Abstract
Covalent modifications of histone proteins play key roles in transcription, DNA repair, recombination, and other such processes. Over a hundred histone modifications have been described, and a popular idea in the field is that the function of a single histone mark cannot be understood without understanding its combinatorial co-occurrence with other marks, an idea generally called the “histone code hypothesis.” This idea is hotly debated, with increasing biochemical evidence for chromatin regulatory factors that bind to specific histone modification combinations, but functional and localization studies finding minimal combinatorial complexity in histone modification patterns. This review will focus on these contrasting results, and will briefly touch on possible ways to reconcile these conflicting views.
INTRODUCTION
The great variety and number of covalent modifications found on proteins provides one of the great challenges of the post-genome era – understanding the functions of the tens of thousands of proteins encoded in a given genome is difficult enough without having to contend with billions of possible combinatorial modification states on individual proteins. Yet this is exactly the problem we are faced with in chromatin – the highly-conserved histone proteins are extensively modified post-translationally, with well over one hundred distinct modification sites described in the literature. Histone modifications include lysine acetylation, lysine methylation, arginine methylation, serine phosphorylation, lysine ubiquitination, and many others. This incredible diversity of histone modifications leads naturally to the question of what it all means – why do so many histone modifications occur in the cell? This question only becomes more vexing when considering that even in the past year mass spectrometry studies have identified scores of previously unknown histone modifications [1], vastly increasing the possible state space of combinatorial modifications. Perhaps the most-cited and most-debated concept in histone modifications is something called “the histone code”, and it is the purpose of this review to critically evaluate current evidence for and against the histone code hypothesis.
What is the “histone code”?
One pervasive semantic problem in the histone modification literature is that different investigators mean different things when they refer to the histone code, meaning that in many cases two papers can present nearly-identical results yet claim that their respective results support or refute the histone code. There are multiple reasons for this confusion, ranging from the multiple dictionary definitions of the word “code” to the number of histone modification reviews that use the code metaphor. Hypotheses based on the code metaphor include the idea that histone modifications have binding partners [2], the idea that histone modification patterns are heritable [3], and the suggestion that histone modifications function in combinatorial patterns [4], among others.
I will focus here on the review from Strahl and Allis entitled “The Language of Covalent Histone Modifications” [4]. This review focused on combinations of histone modifications, and has lead to a hotly-contested notion that specific combinations of histone marks would specify unique biological outcomes. Even focusing on this one histone code hypothesis, there is a great deal of confusion and debate in the literature with the same result often being used as evidence either for or against the “histone code”. However, I believe the most clearly implied hypothesis in the review centers on the concept that a given combination of modifications, such as H3K4me3/H4K16ac, would be “read” by a combination-specific protein or protein complex and thereby lead to a downstream event distinct from the readouts resulting from H3K4me3 alone or H4K16ac alone. It is important to note that even this prediction is subject to some level of ambiguity: if H4K5ac leads to a 2-fold increase in transcription, and H4K8ac leads to a 2-fold increase, then if H4K5acK8ac leads to a 4-fold increase in transcription is this a “code”? Or does the effect of combinations in a code need to be qualitatively distinct from a linear combination of the two single effects? Nonetheless, this aspect of the histone code provides fodder for investigation. How many distinct modification combinations occur, and can specific combinations of modifications be matched with specific outcomes such that the code can be deciphered?
At present, an intellectual schism exists between biochemists on one hand, and geneticists and epigenomics researchers on the other. Genome-wide mapping of histone modifications invariably shows that histone modifications occur in groups of multiple highly-correlated modifications, demonstrating that the huge potential space of modification combinations is not utilized in vivo. Conversely, many chromatin regulatory proteins and complexes contain multiple binding domains specific for histone modifications, and in some cases have been shown to bind in vitro to a combination of modifications more strongly than to either single modification, suggesting that these “readers” must respond to some specific combination in vivo. These results are of course not incompatible – in most described cases, histone binding proteins tend to bind to combinations that occur widely in vivo, so binding sites certainly exist for such factors in vivo. The problem lies in the fact that such combinatorial binding does not therefore explain specificity in the function of chromatin regulators – if a factor binds to two marks that globally co-occur, then combinatorial histone modification patterns do not provide any additional insight into why a given histone modification only affects a small subset of genes at which it occurs in vivo. We believe that bridging this divide between localization, biochemistry, and function is the single greatest challenge facing the histone modification field currently.
Multivalent binding by protein complexes
The pro-complexity viewpoint comes from, and finds support in, the field of chromatin modifying enzyme biochemistry. Chromatin-regulating proteins often carry domains that have been shown to bind specifically to a type of covalently-modified amino acid – bromodomains bind to acetylated lysines [5], PHD fingers bind to methylated lysines [6], and so forth. Most of these domains bind to singly-modified histone peptides, although occasionally it appears that a single modification-binding domain can bind to a multiply-modified histone peptide – for example, bromodomain 1 from the mouse TAF1 homolog Brdt binds to H4K5acK8ac peptides with a Kd of 28 µM, but does not bind detectably to either singly modified peptide [7].
Many individual proteins include more than one of these domains, or in some cases protein dimerization leads to complexes with multiple histone-binding domains. For example, the Snf2 homolog and ATP-dependent remodeling protein Chd1 has two chromodomains, while the yeast RSC subunit Rsc1 carries two bromodomains. When considering entire protein complexes this potential for multivalent binding is obviously much greater, with large complexes such as TIP60 carrying as many as 10 distinct histone modification-binding domains.
This potential for multivalent binding is a significant motivator for the pro-complexity argument (Figure 1). Indeed, in some cases, proteins or complexes with multiple modification-specific domains have been shown to bind to combinatorial histone marks [8]. For example, the Arabidopsis CMT3 chromodomain preferentially binds, as a dimer, to the doubly-modified H3K9me3K27me3 but not to either individual methylation state [9]. As an example of an individual protein with multiple histone binding domains, Chd1 carries tandem chromodomains which bind with higher affinity to H3R2me2K4me3 peptides than to H3K4me3 alone [10]. Another protein involved in ATP-dependent chromatin remodeling, BPTF, carries adjacent PHD and bromodomains. The bromodomain binds to peptides carrying one of three different acetylated H4 residues with affinities from 70 to 130 µM [11], whereas the PHD finger binds to H3K4me3 peptides with 2.7 µM affinity [6]. Together, the PHD-Bromo module from this protein was shown to bind with a ~2-fold increased affinity to H3K4me3/H4K16ac mononucleosomes relative to nucleosomes lacking the H4K16ac [11] (as illustrated in Figure 1A). Peptide microarray studies also showed an increase in binding between this PHD-Bromo domain construct when H3K4me3 was combined with many different H3 acetyl states [10] (which were not tested in the mononucleosome-binding assay of Ruthenburg et al), consistent with the relative promiscuity of bromodomains for acetyl-lysines.
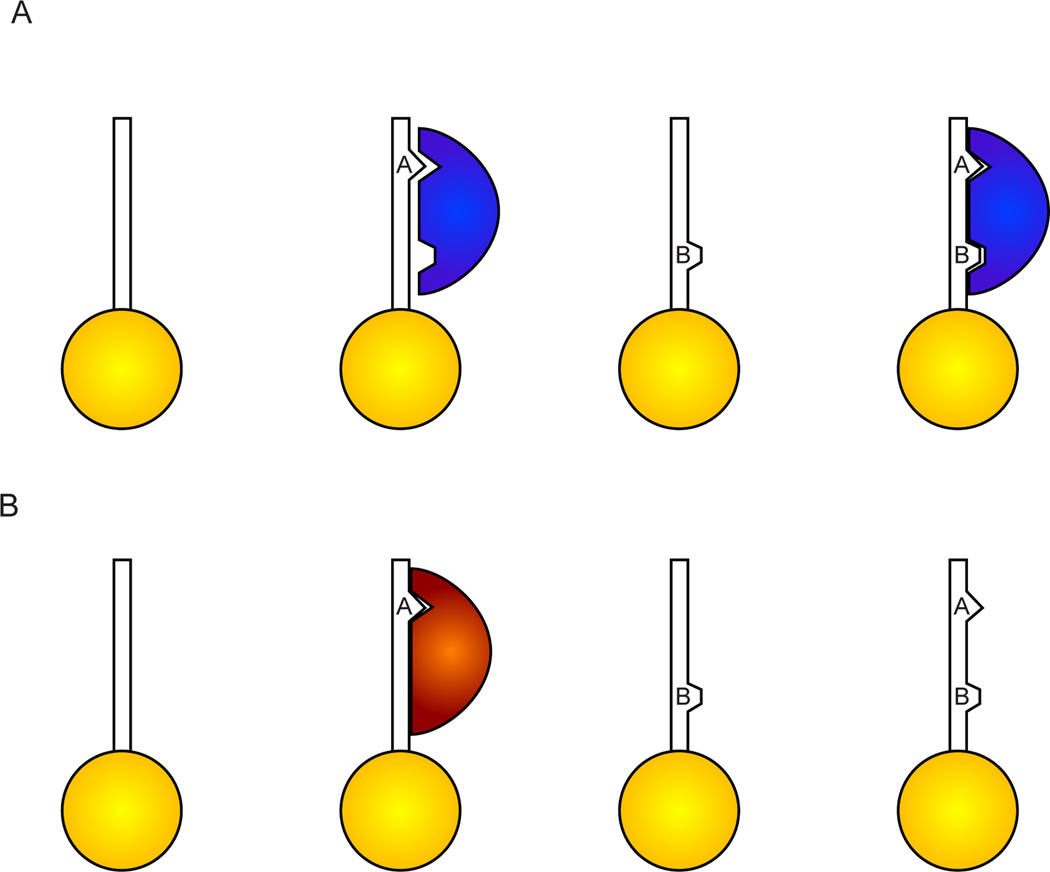
Figure 1. Biochemical evidence for combinatorial histone modification binding.
A) Some proteins (see example of BPTF in the text) show increased affinity for doubly-marked nucleosomes than for single modifications. Here, two modifications are shown generically as A and B. In the case of BPTF, A represents H3K4me3 and B represents H4K16ac. BPTF binding affinity is ~2-fold stronger for AB than for A alone. B) In other cases, a second modification can prevent a protein from binding to a modified histone. This is best-exemplified by the loss of HP1 binding to H3K9me3 when H3S10P is adjacent. Note that only two readout states exist here for the four possible combinations of A and B.
In other (seemingly more numerous) cases, modification-specific binding by a given domain is inhibited by nearby modifications (Figure 1B). Most famously, binding of HP1 to H3K9me3 is inhibited by phosphorylation of the adjacent H3S10 [12]. A similar case was described for a PHD-Bromo cassette in TRIM24 [13], which binds preferentially to H3K23ac, but whose binding is inhibited by methylation at H3K4. Inhibition of modification-specific binding by H3K4 methylation also occurs for the ADD (Atrx, Dnmt3, Dnmt3L) domain of Atrx, which binds to H3 tail peptides carrying H3K9me3 or me2, but whose binding is inhibited by H3K4 methylation [14, 15]. It is worth pointing out in such cases that combinatorial complexity remains low – for HP1 binding, three of four combinations of marks (unmodified H3, H3S10P, and H3K9me3S10P) lead to the same output (no HP1 binding), so the four possible combinations still only yield two biological outputs.
Thus, in an increasing number of cases evidence supports the idea that “multivalent” modification binding can occur in vitro. However, the connections between biochemical binding events (which are generally of disconcertingly low affinities of micromolar and above) and in vivo function are often unclear. Specifically, functional studies in which transcriptional output in specific chromatin mutants is assayed often find little link between the presence of a chromatin regulator and its function (at least in transcription). Moreover, genome-wide mapping studies find little evidence for combinatorial complexity in histone modification patterns, and raise the question of whether combinatorial binding by proteins provides any meaningful biological discrimination (eg if states A and B always co-occur, then an AB-binding protein has no need to discriminate between A and AB). These issues are detailed below.
Gene expression in combinatorial histone mutants
One systematic approach to understand the functions of combinatorial histone modifications has been the whole genome analysis of gene expression changes observed in histone mutants. Such studies typically find that histone mutants exhibit relatively little phenotypic complexity resulting from different combinations of histone mutants. This is seen both in studies focusing on specific histone point mutants and their combinations, as well as in studies focusing on deletion mutants of histone modifying enzymes.
An early study from the Wyrick lab examined all 5 single K→A mutations in the H3 N-terminal tail, as well as the quintuple mutant [16]. Gene expression changes in the single mutants were modest, and were highly correlated with one another. In other words, the loss of H3K9 acetylation appears little different than the loss of H3K18 acetylation to the cell. A similar study on the H4 N-terminal tail systematically examined all 16 possible combinatorial mutations among the 4 lysines in this tail [17]. Here, mutation of three of the residues (lysines 5, 8, and 12) had indistinguishable effects on gene expression, whereas lysine 16 was confirmed to have unique effects on gene expression. Furthermore, gene expression defects in combinatorial mutants were little different from linear combinations of the component mutations – in other words, the effect of the H4K5,16R double mutant on gene expression could be predicted by adding the K5R and K16R datasets together. Similar results are observed when combining deletions of chromatin regulatory factors [18].
Perhaps more troubling, a dramatic mismatch is observed between mapping studies and gene expression analyses. For instance, H3K4me3 occurs universally at the 5’ ends of transcribed genes, yet mutation of H3K4 or of the K4 methylase has almost no effects on gene expression [19, 20]. This observation holds true for virtually all chromatin regulators studied, although it is not routinely pointed out. As one of a multitude of prominent examples, Polycomb group proteins in embryonic stem cells map to the promoters of key developmental regulators, yet various Polycomb knockdowns do not result in upregulation of most of these genes. This conundrum will be discussed below.
Genome-wide modification patterns
How many histone modification combinations occur in vivo? The advent of genome-wide localization analysis has enabled an unprecedented view of histone modifications. The first of these studies to specifically address the question of combinatorial histone modifications came from Schubeler and Groudine, who showed using ~1 kb resolution microarrays that many histone modifications were highly-correlated in vivo [21]. Similar results were obtained by the Grunstein lab for budding yeast, with strong correlations among a variety of histone acetylation states in vivo [22]. Such results were soon obtained at mononucleosomal resolution in yeast [23], where the majority (>81%) of the variance in a set of 12 histone modifications could be captured using two groups of modifications (Figure 2).
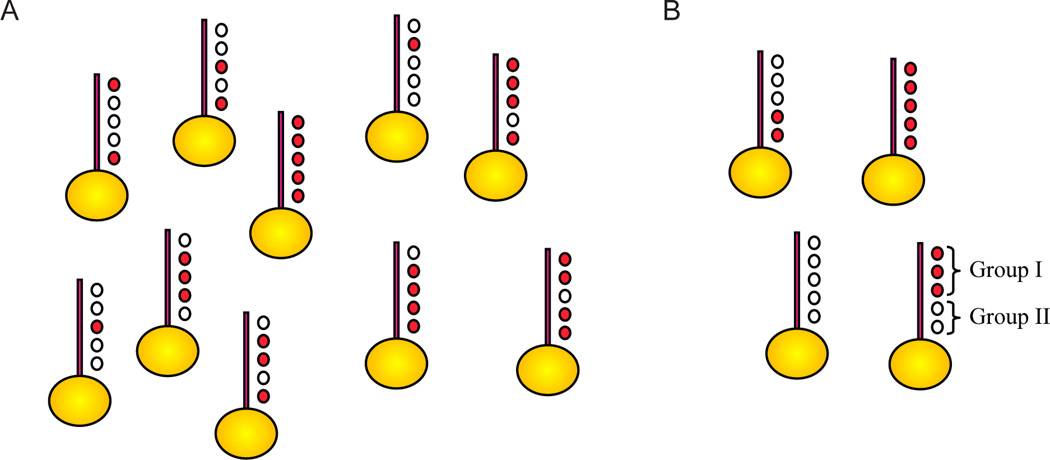
Figure 2. Genomic localization patterns reveal little combinatorial complexity.
A) Cartoon showing one possibility for 5 histone modifications, in which many or all of the 32 possible combinations occur.
B) Cartoon showing limited combinatorial complexity, as actually occurs in vivo. This figure is adapted from Liu et al, 2005.
These results have been seen over and over in ensuing years. In mammals the Zhao laboratory pioneered the genome-wide mapping of many histone methylation and acetylation states in CD4+ T cells [24, 25], and later analysis of these 41 chromatin marks showed that >56% of the variance in this dataset was captured using 3 principal components [26]. Subsequently the ENCODE and reference epigenome projects have extended these results to a wide range of cell lines, with similar conclusions [27]. Similar studies have been published for many species, most extensively D. melanogaster [28], A. thaliana [29], and C. elegans [30]. In all cases, it is found that the potentially enormous space of possible combinatorial modification patterns can be productively compressed with little loss of information. For instance, low resolution analysis of 53 proteins (largely chromatin-related) by DamID in Drosophila revealed that 5 “colors” of chromatin could be used to classify the majority of the genome [31]. At nucleosomal resolution, using 18 chromatin marks in Drosophila [28], the authors identified 9 distinct “combinatorial” histone modifications patterns, dramatically compressing the potential state space down from ~218 (>250,000).
Such results are possible because of the strong co-occurrence patterns of histone modifications, a result also observed (but seldom remarked upon) in mass spectrometry studies. When individual chromatin “states” are defined from mapping studies, these typically describe a handful of genomic locations – promoters, introns, exons, enhancers, etc. At each location, many histone modifications co-occur. Most obviously, actively transcribed promoters, in all organisms studied, are associated with H3K4me3 as well as a range of histone tail acetylation marks such as H3K9ac and many others. Many reasons exist for such co-occurrence patterns, including low substrate specificity for some histone-modifying enzymes (particularly acetyltransferases), recruitment of multiple modifying factors to a genomic locus (as in, eg, multiple histone modifying enzymes traveling with elongating RNA Pol II), and the recruitment/regulation of histone modifying enzymes by other histone modifications (histone crosstalk).
The simplicity of histone modification patterns in vivo therefore raises questions about the meaning of combinatorial histone modification binding observed so often in vitro. Most relevant in my view is the question of whether combinatorial binding by chromatin regulators leads to any biological discrimination between genomic regions. The ability of a protein to bind to combination AB does not mean that the protein’s in vivo function is to distinguish between A and AB (or between B and AB). In other words, if two modifications universally co-occur in vivo and are located close to one another on a peptide, then a protein selected to bind to A will most likely bind to AB. But this does not mean that the protein plays a role in distinguishing A from AB in vivo.
For instance, BPTF binding to H3K4me3 is enhanced by various H3 or H4 acetylation states [10, 11]. Since H3K4me3 typically co-occurs with a large number of H3/H4 acetylation marks in vivo [23, 27, 28], the role of BPTF’s bromodomain in genome-wide localization is likely to be minimal. Furthermore, other factors must play key roles in localization of chromatin factors such as BPTF – Ruthenburg et al compared BPTF localization genome-wide to H3K4me3, H4K12ac, and H4K16ac, and not only are the predicted overlaps observed, but even in the examples presented in Figure 6 from Ruthenburg et al [11] several BPTF binding peaks are observed that cannot be explained by these histone marks. Many other examples of combinatorial histone modification binding proteins or complexes raise similar questions – the Atrx ADD domain binds to H3K9me3, a heterochromatin mark, and this binding is inhibited by methylation at H3K4, which typically occurs in euchromatin. Since these marks seldom if ever co-occur, it remains to be seen whether H3K4 methylation is ever used in vivo to evict Atrx from a locus previously carrying H3K9me3 without K4 methylation.
Taken together, we find little evidence at present to support the hypothesis that recruitment or activation of chromatin regulatory proteins by a specific combination of modifications leads to unique outcomes. Yet there is little question that some proteins do bind in vitro to specific modification combinations. How can we reconcile these views: why do most confirmed combination-specific binding proteins have in vivo functions that appear not to rely on combinatorial modifications? Below, we note several potential reasons for the disconnect between chromatin biochemistry and in vivo function.
Histone modification binding partners – recruiting modules or allosteric regulators?
As noted above, one prediction that might be made for the histone code is that proteins that bind strongly to a specific combination of modifications should be specifically found at genomic loci where this combination occurs. This prediction often fails to hold, as noted for BPTF above. Similarly, Chd1 binds preferentially to H3R2me2K4me3 in vitro, yet localizes very close to the TSS in embryonic stem cells [32], where H3K4me3 likely occurs without concomitant R2 methylation (which is limited to gene bodies). What role, then, does R2 methylation play in Chd1 biology?
An emerging theme in the biology of histone modification binding domains is that these domains may not be involved in recruiting chromatin regulators to specific genomic loci, but may instead play roles in allosterically regulating complex behavior. One of the clearest examples is the Rpd3S complex, a deacetylase complex conserved from yeast to humans. This complex carries a chromodomain protein Eaf3 (yeast), which has been shown to specifically bind to H3K36me3 over the middle and 3’ ends of coding regions [33, 34]. Deletion of the H3K36 methylase Set2 in yeast, or deletion of the Eaf3 chromodomain, both lead to increased histone acetylation over the bodies of transcribed genes [35]. This suggested that H3K36me3 served to recruit this complex to coding regions, where the complex functioned to deacetylate histones. Surprisingly, it has recently been shown that deletion of Set2, or of the Eaf3 chromodomain, has no effect on localization of the Rpd3S complex [36]. Instead, this complex is localized by binding to the S2P form of the Pol2 CTD (the same mark that recruits Set2). Thus, localization of the complex is unaffected by H3K36 methylation, while function of the complex is completely dependent on this mark, suggesting that H3K36me3 serves to activate the Rpd3S complex, rather than recruit it.
A clear example of allosteric regulation of chromatin complexes by modifications comes from elegant work from the Cairns lab on the RSC ATP-dependent chromatin remodeling complex. This complex contains several bromodomains, including a tandem bromodomain in Rsc4, giving it the combinatorial potential to bind to a very specific pattern of lysine acetylation in vivo. However, a crystal structure of Rsc4 found that while the first bromodomain could bind to H3K14ac, the second bromodomain bound to Rsc4 K25ac [37]. Acetylation of this lysine by Gcn5 was shown to inhibit Rsc4 binding to histone peptides. This provides a mechanistically detailed example of allosteric regulation of chromatin regulatory complex function, rather than localization, by lysine modification, although in this case the modification occurs on the complex itself rather than on the histone proteins.
Regulation of other ATP-dependent remodeling enzymes by histone marks appears to be at the level of function rather than localization – the tandem chromodomain Chd1 regulates its nucleosome binding and function, but not its genomic localization [38, 39]. Similarly, regulation of Drosophila ISWI by acetylation of H4K16 appears to occur largely at the level of ATPase activity rather than localization [40, 41].
Together, these examples provide one potential explanation for the disconnect between modification binding in vitro and in vivo localization of specific proteins. If a combinatorial binding protein is activated, rather than recruited, by one or more of its preferred combinations of histone modifications, this could explain why binding proteins with characterized histone binding specificities do not exhibit predicted localization patterns.
What do histone modifications even do? Chromatin mutants have unpredictable effects
We next come to the question of why genetic analyses of histone mutants do not provide any evidence supporting a requirement for combinatorial modification patterns for the in vivo functions of chromatin regulators. In general, there is a vexing mismatch between histone modification occurrence and importance, even for single modifications [42]. In other words, H3K36me3 is a universal mark found over coding regions and has been termed an “elongation” mark, yet H3K36A mutants, or set2Δ mutants, affect a very small subset of genes in yeast (and do not exhibit general defects in transcriptional elongation).
In perhaps the most egregious example, H3K4me3 is universally found at the 5’ ends of genes [23, 24, 43–45], at levels that scale with transcription of the associated gene (or, at genes with paused RNA polymerase, at levels that scale with polymerase abundance). H3K4me3 is linked to gene activation, as Trithorax group proteins in Drosophila required for maintenance of a gene’s active state during development play roles in H3K4 methylation, or are recruited by H3K4 methylation [46]. Yet in budding yeast, deletion of the K4 methylase Set1 is well-tolerated, resulting primarily in derepression of ~50–100 genes such as midsporulation genes [19]. In mammalian ES cells, knockdown of DPY30 results in a global 80% decrease in H3K4me3, yet remarkably few genes change expression [20]. The central question raised is: Why do so few genes “care” about H3K4me3, and what is special about those that do?
Several explanations for this behavior have been proposed. These include, first, the possibility that transcription is not the relevant signal generated by a given mark, which is no doubt often true. Second, in the case of H3K4me3, it was suggested that this mark “poises” genes for future activation, which is plausible in some cases, although alternative hypotheses can explain much of the current evidence for this idea. Third, the gene-specific effects of mutants in the relevant modifying enzymes, provides much of the motivation for belief in the histone code. For example, if a given protein such as BPTF only functions at H3K4me3/H4K16ac promoters then this might explain why not all H3K4me3 promoters are affected by BPTF knockdown. However, based on the widespread co-occurrences noted above, this explanation is unlikely in most cases (although most extant modification mapping studies are carried out in near-optimal growth conditions, leaving the possibility that certain combinatorial modification patterns occur in alternative growth conditions).
One of the most instructive examples for thinking about the context-dependence of histone modifications comes from the yeast RPD3S complex, which as noted above is activated by H3K36me3. The role of this deacetylase complex is to re-compact chromatin loosened by Pol2 passage, and to thereby repress sequences in coding regions (and elsewhere) that would act as promoters if inappropriately opened. These so-called “cryptic” promoters are not universal, occurring within a few hundred open reading frames [47]. Thus, while RPD3S acts universally over coding regions, only genes with cryptic internal promoters are significantly affected by its loss, neatly explaining the specificity of action of this near-universal mark. Recently, a conceptually similar explanation was offered for the role of the H2A.Z ortholog in S. pombe – mutants lacking this near-universal 5’ chromatin mark carry increased levels of antisense transcripts at genes oriented convergently with respect to a downstream gene [48]. Thus, the role of H2A.Z here is to provide a universal 5’-restricted signal, so that the passage of elongating Pol2 must result from an antisense transcript, which can then be degraded. It seems likely that additional examples like this will explain context dependence of histone modifications in the future – in budding yeast, loss of the K4 methylase Set1 is increasingly seen to specifically affect the expression of genes that are regulated by antisense transcripts produced from a different locus (eg in trans) [49–51].
CONCLUSIONS
Bridging the intellectual divide between chromatin biochemistry, functional genetics, and epigenomics is a key goal for histone modification biology. Many chromatin regulatory proteins bind (slightly) more tightly to doubly-modified histones or nucleosomes than to nucleosomes with either single modification, suggesting that combinatorial complexity is meaningful for chromatin biology. Yet histone modification patterns are simple in vivo, occurring in few combinations. Moreover, chromatin regulatory proteins often associate with genomic loci in vivo that lack the modification patterns to which they bind in vitro. Finally, chromatin regulators typically only have measurable effects at a subset of the locations to which they are recruited. These considerations will need to be addressed for a full understanding of the regulation of chromatin function by histone modification-binding factors.
I believe that several avenues of investigation will be fruitful for reconciling these observations. Allosteric activation of proteins by histone marks will help explain the disconnect between in vivo protein localization and the occurrence of relevant histone modifications, and careful functional analysis of gene-specific effects of widespread chromatin regulators has begun to explain some cases of “context dependence” for histone modifications. To fully understand the role of combinatorial binding, such in vivo studies must be extended to carefully chosen mutants in chromatin regulators with altered modification binding behavior in vitro – are any genes transcriptionally affected in BPTF mutants that bind solely to H3K4me3, rather than H3K4me3/H4K16ac?
Importantly, what we learn from the histone modification field will provide unique insights for the many other biological systems that involve large numbers of covalent modifications, as the physical separation of covalently modified histones along the genome provides insights that are difficult to glean from multiply-modified proteins that freely diffuse in solution. To (over)extend the language metaphor, the physical linkage between the language of histone modifications and the “book of life” may give us the Rosetta stone for understanding covalent modifications throughout biology.
ACKNOWLEDGEMENTS
I thank S. Henikoff, C. Peterson, P. Kaufman, T. Fazzio, and especially B. Cairns for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1. Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. In this recent study, authors used mass spectrometry to identify covalent histone modifications, nearly doubling the number of described modifications.
- 2.Turner BM. Decoding the nucleosome. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- 3.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 4.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 5.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. An example of binding by a single domain to a doubly-modified histone tail.
- 8.Wang Z, Patel DJ. Combinatorial readout of dual histone modifications by paired chromatin-associated modules. J Biol Chem. 2011;286:18363–18368. doi: 10.1074/jbc.R111.219139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. Embo J. 2004;23:4286–4296. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol. 2011;21:53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. This manuscript shows BPTF binding to a combinatorial pattern of H3K4me3/H4K16ac on mononucleosomes. Importantly, BPTF binds to H3K4me3 with ~2-fold lower affinity than to the doubly-modified nucleosome, raising the question of the importance of combinatorial binding.
- 12.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 13. Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. Another solid example of combinatorial binding by a histone-binding protein, TRIM24.
- 14. Eustermann S, Yang JC, Law MJ, Amos R, Chapman LM, Jelinska C, Garrick D, Clynes D, Gibbons RJ, Rhodes D, et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol. 2011;18:777–782. doi: 10.1038/nsmb.2070. These studies provide an example of one histone modification (H3K4 methylation) preventing binding of a protein partner to its preferred modification elsewhere on the histone tail.
- 15. Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD, Picketts DJ, Patel DJ, Li H, et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol. 2011;18:769–776. doi: 10.1038/nsmb.2062. These studies provide an example of one histone modification (H3K4 methylation) preventing binding of a protein partner to its preferred modification elsewhere on the histone tail.
- 16.Martin AM, Pouchnik DJ, Walker JL, Wyrick JJ. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics. 2004;167:1123–1132. doi: 10.1534/genetics.104.026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci U S A. 2005 doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindstrom KC, Vary JC, Jr, Parthun MR, Delrow J, Tsukiyama T. Isw1 functions in parallel with the NuA4 and Swr1 complexes in stress-induced gene repression. Mol Cell Biol. 2006;26:6117–6129. doi: 10.1128/MCB.00642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. This paper provides a thorough dataset of gene expression changes in chromatin-related mutants in yeast. Notably, many enzymes responsible for widespread marks, such as Set1, have minimal effects on gene expression when deleted.
- 20.Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 23.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-Nucleosome Mapping of Histone Modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol. 2010;17:1495–1499. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. Genome-wide datasets describing the localization patterns of a large number of histone modifications in many cell types. As with all such studies, these papers reveal limited combinatorial complexity to histone modification patterns in vivo.
- 28. Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. Genome-wide datasets describing the localization patterns of a large number of histone modifications in many cell types. As with all such studies, these papers reveal limited combinatorial complexity to histone modification patterns in vivo.
- 29.Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. Embo J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2011;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 34.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 35.Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 2010;6:e1001173. doi: 10.1371/journal.pgen.1001173. This paper shows that binding of H3K36me3 by the RPD3S complex is not required for RPD3S localization – instead, this binding appears to be required for deacetylase function.
- 37.VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hauk G, McKnight JN, Nodelman IM, Bowman GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39:711–723. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morettini S, Tribus M, Zeilner A, Sebald J, Campo-Fernandez B, Scheran G, Worle H, Podhraski V, Fyodorov DV, Lusser A. The chromodomains of CHD1 are critical for enzymatic activity but less important for chromatin localization. Nucleic Acids Res. 2011;39:3103–3115. doi: 10.1093/nar/gkq1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corona DF, Clapier CR, Becker PB, Tamkun JW. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 2002;3:242–247. doi: 10.1093/embo-reports/kvf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 45.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 46.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 47.Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. Chromatin-and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, Grewal SI. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature. 2009;461:419–422. doi: 10.1038/nature08321. An elegant paper explaining the gene-specific importance of widespread histone variant H2A.Z – loss of this variant in fission yeast largely affects genes oriented convergently, and appears to be required for degradation of readthrough antisense transcripts.
- 49.van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 50.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F. Transacting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009;23:1534–1545. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]