. We took in vivo and in vitro approaches to investigate this possibility.
In vivo evidence of PanD-PanM interactions
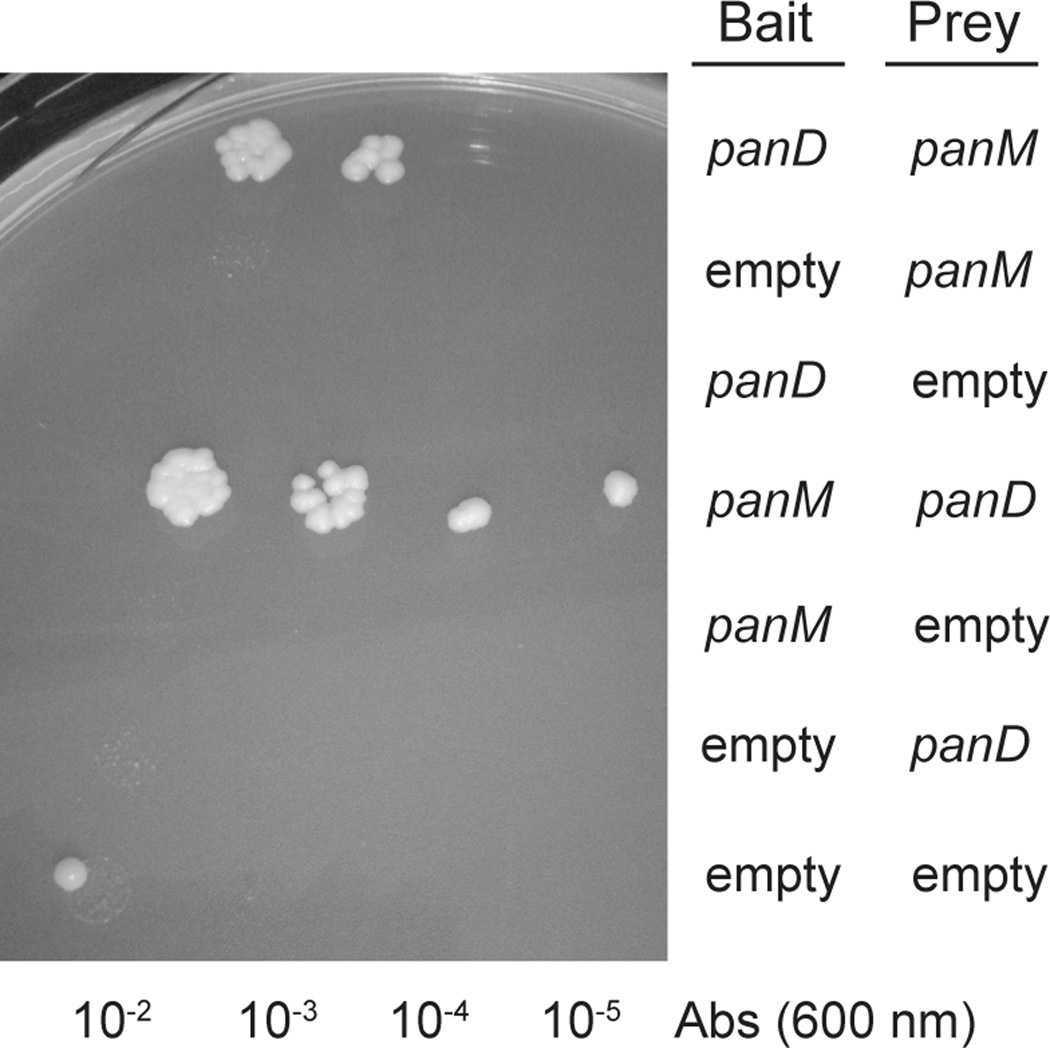
To determine whether or not PanD and PanM interacted, we preformed yeast two-hybrid analysis using the Matchmaker™ Yeast Two Hybrid System (Clonetech). This system places the yeast HIS3 gene for histidine biosynthesis under the control of the Gal4 transcription factor. The bait vector expresses the DNA-binding domain of Gal4 fused to one of the putative interactive partners, whilst the prey vector expresses the activation domain of Gal4 fused to the second putative partner. In this system, the activation and DNA-binding domains are necessary to form active Gal4 needed to transcribe HIS3. We posited that if PanD and PanM interacted, the two domains of Gal4 would be brought together, allowing expression of HIS3 thus growth in the absence of histidine. Yeast strains containing PanD as the bait and PanM as the prey grew in the absence of exogenous histidine, as did strains harboring the bait/prey swapped pair (Fig. 3). Collectively, these results showed direct interactions between PanD and PanM. Control experiments that employed vectors with non-fused Gal4 domains failed to grow, indicating that growth in the absence of histidine depended on PanD-PanM interactions.
Figure 3. Yeast-two hybrid analysis.
Growth of yeast two-hybrid reporter strains on SD medium devoid of histidine. Each row is a spotted serial dilution of a culture of the reporter strain carrying bait and prey plasmids into which panM or panD were cloned.
In vitro evidence of PanD-PanM interactions
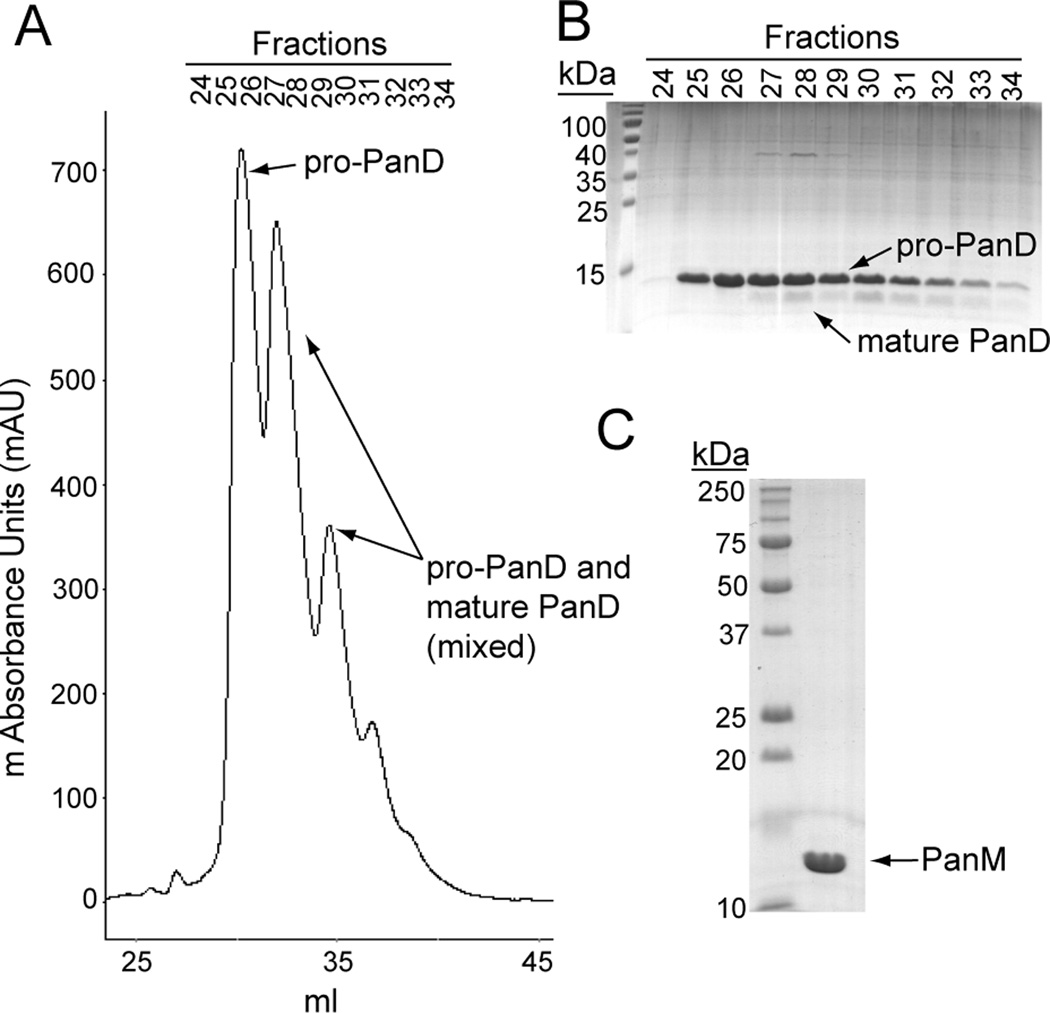
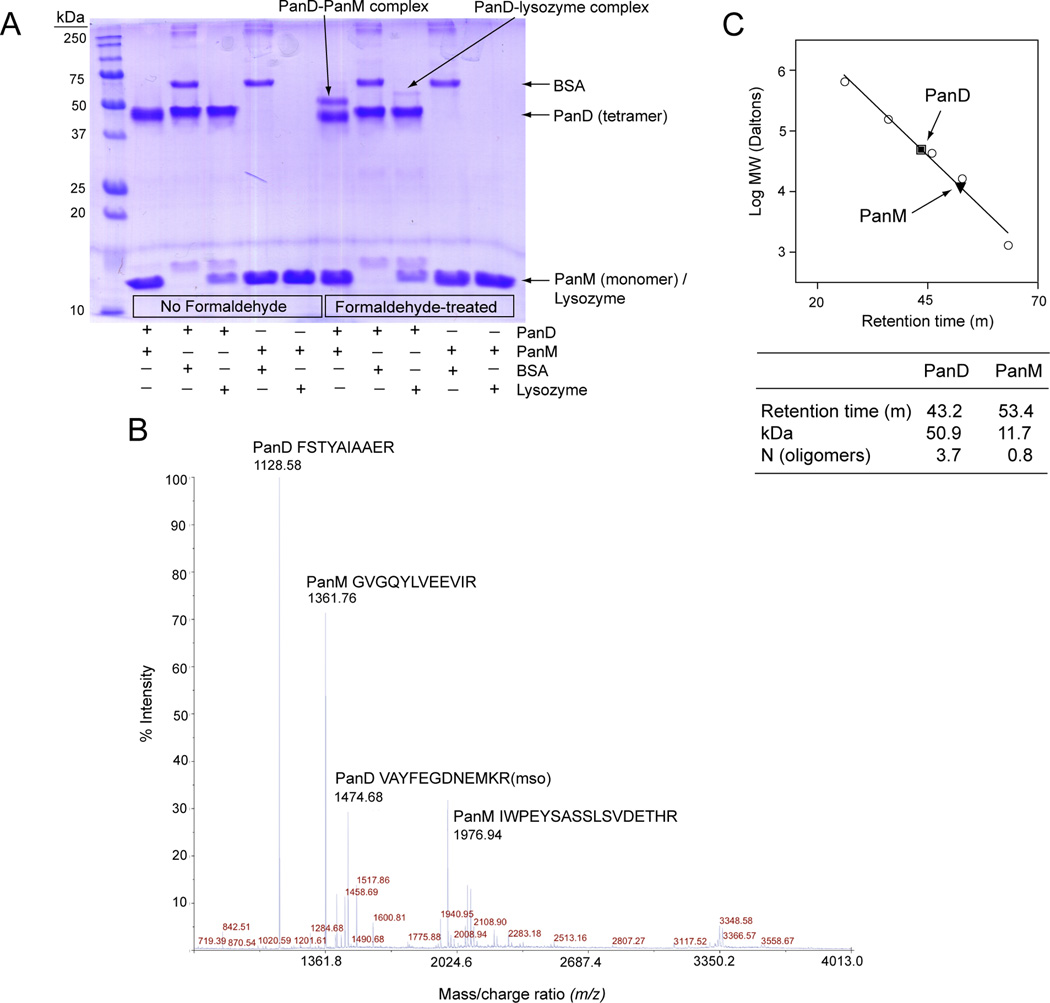
That PanD and PanM interact was confirmed in vitro using formaldehyde crosslinking followed by mass spectrometry. The genes encoding PanD and PanM were each cloned into expression vectors and proteins were purified to >95 homogeneity (Fig. 4). To facilitate our in vitro work, we purified PanD from S. enterica lacking panM (strain JE13153, Table S1). PanD eluted from the final mono-Q column in overlapping peaks (Fig. 4A) composed of pure pro-PanD followed by a mixture of cleaved and pro-PanD (Fig. 4A,B). The pro-PanD fractions were collected for use in cleavage studies. After treating a mixture of pro-PanD and PanM with formaldehyde, the proteins were resolved by denaturing gel electrophoresis (Laemmli, 1970). PanD migrated as a tetramer since the samples could not be boiled without breaking the formaldehyde crosslinks. A unique higher molecular weight band appeared in the formaldehyde-treated sample compared to the mock control (Fig. 5A). This band was excised, trypsinized and analyzed by MALDI TOF-TOF mass spectrometry. Ions corresponding to both PanD and PanM were identified (Fig. 5B), indicating that the band present in the formaldehyde treated sample (Fig. 5A) was a complex of PanD and PanM. These data lent support to the idea of direct interactions between PanM and PanD. It is unclear why only a fraction of the PanD and PanM in the reaction were crosslinked. It is possible that the conditions for crosslinking were sub-obtimal. To control for crosslinking specificity, PanD and PanM were each formaldehyde-treated in the presence of control proteins for which no interaction was expected, bovine serum albumin (BSA) and lysozyme. No interaction was observed between PanM and either control protein or between PanD and BSA. A weak interaction was detected between PanD and lysozyme, which we considered to be non-specific. The detection of the PanD-PanM complex above the non-specific interaction level lends support to the above-mentioned in vivo results.
Figure 4. Purification of pro-PanD and PanM.
A. Chromatogram of final mono-Q step of PanD purification. B. SDS-PAGE of fractions from mono-Q purification shown in panel A. Lane 1 contained PageRuler™ prestained protein ladder (Fermentas). C. SDS-PAGE of purified PanM. Lane 1 contained Precision Plus Protein™ All Blue Standards (Bio-Rad). Lane 2 contained purified PanM protein. The N-terminal β-subunit of PanD was not visible due to its small size and inefficient staining.
Figure 5. Interaction between PanD and PanM.
A. SDS-PAGE of formaldehyde crosslinking reactions. Lane 1 contained the Precision Plus Protein™ All Blue Standards (Bio-Rad). Lanes 2–6 contained untreated proteins, while lanes 7–11 contained formaldehyde-treated proteins (2 and 7: PanD+PanM, 3 and 8: PanD + BSA, 4 and 9: PanD + lysozyme, 5 and 10: PanM + BSA, 6 and 11: PanM + lysozyme). PanD appears as a tetramer in the denaturing gel because the samples could not be heated prior to running gel, which is necessary for tetramer denaturation. B. MALDI-TOF-TOF mass spectrum of the PanD-PanM complex shown in panel A. The amino acid composition and source of the four most abundant ions are indicated. Mso, methionine sulfoxide. C. Gel filtration analysis of PanD (square) and PanM (triangle). Molecular mass standards (circles) are thyroglobulin (bovine; 670 kDa), γ-globulin (bovine; 158 kDa), ovalbumin (chicken; 44 kDa), myoglobin (horse; 17 kDa) and vitamin B12 (1.35 kDa).
PanM interacts with PanD as a monomer
Formaldehyde crosslinking of PanM and PanD yielded a complex with a molecular weight shifted only 10–15 kDa above the band for the PanD tetramer (Fig. 5A). This suggested that one PanM monomer (14.5 kDa) interacted with one PanD tetramer (55.6 kDa). To explore this further, the oligomeric states of both PanD and PanM were determined by gel filtration. Consistent with previous studies (Ramjee et al., 1997);{lbert, 1998 #16628;Williamson, 1979 #20243} PanD behaved as a tetramer (Fig. 5C). PanM behaved as a monomer, with an experimentally-determined molecular mass of 11.7 kDa (Fig. 5C). These data supported the formaldehyde crosslinking results by suggesting that PanM interacted with PanD as a monomer, with a 1:1 PanM(monomer):PanD (tetramer) stoichiometry.
PanM accelerates pro-PanD cleavage
In light of the PanD-PanM interaction data (see above), we hypothesized that PanM directly affected PanD activity, either by controlling pro-PanD cleavage, or by somehow enhancing the activity of mature PanD.
In vitro cleavage studies
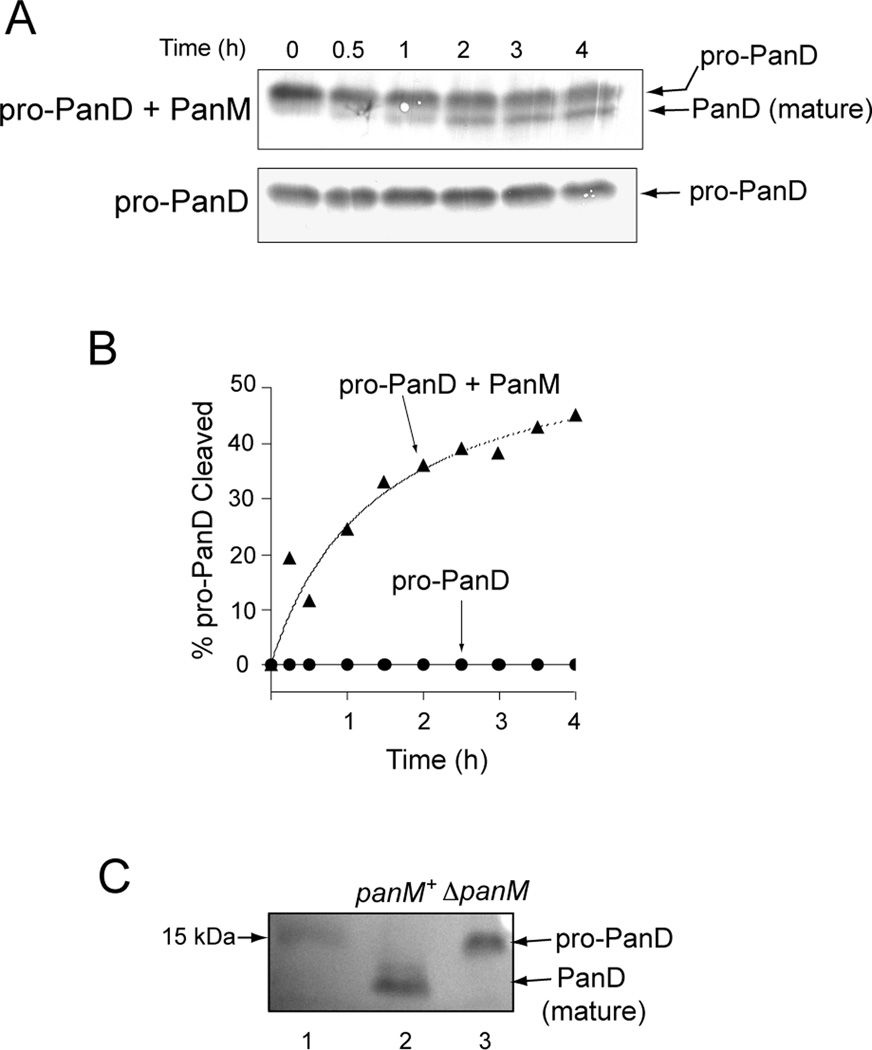
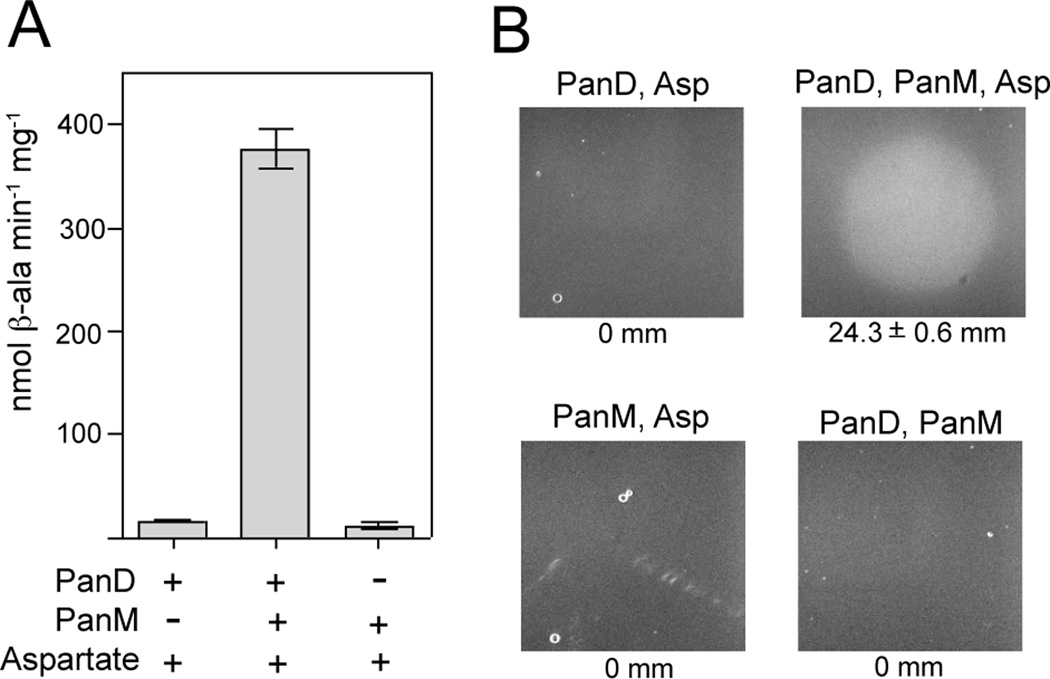
To assess cleavage, pro-PanD was incubated alone or with PanM. At various time points, samples were removed and proteins were resolved by SDS-PAGE followed by Western blot analysis using α-PanD rabbit polyclonal antibodies. Maturation of pro-PanD was only observed in reaction mixtures containing PanM (Fig. 6A,B), showing that PanM was required for pro-PanD maturation in vitro. Specific activities for PanD were determined after pre-incubation with or without PanM. Pro-PanD incubated with PanM was 24-fold more active than pro-PanD incubated without PanM (Fig. 7A). Production of β-alanine by PanD was verified using a bioassay that measured growth of a panD strain (JE13233) on minimal glycerol plates. Reaction mixtures containing pro-PanD incubated with PanM allowed growth of strain JE13233 (Fig. 7B). When PanM, pro-PanD or aspartate was removed from the reactions, no growth was observed, indicating that PanM was required for pro-PanD maturation. Taken together, these results indicated that PanM was necessary for pro-PanD cleavage into active aspartate decarboxylase in vitro.
Figure 6. PanM-dependent cleavage of pro-PanD.
Bands were visualized by Western blot analysis with rabbit polyclonal antibodies against PanD. A. Time course of the incubation of pro-PanD with PanM (top gel) or without PanM (bottom gel). B. Percent PanD cleavage as a function of time; PanD incubated alone (circles), or with PanM (triangles). Percentages were calculated by densitrometry analysis. The r2 for the PanD with PanM curve fit line was 0.87. C. Immunoprecipitation of PanD from S. enterica wildtype (lane 2) and ΔpanM strains (lane 3). Lane 1 contained PageRuler™ pre-stained protein ladder (Fermentas), with the 15 kDa standard indicated on the blot.
Figure 7. PanD activity.
A. PanD specific activities were determined by HPLC analysis of the reaction products from mixtures containing pro-PanD alone (bar 1), pro-PanD with PanM (bar 2), or PanM alone (bar 3). B. Bioassay for β-alanine production measuring growth of a strain carrying a panD deletion in minimal glycerol medium. A 1-µl sample from the specific reaction mixtures were spotted on plates overlayed with the S. enterica panD strain (JE13233). Growth on reaction products present in a complete reaction mixture is shown in the upper right panel. Control reactions omitted PanM (upper left), PanD (bottom left) or aspartate (bottom right). Zones of growth are indicated below each plate image (mm ± SEM).
In vivo validation
On the basis of the above results we predicted that pro-PanD would accumulate in a panM strain. We approached this question using Western blot analysis that employed α-PanD rabbit polyclonal antibodies to detect different forms of PanD. The PanD protein (either as pro-PanD or PanD) was immunoprecipitated from cell-free extracts of a panM strain (JE12555) and the panM+ parental strain (DM10310). As seen in figure 6C, the only detectable form found in the panM+ strain (DM10310) was PanD. In contrast, the panM strain (JE12555) accumulated only pro-PanD, confirming that PanM was required for pro-PanD maturation in vivo.
PanM does not act on PanD catalytically
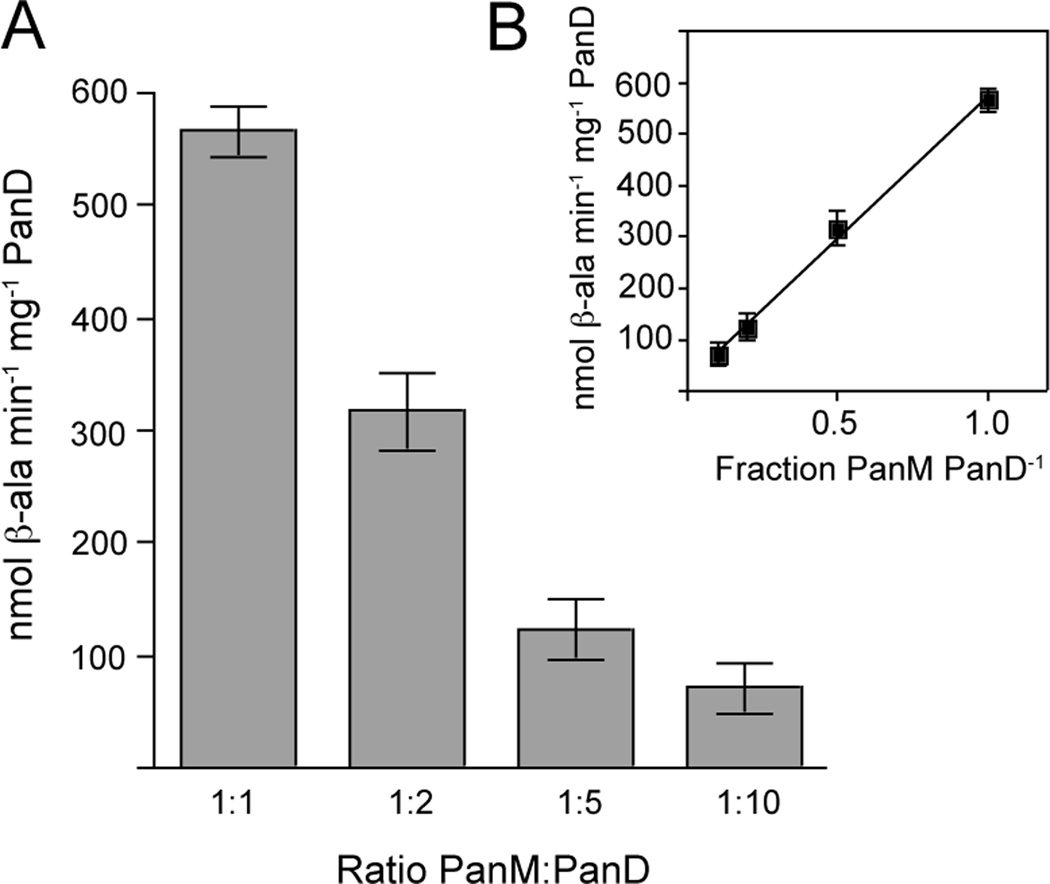
To assess the stoichiometry of the PanD-PanM interaction, PanD activity was measured as a function of PanM concentration. As the ratio of PanM to PanD was decreased, PanD activity also decreased (Fig. 8A). The relationship between PanD activity and the fraction of PanM in the reaction was linear with an r2 of 0.98 (Fig. 8B). Optimal PanD activity was observed at a 1:1 ratio of PanM to PanD, suggesting that PanM was not acting catalytically on PanD.
Figure 8. PanD activity as a function of PanM concentration.
A. PanD specific activities were determined from mixtures containing pro-PanD with decreasing concentrations of PanM, from 1:1 to 1:10 ratios of PanM to pro-PanD. B. Linear regression of L-aspartate decarboxylase activity as a function of the ratio of PanM to por-PanD, with an r2 of 0.98.
High expression of S. enterica panD can restore growth of a panM strain
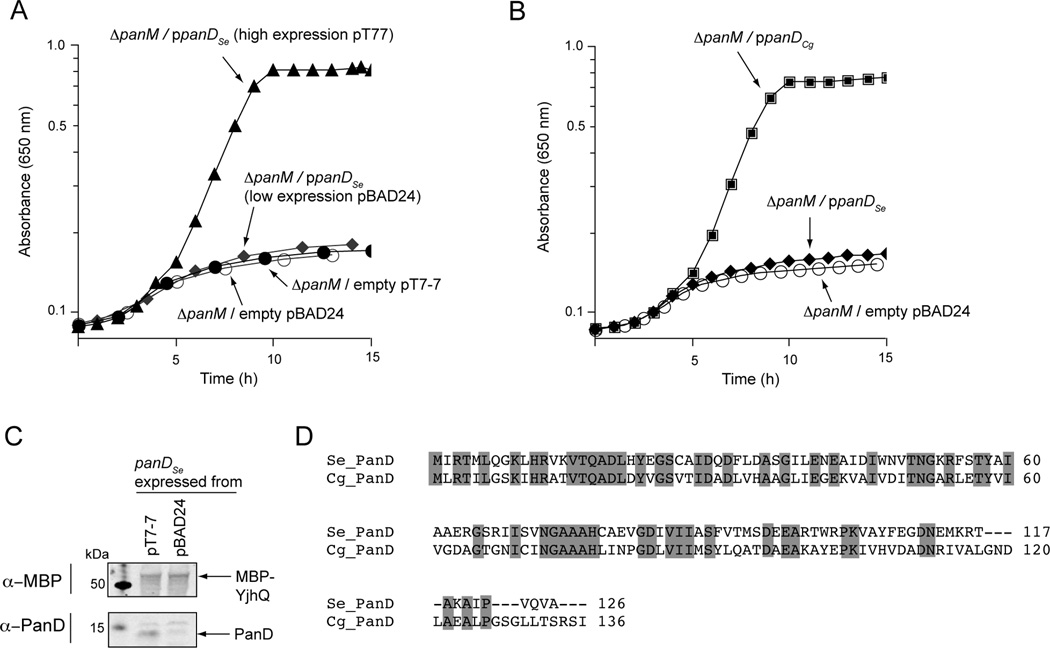
PanM is necessary for PanD cleavage, but it is unclear whether PanM participates directly or indirectly in the cleavage chemistry. Like other pyruvoyl enzymes, PanD cleavage is proposed to be autocatalytic (van Poelje & Snell, 1990, Albert et al., 1998, Ramjee et al., 1997). This is supported by the fact that a small amount of PanD purified from cells lacking panM is in its cleaved form (Fig. 4B). To further test if PanD from S. enterica could self-cleave, we expressed panD from plasmids with varying levels of expression; low expressing pBAD24 (Guzman et al., 1995) and high expressing pT7-7 (Tabor & Richardson, 1985). When S. enterica panD was expressed at low levels (pPAN7; Table S2), the strain carrying a panM deletion did not grow in minimal glycerol medium (Fig. 9A; diamonds). However, expression at higher levels from pPAN9 (Table S2) was able to restore growth in the absence of panM (Fig. 9A; triangles). Expression levels were verified using western blot analysis with α-PanD polyclonal antibodies, which showed higher PanD production when panD was expressed from pT7-7 compared to pBAD24 (Fig. 9C). These data suggest that PanD contains all of the determinants necessary for self-cleavage in vivo and PanM is necessary to accelerate the rate of cleavage.
Figure 9. Growth of a ΔpanM S. enterica strain in minimal glycerol medium containing ampicillin.
A. Complementation of a ΔpanM S. enterica with panD+ expressed at low levels (pPAN7; diamonds) or high levels (pPAN9; triangles). Empty vector controls for pBAD24 (open circles) and pT7-7 (closed circles) are shown. B. Complementation of a ΔpanM S. enterica strain with S.e. panD+ (pPAN7; diamonds) or C. glutamicum panD+ (pPAN14; squares); empty vector (pBAD24; open circles). SEM for all data was less than 0.03 absorbance units. C. Western blot analysis of cell lysates of a S. enterica ΔpanM strain expressing S.e. panD+ from either pT7-7 (lane 2) or pBAD24 (lane 3). The loading control, MBP-YjhQ, was added to lysates and 25 µg of total protein were loaded into each lane. Lane 1 contained Precision Plus Protein™ All Blue Standards (Bio-Rad). MBP-YjhQ was detected using mAb-MBP monoclonal antibodies (top panel) and the PanD protein was detected using rabbit polyclonal α-PanD antibodies (bottom panel). D. Alignment of S. enterica and C. glutamicum PanD proteins. Conserved residues are highlighted in gray.
PanD from C. glutamicum does not require PanM for maturation
Unlike E. coli or S. enterica PanD, Corynebacterium glutamicum PanD is completely cleaved when overexpressed (Dusch et al., 1999). To test whether C. glutamicum PanD still required PanM for cleavage, we cloned the C. glutamicum panD gene into pBAD24 (pPAN14; Table S2). An S. enterica strain carrying a chromosomal panM deletion and plasmid pPAN14 grew well in minimal glycerol medium (Fig. 9B), indicating that C. glutamicum PanD did not require PanM for cleavage. An alignment of C. glutamicum and S. enterica PanD proteins revealed only 41% sequence identity (Fig. 9D) making difficult to identify features of the PanD proteins that may be responsible for the differences in cleavage requirements.
Discussion
We have a good understanding of the biochemistry underpinning CoA biosynthesis in prokaryotes (Genschel, 2004). In S. enterica and E. coli, nine enzymes comprise the CoA biosynthetic pathway, one of which is aspartate decarboxylase (PanD) (Fig. 1). Notably, PanD is synthesized as an inactive precursor (pro-PanD) because it lacks the pyruvoyl moiety critical for activity. Maturation requires a self-cleavage event at residue Ser25. Although pro-PanD cleavage has been studied in some detail (Ramjee et al., 1997), we still know very little about what triggers it. Over 10 years ago, Ramjee et al. reported that heating of pro-PanD to 50°C for 48 h yielded active PanD, showing that pro-PanD contains all the determinants needed for its conversion to mature, active PanD (Ramjee et al., 1997). However, the high temperature and long incubation times made this an unlikely method for in vivo cleavage. Results of phenotypic analysis of E. coli panM strains reported >21 years ago (Adams et al., 1990) indicated that PanM (formerly YhhK) function was linked to CoA biosynthesis.
PanM is the missing link in CoA biosynthesis
Our data show that PanM is necessary for pro-PanD cleavage under physiological conditions in vitro (Fig. 6A,B). PanM is also required for pro-PanD maturation in vivo, as shown by the results of pull down experiments using α-PanD antibodies (Fig. 6C). A PanM-deficient strain accumulated pro-PanD, while the PanM-proficient strain accumulated only mature PanD. It is unclear why the cleavage efficiency was slower in vitro compared to the complete cleavage observed in vivo. The slow rate of pro-PanD cleavage in vitro may be due to the involvement of as-yet-unidentified cellular factors. Regardless, the identification of PanM as a protein involved in the maturation of pro-PanD fills the gap of knowledge in CoA biosynthesis that has existed in the literature for decades.
Not all pro-PanD enzymes require PanM to mature
PanM-aided maturation of pro-PanD in S. enterica may not be a strictly conserved trait. For example, Corynebacterium glutamicum pro-PanD appears to efficiently mature without the aid of other factors. When panD was overexpressed in E. coli, only ca. 10% of the pro-PanD made was converted to its mature form (Ramjee et al., 1997); we obtained similar results in S. enterica (Fig. 2B). In contrast, no pro-PanD can be detected in C. glutamicum panD over-expressions (Dusch et al., 1999) and C. glutamicum panD+ can compensate for the lack of PanD in a S. enterica carrying a chromosomal panM deletion, suggesting that not all pro-PanD proteins require PanM to mature. A BLAST® search (Altschul et al., 1997) of the S. enterica PanM protein against the C. glutamicum genome resulted in only weak (<30% identity) matches to several putative C. glutamicum acetyltransferases. This is similar to the sequence identities between different acetyltransferases in E. coli, which range between 2–24%. The search is further complicated by the fact that sequence identity between acetyltransferases of different organisms that carry out the same cellular function is often very weak. For example, the Pat protein of S. enterica and the AcuA protein of Bacillus subtilis both regulate the activity of acetyl-CoA synthetase (Gardner et al., 2006, Starai & Escalante-Semerena, 2004), yet their protein sequence identity is only 18%. This lack of conservation makes it difficult to determine the distribution of PanM based solely on bioinformatics analyses.
Maturation of other pyruvoyl enzymes
At the time of publication, there was one other example of a pyruvoyl enzyme requiring an ancillary protein for maturation. The histidine decarboxylase (HdcA; EC 4.1.1.22) of Streptococcus thermophilus was shown by Trip et al. to require HdcB for cleavage (Trip et al., 2011). HdcB is annotated as a domain of unknown function and the mechanism by which HdcB participates in HdcA cleavage was not determined. We believe PanM and HdbC may have evolved convergently, since the proteins are not homologous. The PanM protein does have homology to the GCN5-like N-acetyltransferases, a family of enzymes that post-translationally regulate proteins by acetylation (Vetting et al., 2005). However, preliminary experiments in our laboratory have failed to detect acetyltransferase activity by PanM (data not shown).
How does PanM contribute to pro-PanD maturation?
Pro-PanD cleavage is self-catalyzed under conditions that provide external energy, such as high temperature (Ramjee et al., 1997). Such input of energy may be needed to maintain pro-PanD in a conformation in which the maturation process can be triggered. Our data show that PanM and pro-PanD interact directly (Fig. 3,5), and that such an interaction is required for pro-PanD cleavage (Fig. 6). Since we have not detected acetyltransferase activity associated with PanM, it is plausible that the role of PanM is to stabilize the conformation of pro-PanD needed for cleavage under physiological conditions. This model is supported by the fact that PanM acts on PanD in a non-catalytic manner (Fig. 8). The mechanism of PanM function is the subject of future work.
Experimental procedures
Bacterial strains, media, and growth conditions
All strains and plasmids used in this study are listed in Table S1. Salmonella strains are derivatives of S. enterica serovar Typhimurium LT2. All growth analyses were preformed in biological triplicate and data presented are averages of at least two independent experiments. All cultures were grown aerobically at 37°C (bacteria) or 30°C (yeast) with shaking at 200 rpm unless otherwise stated. Bacterial strains were grown on either lysogeny broth (LB) (Bertani, 1951, Bertani, 2004) or no-carbon E (NCE) medium (Berkowitz et al., 1968) containing glycerol (20 mM) as the source of carbon and energy. Bacterial growth was monitored by absorbance at 650 nm using an EL808 plate reader (Biotek). Yeast strains were grown on either yeast extract peptone dextrose (YPD) or synthetic minimal (SD) medium (Amberg et al., 2005). Pantothenate (or its precursors) was present in the culture medium at a final concentration of 20 µM, ampicillin was used at 100 µg ml−1, and kanamycin at 50 µg ml−1. Pantoate and keto-pantoate were gifts from Diana Downs (University of Wisconsin-Madison). All other chemicals were purchased from Sigma.
Strain and plasmid construction
A chromosomal deletion of the S. enterica panM gene was constructed by replacing the panM coding sequence (leaving only the start and stop codons) with a kanamycin resistance cassette using established protocols (Datsenko & Wanner, 2000) followed by PCR verification. The panD-lacZ+ transcriptional reporter and panD over-expression strains were constructed by transduction using the high-frequency-of-transduction, generalized transducing phage P22 HT105/1 int-210 as described (Davis et al., 1980, Schmieger & Backhaus, 1973). All plasmids were constructed using the Clontech In-Fusion® Cloning Kit with Applied Biosystems (AB) GeneAmp High Fidelity PCR System. Inserts were amplified from either S. enterica LT2 or C. glutamicum T2 genomic DNA and were confirmed by DNA sequence analysis at the Biotechnology Sequencing Center (University of Wisconsin, Madison) using AB BigDye® methodology. Primers used in this study are listed in Table S2.
β-Galactosidase assays
Triplicate overnight LB cultures of panD-lacZ+ panM+ (JE13233) and panD-lacZ+ panM (JE13234) reporter strains were sub-cultured (1:100) into 5 ml of minimal glycerol medium containing β-alanine. Cultures were grown to mid-log phase (~0.5 A650), and β-galactosidase activities were determined as described (Miller, 1972).
Yeast two-hybrid analysis
The panD and panM genes were each cloned into the Clontech Matchmaker™ bait and prey plasmids. Bait and prey plasmids were transformed into the Matchmaker™ reporter strain AH109, and grown by auxotrophic selection on SD medium lacking leucine and tryptophan. Overnight cultures were diluted to A600 of 0.1, then serially diluted and spotted onto SD plates lacking histidine to detect bait-prey interactions. Plates were imaged after 45 h of growth at 30°C using a Fotodyne digital imaging system.
Purification of PanM
The panM gene was cloned into the pKLD66 expression vector (Rocco et al., 2008). The encoded PanM protein had a His6-MalE (H6-MBP) tag at its N-terminus. The H6-MBP tag was removed using a recombinant tobacco etch virus protease fused at its N-terminus to a His7 tag (His7-rTEV) protease (Blommel & Fox, 2007). The resulting pPAN68 plasmid was transformed into E. coli C41 (DE3), then grown to an OD600 of 0.9 in 8 L of Terrific Broth (TB). At that point, exression of his6-malE-panM was induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG), and the culture was grown overnight at 28°C. Cells were collected by centrifugation at 6,000 × g, and the cell pellet was re-suspended in 140 ml of buffer A [HEPES buffer (100 mM, pH 7.5), tris-2-carboxyethylphosphine (TCEP, 1 mM), NaCl (0.5 mM), imidazole (10 mM), glycerol (1.1 M)]. Cells were lysed by sonication (3.5 min on ice; setting 10) using a 550 Sonic Dismembrator (Fisher Scientific). Lysates were clarified by centrifugation at 40,000 × g for 30 min at 4°C. Clarified lysates were applied to a 5-ml HisTrap HP Ni Sepharose column (GE Healthcare). H6-MBP-PanM was eluted off the column using a linear gradient of imidazole in buffer A from 10 to 500 mM. The H6-MBP tags were removed by overnight incubation at 4°C with His7-rTEV protease at a molar ratio of 1:50 His7-rTEV:H6-MBP-PanM. After dialysis against buffer A, a second purification step using a Ni-NTA Sepharose column (10 ml) was preformed to separate PanM from His7-rTEV and H6-MBP. Purified PanM was dialyzed against PanM storage buffer [HEPES (100 mM, pH 7.5) containing NaCl (50 mM), TCEP (1 mM), glycerol (1.1 M)] and stored at −80°C until used. All dialyses exerted at least a 108 dilution factor.
Purification of aspartate decarboxylase
The panD gene was cloned into vector pT7-7 (pPAN9, Table S2) for the overproduction of pro-PanD in strain JE13153, in which panM was inactivated (Table S1). The pPAN9 plasmid was transformed into JE13153, then grown overnight at 37°C in 20 L of NCE glycerol medium containing ampicillin, β-alanine and histidine (2 mM). panD expression was induced for 2 h at 37 °C) after the addition of IPTG (1 mM) to the culture. Cells were pelleted by centrifugation, re-suspended in 100 ml of Tris-HCl buffer (20 mM, pH 7), and disrupted by sonication as described for PanM. Lysates were clarified by centrifugation and purified on a 25 ml Q FF column (GE Healthcare) using an ÄKTA explorer FPLC system (GE Healthcare). The column was equilibrated with Tris-HCl buffer (20 mM, pH 7), and protein was eluted off the column using a linear gradient of NaCl (0–1 M). Any remaining PanD present in flow-through fractions was re-applied to the Q FF column and purified as described. Fractions from all Q purifications containing PanD were pooled and dialyzed into Tris-HCl buffer (20 mM, pH 7) containing ammonium sulfate (1 M), then passed through a 20 ml Phenyl FF column (GE Healthcare) to remove contaminating proteins. Flow-through fractions containing PanD were desalted using a 20-ml bed volume Desalting Column (GE Healthcare) equilibrated with Tris-HCl (20 mM, pH 8). Desalted PanD fractions were applied to a 1-ml Mono-Q 5/50 column (GE Healthcare). PanD was eluted off the column using a shallow gradient from 0–30% B (Tris-HCl buffer, 20 mM, 1M NaCl, pH 8). Elution fractions containing pure, un-cleaved PanD were pooled and dialyzed against PanD storage buffer [HEPES buffer (20 mM), glycerol (1.1 M), pH 7), then stored at −80°C until used.
Chemical crosslinking
Reactions (20 µl) containing pro-PanD and PanM (0.2 mg ml−1 each) in HEPES buffer (50 mM, pH 7) were incubated either alone or in the presence of formaldehyde (25 mM) for 30 min at 37°C. Control reactions included PanD and PanM each incubated with either BSA or lysozyme. Crosslinking reactions were stopped by the addition of an equal volume of SDS buffer [Tris-HCl (0.125 M, pH 6.8), glycerol (2.2 M), SDS (4%, w/v), Coomassie Blue (0.003%, w/v]. Samples (10 µl) of each reaction were resolved on a denaturing 12% acrylamide gel; proteins were visualized using Coomassie Blue staining. The brightness and contrast of gel images was adjusted in Photoshop vCS2 (Adobe).
Mass spectrometry
The unique band present in the formaldehyde-treated sample was excised from the gel and submitted to the Proteomics Facility (University of Wisconsin-Madison Biotechnology Center) for mass spectrometry analysis. Briefly, the gel slice was de-stained twice for 5 min in 50% (v/v) methanol containing NH4 HCO3 (100 mM) followed by a three-step dehydration: (i) 5 min in 50% (v/v) acetonitrile [ACN] containing NH4 HCO3 (25 mM); (ii) 1 min in 100% ACN; (iii) dried 1 min in under vacuum. The dried sample was reduced for 30 min (dithiothreitol (25 mM) in NH4 HCO3 (25 mM) at 56°C), alkylated for 30 min using iodoacetamide (55 mM) in NH4HCO3 (25 mM) at room temperature), then dehydrated once more. The proteins in the gel slice were cut into peptides by rehydrating the sample with 20 µl of digestion solution (0.01% w/v ProteaseMAX [Promega] in NH4 HCO3 (25 mM) containing 10 ng µl−1 Trypsin Gold (Promega). An additional 30 µl of the digestion solution was added once the gel was hydrated and the sample was incubated for 3 h at 42°C. Peptides generated were acidified to a final concentration of 0.3% trifluoroacetic acid (TFA), centrifuged at maximum speed in a table top microfuge for 10 min, then applied to a ZipTip® C18 column for purification. Peptides were eluted with 60% ACN containing 0.1% TFA into a Protein LoBind tube (Eppendorf).
Eluted peptides (0.5 µl) were deposited onto an Opti-TOF™ 384 well plate (Applied Biosystems) and re-crystalized with 0.5 µl of matrix (10 mg ml−1 α-cyano-4-hydroxycinnamic acid in ACN (75%, v/v) containing TFA (0.1% w/v). Peptide map fingerprint result-dependent MS/MS analysis was performed on a 4800 Matrix-Assisted Laser Desorption/Ionization-Time of Flight-Time of Flight (MALDI-TOF-TOF) mass spectrometer (AB Sciex). Briefly, a peptide fingerprint was generated by scanning the 700–4,000 Da range using 1,000 shots acquired from 20 randomized regions of the sample spot (OptiBeam™ intensity of 4,000; on-axis Nd:YAG laser with 200 Hz firing rate; 3–7 ns pulse width; positive reflectron mode). The 15 most abundant peptides were selected for subsequent tandem MS analysis (1,200 total shots; 4,400 laser intensity; 2 kV collision induced activation using air). Post-source decay fragments were isolated by timed-ion selection and reaccelerated into the relectron to generate the MS/MS spectrum. Raw data Raw data was deconvoluted using GPS Explorer™ software and mapped against the NCBI Salmonella database using Mascot search engine.
Gel filtration analysis
Homogeneous PanD and PanM were run on a Superdex 200 HR 10 (GE Healthcare) column pre-equilibrated with Buffer A (50 mM HEPES pH 7 + 150 mM NaCl). Samples of each protein (50 µg) were injected, and the resin was developed isocratically using 150 ml of Buffer A at a rate of 0.3 ml min−1. Molecular mass standards (Bio-Rad) used to estimate the mass of PanM and PanD. Data were analyzed and graphed using Prism 4.0 (Graphpad).
Pro-PanD cleavage assays
Pro-PanD was incubated either alone or with equimolar PanM at 37°C in HEPES buffer (50 mM, pH 7) for 4 h. At 30 min intervals, 40-µl samples (containing pro-PanD and PanM, 3 µg each) were removed, mixed with an equal volume of Laemmli Loading Buffer (BioRad) and frozen at −20°C to stop the reactions. Aliquots of each sample (0.5 µg PanD) were run on a TGX Any kD gel (Bio-Rad), then transferred to an Immobilon-P membrane (Millipore). The membrane was incubated with α-PanD rabbit polyclonal antibodies (Harlan) at a 1:5,000 dilution for 1 h followed by a 1:10,000 dilution of the secondary antibody (Pierce Immunopure Alkaline Phosphatase-conjugated goat anti-rabbit Immunoglobulin G) for 30 min. Western blots were developed with NBT/BCIP reagent (ThermoFisher) and analyzed by densitometry using a computer-controlled Fotodyne imaging system with Foto/Analyst v5.00 software (Fotodyne, Inc.) for image acquisition and TotalLab v2005 software for image analysis (Nonlinear Dynamics). Non-linear regression analysis was preformed on data from two independent experiments with Prism v 4.0 (Graphpad).
Immunoprecipitation
panM+ (DM10310) and panM (JE12555) strains were grown overnight in LB and sub-cultured into 200 ml of fresh minimal glycerol medium containing β-alanine. Cultures were grown to late log phase, centrifuged at 5,000 × g for 15 min and re-suspended in 0.5 ml of HEPES buffer (50 mM, pH 7). Cells were lysed by sonication for 30 s as described above, cell-free extracts were clarified by centrifugation at 14,000 × g for 30 min at 4°C. Protein concentration was determined using the Bradford Assay (Bio-Rad), and 9 mg of protein from each sample was mixed with 100 µl of α-PanD rabbit polyclonal antibodies (Harlan). The protein-antibody samples were applied to Nab™ Protein G 0.2 ml spin columns (Thermo Scientific) and PanD-antibody complexes were purified according to manufacturer’s instruction. For each strain, duplicate cultures were grown and purified; samples were combined after elution to increase the final yield. Samples were exchanged into HEPES buffer (50 mM, pH 7) and concentrated to 50 µl using Amicon Ultra 0.5 ml concentrators (Millipore) with a 30-kDa molecular mass cut off. Samples were run on denaturing acrylamide gels and PanD was detected by Western blot as described above. Images were taken with a Fotodyne imaging system with Foto/Analyst v5.00 software (Fotodyne, Inc.).
PanD activity assays
Aspartate decarboxylase activity was determined using a modified assay of Dusch et al (Dusch et al., 1999). Cleavage assays described above were incubated for 2 h, then diluted to a final volume of 140 µl in PanD Assay Buffer (100 mM potassium phosphate pH 6.8, 0.1 mM ethylenediaminetetraacetic acid [EDTA]). Reactions were started by adding aspartate at 2 mM, then incubated at 37°C for 1 hour. An equal volume of Stop Buffer (3% perchloric acid containing the internal standard 4-amino-butyrate at 0.1 mM) was added and precipitated proteins were removed by filtration (0.4 micron SpinX; Corning). The amount of β-alanine in each sample was determined by Phthaldialdehyde derivatization (Sigma) followed by reverse phase HPLC using a 150 × 4.6 mm Synergi 4u Hydro-RP C18 column (Phenomenex). Aliquots (50 µl) were injected and eluted in three steps: (i) 4 min with Buffer A (0.1 M sodium phosphate, 0.1 mM EDTA, pH 6.4) and 30% methanol, (ii) gradient to 40% methanol over 1 min, (iii) Buffer A containing 40% methanol over 10 min. Results were normalized to the internal standard by dividing the area under the β-alanine curve by the area under the 4-amino-benzoate curve. Normalized areas were compared to a curve of known β-alanine standards to determine β-alanine concentrations. Data from at least two independent experiments were graphed using Prism v4.0 (Graphpad).
β-alanine bioassays
Cleavage and activity assays were preformed as described above. Samples (1 µl) of each reaction were spotted onto minimal glycerol plates overlayed with washed panD (JE13233) cells. Plates were imaged after 20 h of growth at 37°C and zones of growth (diameter) were measured. Representative plate images are shown with measurements averaged for three separate experiments preformed in biological triplicate. The brightness and contrast of plate images were adjusted using Photoshop vCS2 (Adobe).
PanD over-expression western blots
The JE12555 (panM) strain was transformed with pPAN7 (panD in pBAD24) or pPAN14 (panD in pT77) and grown to late log (A600 of 0.9) in 200 ml minimal glycerol medium containing ampicillin and β-alanine. Cells were centrifuged, resuspended in 50 mM HEPES pH 7, lysed by sonication and clarified as described for immunoprecipitaion above. Protein concentration was determined by Bradford assay (Bio-Rad). Lysates were diluted to 25 mg ml−1 and MBP-YjhQ pure protein was added to each lysate at 50 µg ml−1 as a loading control. A total of 25 µg of protein from each lysate was loaded onto an Any Kd TGX gel (Bio-Rad), then transferred to a nitrocellulose Hybond ECL membrane (GE Healthcare). PanD was detected by western blot with α-PanD rabbit polyclonal primary antibodies (Harlan; 1:2,000 dilution) and IR Dye 680CT goat anti-rabbit secondary antibodies (Li-COR Biosciences; 1:5,000 dilution). The MBP-YjhQ loading control was detected using mAb-MBP mouse monoclonal primary antibodies (Invitrogen; 1:2,000 dilution) with IR Dye 800CW goat anti-mouse secondary antibodies (LI-COR Biosciences; 1:5,000 dilution). Washes were preformed using the Odyssey® Western blotting kit (LI-COR Biosciences) and SNAP i.d.® system (Millipore). The blot was scanned using the Odyssey infrared imaging system (LI-COR Biosciences) with a gain of 3.5 for the 700 nm channel and 7 for the 800 nm channel. The image was converted to grayscale, contrast adjusted and cropped in Photoshop vCS2 (Adobe).
Protein sequence alignment
The S. enterica serovar typhimurium LT2 (NP_459185) and C. glutamicum ATCC 13032 (NP_599388) PanD sequences were aligned using ClustalW2 (www.ebi.ac.uk/Tools/msa/clustalw2).
Protein acetylation assays
Conditions used to assess protein acetylation have been described (Thao et al., 2010), and were used without modifications. Reaction mixtures contained pro-PanD, PanM, and [14 C, C-1]Ac-CoA (50 µM, 5.8 mCi/mmol; Moravek). After an incubation period (2 h), proteins were resolved by SDS-PAGE and the gel was imaged using a Typhoon FLA9000 phosphor imager (GE Healthcare).
Supplementary Material
Acknowledgments
This work was supported by the USPHS grant R01-GM62203 (to J.C.E.-S.) and NIH Molecular Biosciences Training Grant T32-GM07215 (to T.N.S.). We thank Jannell Bazurto and Diana Downs for the kind gift of pantoate, keto-pantoate, and strains, Hazel Holden for C. glutamicum DNA, and Grzegorz Sabat for assistance with mass spectrometry.
Footnotes
The authors do not have any conflicts of interest to declare.
References
- Abiko Y. Metabolism of coenzyme A. In: Greenburg DM, editor. Metabolic Patways: Metabolism of sulfur compounds. New York: Academic Press; 1975. pp. 1–25. [Google Scholar]
- Adams DM, Wagner LM, Graddis TJ, Landick R, Antonucci TK, Gibson AL, Oxender DL. Nucleotide sequence and genetic characterization reveal six essential genes for the LIV-I and LS transport systems of Escherichia coli. J. Biol. Chem. 1990;265:11436–11443. [PubMed] [Google Scholar]
- Albert A, Dhanaraj V, Genschel U, Khan G, Ramjee MK, Pulido R, Sibanda BL, von Delft F, Witty M, Blundell TL, Smith AG, Abell C. Crystal structure of aspartate decarboxylase at 2.2Å resolution provides evidence for an ester in protein self-processing. Nat. Struct. Biol. 1998;5:289–293. doi: 10.1038/nsb0498-289. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Miller W, Lipmann DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Burke D, Strathern JN. Methods in Yeast Genetics. Cold Spring Harbor, N. Y.: Cold Spring Harbor Press; 2005. [Google Scholar]
- Berkowitz D, Hushon JM, Whitfield HJ, Jr, Roth J, Ames BN. Procedure for identifying nonsense mutations. J. Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 2004;186:595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr. Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho BA, Olfson P, Casadaban MJ. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J. Bacteriol. 1984;158:488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE, Jr, Littel KJ, Jackowski S. Genetic and biochemical analyses of pantothenate biosynthesis in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 1982;149:916–922. doi: 10.1128/jb.149.3.916-922.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RW, Botstein D, Roth JR. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1980. [Google Scholar]
- Dusch N, Puhler A, Kalinowski J. Expression of the Corynebacterium glutamicum panD gene encoding L-aspartate-alpha-decarboxylase leads to pantothenate overproduction in Escherichia coli. Appl. Environ. Microbiol. 1999;65:1530–1539. doi: 10.1128/aem.65.4.1530-1539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JG, Grundy FJ, Henkin TM, Escalante-Semerena JC. Control of acetyl-coenzyme A synthetase (AcsA) activity by acetylation/deacetylation without NAD(+) involvement in Bacillus subtilis. J. Bacteriol. 2006;188:5460–5468. doi: 10.1128/JB.00215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschel U. Coenzyme A biosynthesis: reconstruction of the pathway in archaea and an evolutionary scenario based on comparative genomics. Mol. Biol. Evol. 2004;21:1242–1251. doi: 10.1093/molbev/msh119. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CE, Brook JM, Buck D, Abell C, Smith AG. Cloning and sequencing of the Escherichia coli panB gene, which encodes ketopantoate hydroxymethyltransferase, and overexpression of the enzyme. J. Bacteriol. 1993;175:2125–2130. doi: 10.1128/jb.175.7.2125-2130.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi T, Katayama T, Suzuki H, Kumagai H. Identification of the LIV-I/LS system as the third phenylalanine transporter in Escherichia coli K-12. J. Bacteriol. 2004;186:343–350. doi: 10.1128/JB.186.2.343-350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leonardi R, Jackowski S. Biosynthesis of pantothenic acid and coenzyme A. Washington, D.C.: American Society for Microbiology; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. Assay of beta-galactosidase. In: Miller J, editor. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- Miyatake K, Nakano Y, Kitaoka S. Pantothenate synthetase from Escherichia coli [D-pantoate: beta-alanine ligase (AMP-forming), EC 6.3.2.1] Methods Enzymol. 1979;62:215–219. doi: 10.1016/0076-6879(79)62221-8. [DOI] [PubMed] [Google Scholar]
- Ramjee MK, Genschel U, Abell C, Smith AG. Escherichia coli L-aspartate-alpha-decarboxylase: preprotein processing and observation of reaction intermediates by electrospray mass spectrometry. Biochem. J. 1997;323:661–669. doi: 10.1042/bj3230661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid. 2008;59:231–237. doi: 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H, Backhaus H. The origin of DNA in transducing particles in P22-mutants with increased transduction-frequencies (HT-mutants) Mol. Gen. Genet. 1973;120:181–190. doi: 10.1007/BF00267246. [DOI] [PubMed] [Google Scholar]
- Smith CM, Song WO. Comparative nutrition of pantothenic acid. J. Nutrition. Biochem. 1996;7:312–321. [Google Scholar]
- Starai VJ, Escalante-Semerena JC. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 2004;340:1005–1012. doi: 10.1016/j.jmb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Tabor S, Richardson CC. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. U.S.A. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teller JH, Powers SG, Snell EE. Ketopantoate hydroxymethyltransferase. I. Purification and role in pantothenate biosynthesis. J. Biol. Chem. 1976;251:3780–3785. [PubMed] [Google Scholar]
- Thao S, Chen CS, Zhu H, Escalante-Semerena JC. N(epsilon)-Lysine acetylation of a bacterial transcription factor inhibits its DNA-binding activity. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0015123. e15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trip H, Mulder NL, Rattray FP, Lolkema JS. HdcB, a novel enzyme catalysing maturation of pyruvoyl-dependent histidine decarboxylase. Mol. Microbiol. 2011;79:861–871. doi: 10.1111/j.1365-2958.2010.07492.x. [DOI] [PubMed] [Google Scholar]
- van Poelje PD, Snell EE. Pyruvoyl-dependent enzymes. Annu. Rev. Biochem. 1990;59:29–59. doi: 10.1146/annurev.bi.59.070190.000333. [DOI] [PubMed] [Google Scholar]
- Vetting MW, Carvalho LPSd, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Williamson JM, Brown GM. Purification and properties of L-Aspartate-alpha-decarboxylase, an enzyme that catalyzes the formation of beta-alanine in Escherichia coli. J. Biol. Chem. 1979;254:8074–8082. [PubMed] [Google Scholar]
- Zheng R, Blanchard JS. Kinetic and mechanistic analysis of the E. coli panE-encoded ketopantoate reductase. Biochemistry. 2000;39:3708–3017. doi: 10.1021/bi992676g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.