Abstract
Intron removal is at the heart of mRNA synthesis. It is mediated by one of the cell's largest complexes, the spliceosome. Yet, the fundamental chemistry involved is simple. In this review we will address how the spliceosome acts in diverse ways to optimize gene expression in order to meet the cell's needs. This is done largely by regulating the splicing of key transcripts encoding products that control gene expression pathways. This widespread role is evident even in the yeast Saccharomyces cerevisiae, where introns appear to have been lost; yet how this control is being achieved is known only in a few cases. Here we explore the relevant examples and posit hypotheses whereby regulated splicing fine-tunes gene expression pathways to maintain cell homeostasis.
Keywords: spliceosome, Yra1, Sus1, L30, Mer1, export, histone modification
Introduction
In general terms, prokaryotic genes are compact, can be grouped based on their functions, and are translated while still being transcribed. Gene expression in prokaryotes has evolved towards speed and efficiency. In contrast, most eukaryotic genes are isolated and, moreover, have their sequences interrupted, such that the coding pieces must be spliced together to produce a meaningful transcript or mRNA. Transcripts undergo further processing before being transported to the cytoplasm. Accordingly, mRNAs exist in the cell as RNPs (ribonucleoprotein) particles that act as both executors and bearers of their fates. In light of this complex mRNA processing, it is clear that eukaryotic gene expression is under a set of selection rules whereby speed and efficiency are only part of the story.
This review will focus on the diverse roles that pre-mRNA splicing plays in the control of the processes underlying gene expression. Elegant genetic, biochemical, and cell biological studies in the yeast Saccharomyces cerevisiae have made it possible to glean mechanistic insights into these processes and their interplay. As such, these will be the basis of this review. Nonetheless, many of the guiding principles learned from S. cerevisiae are likely to inform our general understanding of how regulation of splicing and the integrated gene expression networks that involve splicing contribute to the cellular homeostasis.
pre-mRNA splicing
Pre-mRNA splicing is the process by which the coding segments of a gene (exons) are spliced together, and the interrupting fragments (introns) are removed and discarded. Splicing is the task of the spliceosome, one of the most complex macromolecular machines in the cell [1]. This evolutionarily conserved assembly starts anew in each round of splicing (reviewed in [2]). First, once the intronic 5' end (5' splice site or 5'ss) is transcribed, it is recognized by the U1 snRNP (small nuclear RNP). This interaction is thought to be driven by the complementarity between the U1 snRNA and the 5'ss, although its protein components play important roles as well [3]. Subsequently, the 3'end of the intron, including the branch-site (BS) and the 3' splice site (3'ss) are identified by the factors BBP and Mud2 in yeast or their homologs SF1 and U2AF in other systems (including human). These factors play a role in the initial recognition of the BS by the U2 snRNP. Early interactions between the U1 and U2 snRNPs on the same intron are thought to be essential to define the intron to be removed and, therefore, the exons to be joined. There is an alternative way to define the substrate for the spliceosome, based on “exon definition,” which posits that interactions between an upstream U2 snRNP and a downstream U1 snRNP are critical for the further assembly of the nascent spliceosome, marking the pre-mRNA in between as an exon. This appears to be particularly important for genes that contain multiple, large introns, as is usually the case in metazoans [4] As we will further examine, recent studies in Saccharomyces cerevisiae support the notion that cross-exon interactions are also important in yeast.
In a subsequent step, which remains somewhat obscure, molecular rearrangements lead to the interaction between U1 and U2 of the same intron (as in the previous “intron definition”), and spliceosome assembly proceeds with recruitment of the U4/U6.U5 tri snRNP. Subsequent remodeling steps, catalyzed by RNA-driven ATPases, displace U1 and U4 and facilitate the formation of the catalytically active spliceosome, which will form a lariat structure via a 2’-5’ phosphodiester linkage between the adenosine at the BS and the 5’ end of the intron, followed by exon joining and intron removal. Thus, a sophisticated molecular ballet culminates in two chemically simple transesterifications. That this rather straightforward chemistry involves such a complex cellular machinery is a testament to the relevance of spliceosome dynamics. By modulating spliceosomal function to produce a variety of splicing outcomes from a single pre-mRNA, evolution has developed a formidable tool to enlarge the coding potential of genomes. The initial recognition of the pre-mRNA by U1 and U2 is an important target for regulation. Regulatory proteins can interfere with either the recognition of intronic sequences (leading to intron splicing or retention); or the interactions that occur during exon definition (promoting exon inclusion or skipping). Switching the 3'ss after spliceosome assembly has been shown in metazoans [5], indicating that regulation of the spliceosome goes beyond controlling initial splice site recognition. In some instances the pre-mRNA structure plays a role in intron recognition, and in many yeast introns it is critical for proper 3'ss selection [6, 7]. Pre-mRNA intronic and exonic sequences, known as splicing enhancers or silencers, participate in the recruitment of splicing factors, such as SR proteins and hnRNPs. As is the case with many of the pre-mRNA sequences involved in splicing, the information content of silencers and enhancers is rather low, and the splicing outcome will also depend on the relative concentration of the splicing regulatory proteins. Thus, alternative splicing is a function of the pre-mRNA sequence (including exonic structure and RNA folding) and the concentration of splicing factors, which together make up a putative “splicing code”. Although regulated alternative splicing has been best characterized in metazoans, studies of the yeast S. cerevisiae, in which regulated splicing primarily is seen in the form of intron retention, offers an opportunity to glean basic insights into the reaction.
Chromatin, transcription, and splicing converge to modulate gene expression
About 20 years ago, evidence emerged indicating that intron removal could occur on nascent transcripts. Elegant imaging studies by Beyer and co-workers revealed nascent pre-mRNA shortening caused by splicing [8, 9]. Subsequently, a number of important studies using Saccharomyces cerevisiae also showed that spliceosome assembly (beginning with intron recognition) occurred on the nascent transcript, although it was less clear whether the actual chemistry of splicing occurred cotranscriptionally [10-14]. Recently, RNA polymerase II has been reported to pause in a manner that facilitates both co-transcriptional assembly of the spliceosome as well as splicing [15, 16], supporting the notion that most splicing occurs co-transcriptionally [16].
In light of this close spatial and temporal relationship between transcription and splicing, it is not surprising that polymerase activity can have important effects on splicing. For example, in mammals as in yeast, the speed of the polymerase influences recognition of splice sites, such that an exon that is normally excluded (exon skipping) is recognized and included when the polymerase rate is slowed [17, 18]. Additionally, in mammals RNA polymerase pausing has been shown to affect exon recognition [19]. A more extensive survey of co-transcriptional splicing mechanisms can be found in a number of excellent recent reviews (see for example [20-22]). Nonetheless, these studies demonstrate a growing appreciation that in order to understand splicing and its regulation, it is important to view splicing within its co-transcriptional context.
More recently, it has become clear that one of the important mechanistic consequences of transcription-coupled splicing is that the state of the chromatin influences splicing outcomes. Analysis of genome-wide surveys from Drosophila and mammals have revealed that nucleosomes and, according to several of these studies, specific histone modifications are enriched in exon sequences, suggesting that there may be specific histone “marks” that are associated with splicing [23-29]. Additionally, proteins that bind to histones have been shown to facilitate the recruitment of snRNPs to the nascent transcript and influence efficiency of splicing and alternative splicing (Reviewed in [30, 31]). The ability of chromatin to influence pre-mRNA splicing appears to be a conserved feature of co-transcriptional splicing. For example, Gcn5, the histone acetyltransferase that is part of the SAGA complex, has strong genetic interactions with components of the U2 snRNP, and its catalytic activity is required for co-transcriptional spliceosome assembly [32, 33].
In light of the integral relationship between chromatin and splicing, one of the outstanding questions is whether the reverse relationship exists. Namely, can the splicing reaction or specific splicing factors influence transcriptional regulation through effects on chromatin--either by direct coupling of histone modification with intron recognition or via regulated splicing of components of the histone modifying machinery? Several recent mammalian studies report that histone H3K36 methylation is mediated through splicing signals, although the mechanism for this is unclear [34, 35], and it is not clear whether other histone marks may also be influenced by splicing. In yeast, splicing has been shown to affect histone H2B ubiquitination by regulating the expression of a component of the histone H2B deubiquitinase machinery SUS1. However, thus far, no evidence of a direct relationship between histone H2B ubiquitination and splicing has been reported.
Yeast as a model to dissect regulated splicing
When facing complex scientific problems, a reductionist approach can often be a valuable tactic. The budding yeast Saccharomyces cerevisiae has provided such a tool since the discovery of the ACT1 intron [36]. With its streamlined genome and high functional conservation of the basic machinery [37] it has been particularly useful to uncover the intricacies of spliceosome assembly and catalysis. However, the scarcity of introns (~95% of yeast genes are intronless), and the lack of clear homologues of SR-proteins (ubiquitous splicing modulators in metazoan cells), has encouraged the view that the yeast cell does not use this important instrument to regulate gene output. While it is true that there are quantitatively fewer alternate mRNAs that arise from splicing changes in yeast compared to metazoans, regulation of splicing in yeast is essential to cellular fitness and meiotic differentiation (Figure 1). Given that the fundamental aspects of the spliceosome are conserved in evolution, the study of these regulatory strategies is relevant to understanding splicing across eukaryotes, and likely it will reveal possibilities that may be harnessed by other organisms. In fact, its reduced intronic set simplifies the study of splicing at the genomic level. This, combined with the manageability of yeast as a model system, has helped to advance our understanding of some molecular mechanisms behind regulated splicing.
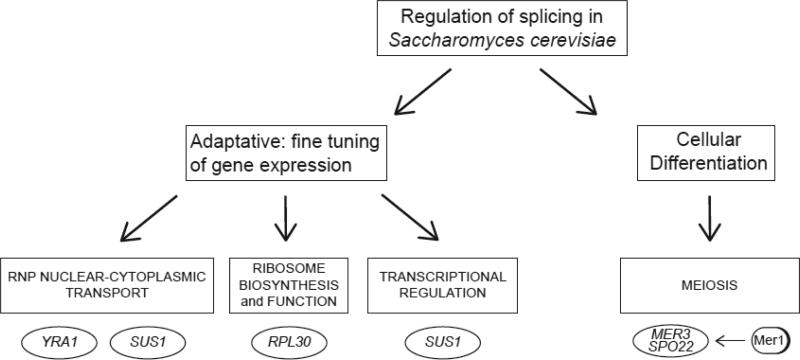
Figure 1.
Pre-mRNA splicing can fine-tune several different gene expression reactions via splicing sensitive regulators, including RNP biogenesis and transport, ribosome biogenesis and function, or transcription itself. The factors shown in the ovals undergo regulated splicing to affect the processes shown in the boxes. It is likely that others are still unknown. Nevertheless, those indicated provide a paradigm for regulatory strategies centered on the control of pre-mRNA splicing.
As a single-celled organism, yeast has been subjected to strong selection for rapid and adaptive growth. A growing yeast cell is under the Sisyphean task of constantly adjusting to an ever-changing environment and one might imagine that regulation of splicing is an important part of this business. Indeed, the cell uses splicing to fine-tune RNA biogenesis, as genes encoding proteins involved in a number of critical steps in RNA biogenesis can undergo regulated splicing—including translation, RNA export, RNA splicing, and transcription. This provides a powerful tool for regulating gene expression as changing the capacity to translate mRNAs from the assembled and transported mRNPs can fundamentally alter gene expression. We will now focus on examples of how splicing affects these critical gene expression reactions.
Regulated splicing to control ribosome biosynthesis and function
Ribosome biosynthesis constitutes the yeast cell's largest energy investment [38]. Hence, it is not surprising that expression of ribosome components is subject to tight regulation at multiple levels, including splicing. Ribosomal proteins are obvious candidates for splicing regulation, as over one third of yeast introns are located in their genes [39]. Furthermore, nearly a third of all mRNAs are produced from intron-containing genes—most of which are ribosomal protein genes (RPGs). This all leads to the obvious prediction that there are likely to be instances of regulation at the level of RPG splicing that may have so far escaped description.
Recent studies that systematically deleted the introns from intron-containing RP genes demonstrated that at least some introns were required for optimal fitness or adaptation to stress conditions, indicating that proper expression is tied to intron-dependent gene expression changes [40, 41]. In light of genome-wide studies indicating that inefficient splicing may be an important and widely used mechanism for downregulating gene expression in yeast [42], it is likely that at least one of the functions of the introns in RP genes is to confer regulatory flexibility and, perhaps, allow the rapid downregulation of ribosome biogenesis.
At a mechanistic level, there are undoubtedly a number of ways by which RPG intron removal is regulated. This includes autoregulation via the binding of the RP to its own pre-mRNA to affect its own splicing [43, 44]}. Remarkably most introns are found within duplicated ribosomal protein genes, and not only does deletion of the intron in a duplicated RP gene affect the host gene (intragenic regulation), but also the expression of the paralog. Interestingly, in about a third of the cases, this intron-dependent extragenic regulation is dependent upon the intron in the affected paralog. These studies support models in which autoregulation and cross-paralog regulation at the level of intron removal play critical roles in controlled expression of ribosomal subunits [41]. In agreement with this, a new mechanism of nuclear RNA degradation, which is driven by introns that are bound by regulatory factors and the poly(A) binding protein, has been described in fission yeast [45]. Some S. cerevisiae RPG's (notably RPS14 [46]) could be regulated in a similar manner, although it is apparent that cytoplasmic decay also plays a role in intron-dependent RNA degradation [47].
Of course an outstanding question raised by these studies is what role there is for differential expression of individual ribosomal proteins, including RPG paralogs? While the data above are suggestive of a link between splicing and ribosome function, the direct mechanism remains to be determined. Intriguingly, studies by Komili et al. demonstrated a differential requirement for paralogs for expression of localized RNAs. Furthermore, these studies also demonstrated that ribosomal protein paralogs have differential requirements for their assembly and localization, suggesting the existence of “specialized ribosomes” [48] [49]. Future experiments will undoubtedly shed light on this provocative possibility.
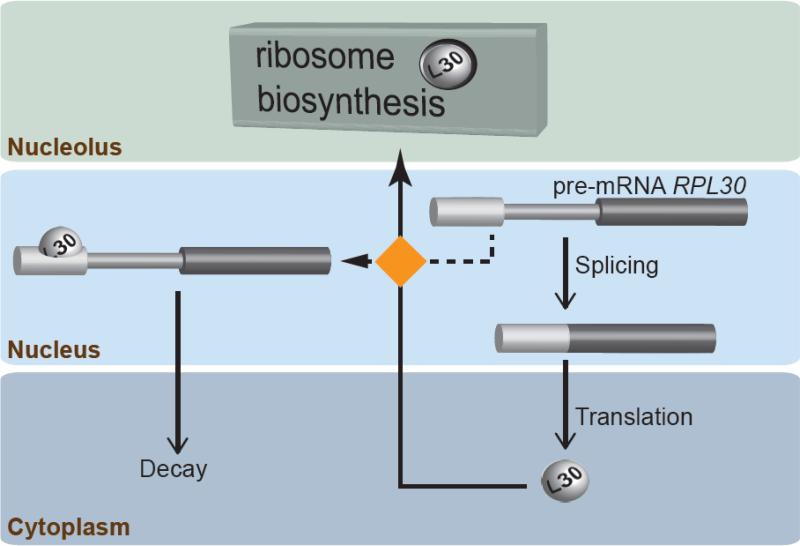
An example of how a specific RPG is regulated (and autoregulated) at the level of splicing to control ribosomal function is RPL30 (Figure 2). Expression of this gene includes an autoregulatory feedback loop, in which excess L30 binds to its own premRNA to stall spliceosome assembly. Thus, L30 can act as either a ribosomal protein or as a splicing factor. Therefore, a newly made L30 molecule is offered two choices: to either participate in ribosome function (ribosomal protein role); or to stop its own synthesis by binding to the pre-mRNA (splicing factor role). This strategy is based on evolving a binding site for the protein in the pre-mRNA plus a delicate balance of the relative affinities of L30 for the RNA and/or proteins it interacts with. Additionally, this binding site must be in a precise location in the pre-mRNA in order for the protein bound to it to block spliceosome assembly [50]. L30 (a mostly cytoplasmic protein, with a fraction in the nucleolus) binds co-transcriptionally to the RPL30 pre-mRNA to prevent full spliceosome assembly [50] and ultimately promote its export to the cytoplasm, where it gets degraded [51].
Figure 2.
Splicing regulation of the RPL30 transcript. The RPL30 gene encodes the essential protein L30. Pre-mRNA splicing contributes to maintaining the protein levels by an autoregulatory feedback loop. The newly made L30 (shown as a ball) must be transported to the nucleolus to join ribosome biosynthesis (top). However, if that is not possible (conditions of excess L30), the protein binds its own pre-mRNA. This prevents RPL30 splicing and triggers the transport and decay of the pre-mRNA in the cytoplasm. The choice for L30 between either ribosome biosynthesis or splicing control (orange square) has been depicted in the nucleus, but its mechanism and location is unknown (for simplicity reasons the binding site of L30 is shown entirely in exon 1, although it includes the 5' ss).
This scheme is essential for the cellular fitness [47] and it must be sensitive and fast to be efficient. In a growing yeast cell there are 105 L30 molecules. Thus, it is likely that regulation of splicing must be responsive to small variations, since the starting L30 levels are already high. How the cell achieves this remains mostly unknown, yet it must involve rapid relocalization of L30, as L30 plays roles in distinct nuclear compartments. Presumably after failing to be recruited to the nascent nucleolar ribosome, L30 is able to rapidly find the site of RPL30 transcription, because every minute, more than one thousand L30 molecules must go through the same ritual (a growing yeast cell makes 105 ribosomes in 1.5 hours). Consistent with the importance of the relative affinity of L30 for its two alternate destinations, unrestricted expression of an extra copy of an L30 version that fails to assemble into the ribosome is lethal. This is a consequence of repression of splicing of the endogenous RPL30 by this additional L30, thus depriving the cell of this essential protein [52].
From the point of view of splicing mechanisms, the fact that a ribosomal protein can block spliceosome assembly is intriguing. A model for this regulation is emerging based on the multiple conformational changes that occur during spliceosome assembly. According to this model, L30 binding to its pre-mRNA plays its inhibitory role by steric hindrance, blocking an early rearrangement. This supposes a novel way of splicing regulation, one that seems well-suited to RNA-binding proteins, and other examples of this strategy will surely follow.
Regulated splicing to control RNP Biosynthesis and mRNA export: SUS1 and YRA1
Just as is the case with ribosomal RNAs, most mRNA molecules spend their lives in a complex of proteins and RNA (RNP). During transcription, factors involved in capping, splicing, termination, and polyadenylation interact with the nascent RNA. This protein-bound RNA is delivered to the correct destination, which requires its interaction with specific factors. Once in the cytoplasm (for the RNAs that go there), the RNPs are further remodeled to prepare the RNA for translation, storage, or further transport. Finally, many of these proteins bound to the RNA will need to be removed, and others recruited, before the RNA is degraded. These molecular rearrangements are each a potential target for regulation, and splicing appears to play a “behind the scenes” role in a number of these.
SUS1 encodes a small, evolutionarily conserved protein that participates in both transcription and RNA export. As part of the SAGA complex, it is involved in histone H2B deubiquitination [53, 54]. During transcription, Sus1 interacts with the export factors Yra1 and Mex67. Sus1 is also at the nuclear pore, participating in the TREX2 complex (TRanscription EXport complex 2). In this capacity it plays a central role in RNA export.
Compared to other yeast genes, SUS1 is atypical because it bears two introns, a feature shared by only a handful of yeast genes, and this gene architecture has been shown to be relevant to its function. Moreover, intron 1 of SUS1 has a non-consensus 5' end (A at position 6 instead of U) and BS (UACUGAC, as opposed to the canonical UACUAAC), and both of these features influence SUS1 splicing. Analysis of SUS1 splicing in WT cells indicates that some retention of the first intron occurs and, in fact, experiments using NMD- strains (deleted of the nonsense-mediated decay machinery) show that intron retention increases under different environmental conditions such as change in carbon source and upon temperature shift [55, 56] Furthermore, recent results support the notion that the ability to produce different SUS1 isoforms is essential for proper RNA export. It is not yet clear whether the change in SUS1 splicing simply achieves a necessary change in the level of mature SUS1 mRNA or if SUS1 splicing changes mediate some other effect on RNA export. However, it is notable that a small peptide can be generated from the intron 1 containing transcript [55]. Further experiments are necessary to determine precisely how this peptide may affect the export machinery.
A second RNA export factor, Yra1, also has features that target it for regulated splicing. Although YRA1 has a more typical structure, with only one intron, this intron is comparatively large and, more importantly, has a non-consensus BS sequence (GACUAAC). As with SUS1, YRA1 splicing is suboptimal, and this decreased splicing efficiency contributes to its regulated intron retention and degradation to maintain the appropriate levels of Yra1 protein. Yra1 and the RNA helicase Sub2 are recruited cotranscriptionally to the nascent RNA, participating in the assembly of the TREX complex involved in RNA transport. When Yra1 levels are high, the Yra1 protein facilitates the export of the unspliced YRA1 message such that it can be targeted to the Xrn1 (but not NMD) -dependent decay pathway in the cytoplasm [57-59], thus forming a tight regulatory loop.
While it is clear that splicing of particular transcripts is necessary for RNA export, an intriguing model is that splicing and transport are, in fact spatially coordinated to allow feedback regulation of the two processes. For example, in light of evidence that export factor recruitment and spliceosome assembly occur co-transcriptionally, it is likely that there is competition between RNA transport and splicing, allowing that poor splicing substrates will tend to be exported as pre-mRNAs; conversely, their splicing will be favored under conditions of poor export (for instance, in export mutants). With this in mind, a simple model is that regulated splicing of export factors provides a mechanism by which the balance between export and splicing can be achieved by a feedback loop between the two pathways. If splicing cannot keep up with the export pipeline, the pipeline must be slowed down, achievable by inefficient production of YRA1 or SUS1. The reverse may also be true. If the export machinery must accommodate high levels of newly made transcripts, then export should be promoted. This could be achieved by more efficient production of export factors via splicing of pre-mRNAs encoding export factors such as Sus1 and Yra1 and a concomitant increase in the capacity for export of newly-synthesized mRNAs. In summary, this model posits that the export machinery is tightly coupled to splicing via the use of splicing sensitized pre-mRNAs. Consistent with the prediction of an important and delicate balance of these factors, overexpression of either Yra1 or Sus1 is toxic to the cell [55, 58].
Overall this model predicts that regulated splicing of specific transcripts could act as a “sensor” of robust RNA processing and a “control switch” to downregulate or upregulate the RNA export machinery. It is intriguing to envision that YRA1 and SUS1 (perhaps through its role in transcription) could play slightly different roles in the delicate fine-tuning of RNA metabolism.
Regulated splicing to fine tune transcription: Regulation of histone modification and the meiotic transcription program
A picture is emerging of how regulated splicing in S. cerevisiae plays an important role in regulating RNA export and fine-tuning ribosome biogenesis and function. Next we will explore how regulated splicing of specific transcripts influences transcriptional programs in response to environmental change. In addition to its role in RNA export via its interactions with the TREX-2 complex, Sus1 is a component of the transcriptional coactivating complex, SAGA. This complex is comprised of two catalytic modules: one that mediates histone acetylation and gene activation, and another module comprised of the histone H2B deubiquitinase Ubp8. Sus1 (along with Sgf11 and Sgf73) is required for the Ubp8-dependent deubiquitination [60, 61].
Since Sus1 contains two introns, the first of which has features that reinforce its retention, it is worth looking more closely at the details underlying removal of the Sus1 introns. Expression of SUS1 pre-mRNA in which the first intron is absent reveals that removal of the downstream intron is facilitated by recognition of the first [56]. While the mechanisms underlying this “cross-exon” effect are not clear, these results suggest that, as is the case in metazoans, removal of the introns from a multi-intron pre-mRNA can be coordinated [62], and it is possible that the cross-exon effects that are observed involve similar interactions as metazoan “exon-definition.” While the number of yeast genes with multiple introns in S. cerevisiae is small (10), non-canonical sequences in the first intron are overrepresented (half of the two-intron genes), suggesting that similar mechanisms observed for SUS1 splicing might influence processing of these transcripts.
As described above, stress conditions (e.g. temperature) lead to preferential retention of the first intron and changes in Sus1 protein levels. Furthermore, at higher temperature cells show increased sensitivity to deletion of SUS1 [55, 56]. Only those cells that express a construct that is able to undergo splicing (and not the cDNA with the introns removed) can restore WT levels of H2B ubiquitination and WT growth in response to temperature change. These data suggest that production of SUS1 isoforms—mature and intron 1-retained RNA—allow proper levels of H2B ubiquitination in response to stress, perhaps by maintaining the delicate balance of Sus1 protein necessary for H2B deubiquitination within genes involved in stress response. Since the retained intron product appears to have two possible fates—degradation through NMD and production of a truncated peptide [55], it remains to be determined precisely how H2B levels are maintained by the production of the retained intron isoform. Nevertheless, these data suggest that cellular response to stress involves control of H2B ubiquitination via SUS1 splicing. Consistent with these observations, we have observed that a number of genes involved in temperature stress response in S. cerevisiae are upregulated under the very conditions under which the first intron of SUS1 is retained (TJohnson, unpublished results). The role of SUS1 splicing regulation in RNA export and histone H2B deubiquitination are summarized in Figure 3.
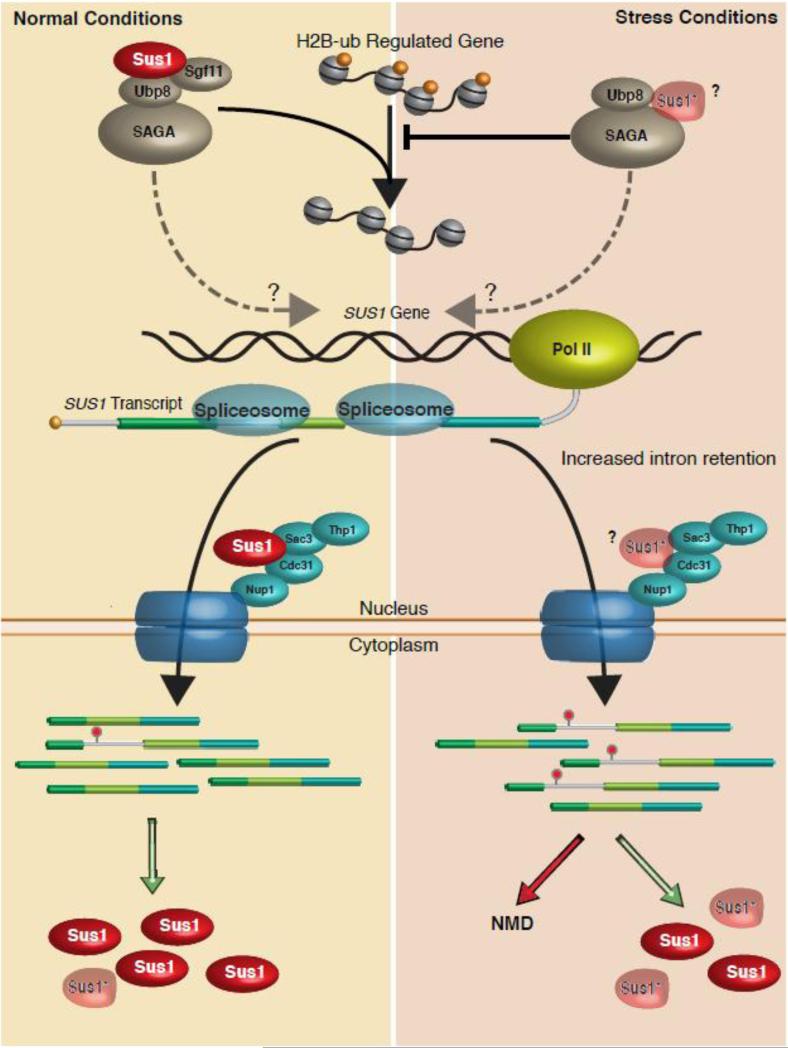
Figure 3.
Regulated splicing of SUS1 fine-tunes RNA export and histone H2B ubiquitination. Under unstressed conditions (Left side of figure) SUS1 pre-mRNA undergoes splicing and is exported to allow production of Sus1 protein that can either participate in histone H2B deubiquitination as part of the SAGA complex (top) or can associate with the TREX-2 proteins at the nuclear pore (Shown in dark blue). Intron 1 is retained in a small fraction of the RNAs and is either targeted to NMD or may make a truncated product. Under stress conditions, intron 1retention is enhanced. However, it is unclear how much of this product is targeted to NMD vs. translation of a truncated product. It is possible that the truncated protein can associate with either the TREX-2 complex or with SAGA (indicated by “?”). Sgf11 is destabilized by the lack of Sus1 [81], so it has been left off out of the SAGA complex that contains less SUS1. It is also possible that Sus1, via its H2B ubiquitination activity or by interacting with the splicing (not shown), may influence its own splicing (Dotted arrow).
Both YRA1 and RPL30 provide mechanisms whereby the protein product regulates splicing of its own transcript. While it is unclear whether Sus1 contributes to its own regulation, it has previously been shown that the acetylation module of the SAGA complex (catalyzed by Gcn5) is able to influence pre-messenger RNA splicing via its acetylation activity. It is appealing to speculate that Sus1-mediated H2B deubiquitination or one of the Sus1 products could mediate changes in the chromatin of the SUS1 gene that affect its own splicing, adding an additional layer of regulation of SUS1 mRNA levels.
In the context of RNA export, SUS1 appears to be a “sensor” of robust RNA processing (indicated by levels of the retained intron), and its gene product can control the fate of the mRNPs through its export activity. Hence the coupling of splicing with export through one protein ensures the fine tuned coordination of multiple processes. A parallel “fine tuning” may occur via Sus1's role in histone modification. In this case, the SUS1 transcript provides a readout for splicing efficiency, with production of the retained intron product indicating a decrease in splicing efficiency. The Sus1 gene product(s) could thereby modulate transcription, perhaps of a critical subset of genes, via effects on H2B-ub. Such a feedback mechanism would ensure a balance between the activity of the processing machinery and the RNA load generated through transcription. These models of feedback between the Sus1 splicing and transcription to maintain homeostasis remain to be tested.
In addition to SUS1, it appears that the entire meiotic regulatory network is under tight control via regulated splicing. Under conditions of nutrient starvation diploid yeast cells enter meiosis and sporulate. As with ribosome biosynthesis, this process is tightly regulated at the transcriptional level [63] as well as at the level of splicing [64, 65]. Consistent with this, a number of intron-containing meiotic transcripts can be detected in vegetative cells, but these remain unspliced [42]. Correct processing of four key meiotic messages requires the meiosis-specific splicing factor Mer1. This factor promotes intron recognition in poor substrates through its ability to interact with a conserved intronic enhancer sequence [66], and the meiotic program cannot proceed without it.
Mer1 dependent splicing bridges two transcriptional regulatory networks in the meiotic gene expression program. First, Mer1, like 3 of the 4 other meiotic genes that it regulates is under the control of Ume6, the activator of early meiotic genes, thus ensuring that the regulation of these transcripts is temporally constrained until there is sufficient accumulation of Mer1p after Ume6-mediated activation. Furthermore, splicing of two Mer1 specific genes MER3 and SPO22 is necessary for expression of NDT80, a meiosis specific transcription factor [64]. Hence, regulated splicing ensures control of a transcriptional program essential for cells to induce meiotic gene expression.
Environment-dependent changes in splicing fine-tune the cellular mRNA landscape
While the focus of this review has been specific examples of regulated splicing, it is likely that these examples, while important for their roles in coordinating multiple gene expression reactions may only be the tip of the iceberg. Recent data indicate that the spliceosome can affect the composition of the transcriptome (understood here as the cellular mRNA landscape) in response to environmental stimuli [67]. For example, changes in amino acid starvation or ethanol stress provoke alterations in the splicing of many genes. These changes are too rapid to be easily explained by the synthesis of new splicing factor(s) to modify the spliceosome activity, and no obvious cis-elements have been identified in the pre-mRNAs that would explain this behavior [67]. Nonetheless, the spliceosome can identify which, and how, pre-mRNAs are to be changed when the yeast cells encounter certain situations. It is possible that the rapid response to environmental change may be mediated by rapid and regulated changes in the core splicing machinery through, for example, post-translational modifications of components of the splicing machinery. Along these lines, large-scale proteomics studies reveal widespread modification of splicing factors [68]. Furthermore, splicing-sensitive microarrays confirm that intron-containing genes are differentially sensitive to the activities of specific splicing factors [69, 70], raising the possibility that modification of individual components of the splicing machinery could “fine-tune” it to target specific introns (or groups of introns).
Intriguingly, detailed studies of meiotic regulation of splicing in fission yeast, support previous mammalian studies that demonstrate that the promoter can dictate splicing regulation [71]. These studies suggest that some RNA-binding proteins, recruited at the promoter level, may tag the transcript in a way that can affect the spliceosome or that the elongation properties of the polymerase, which can be set up during early stages of transcription, affect splicing. Nonetheless, the ability of the cell to to coordinate changes in splicing of the transcripts from many genes is remarkable and illustrates the likely role for regulated splicing as a cellular strategy for adaptation to environment. Transcriptome analyses in upf1Δ and xrn1Δ mutants (where many RNAs that would otherwise be turned over are stabilized) indicate that there is a small population of “aberrant” transcripts, which includes unspliced, partially spliced, and misspliced RNAs [72]. Whether this is part of a regulatory mechanism or simply evidence that the cell efficiently “cleans up” sloppy splicing remains to be determined. However, the examples above of SUS1, RPL30, and YRA1 all illustrate that regulated degradation of differentially spliced transcripts provide a powerful mechanism whereby the cell can respond to changes in environmental demands.
Conclusions
As a critical player during early mRNA synthesis, pre-mRNA splicing can have a profound effect on gene expression. Akin to a limited molecular ghostwriter, the spliceosome modifies a first draft --the nascent transcripts-- to produce a message tailored to the cell's needs and that can affect fundamental cellular pathways. Therefore, the cell must communicate to the spliceosome a wide variety of information, and the spliceosome must be able to respond quickly enough to elicit the appropriate response. Surely this underlies the remarkable complexity of the spliceosome; after all, group II introns, the likely ancestors of spliceosomal introns, can perform the same basic chemistry mostly by themselves [73]. This raises a number of outstanding questions. How is this communication carried out? Who are the key players? How do changes in chromatin mediate the communication between environment and splicing? And can splicing in fact, contribute to establish epigenetic changes that mark the “experience” of the cell? While we still know frustratingly little, recent data on co-transcriptional splicing indicate that the coupling between transcription and splicing can be modulated by transcript features, from secondary structures to the recruitment of trans acting splicing factors [74].
Regulation of splicing acts in two ways: working constantly to keep gene expression fine-tuned (e.g. maintaining proper levels of RNA export); and occasionally, when a new cellular program needs to be launched and maintained (e.g. meiosis). Whether there is crosstalk between these modes of regulation remains to be elucidated. For example, it is not known whether regulated splicing may completely reprogram particular cellular processes when such changes are extremely relevant to the cell (e.g. protein synthesis). Notably, as we have seen more than a third of yeast introns are located in genes related to the ribosome, making ribosomal biogenesis and function highly subject to splicing control. This has been connected to the generation of “specialized” ribosomes [41]. It also may have particular relevance to understanding alterations in ribosome biogenesis, detrimental to health, as variations in the concentration of ribosomal proteins have been linked to disease [75] and development defects [76]. Along these lines, and given the relevance that splicing has to health, it is of great interest to develop strategies to alter the splicing outcome. Some initial successes in this area have been very encouraging [77, 78].
The U12 spliceosome constitutes the obvious example supporting the notion, analogous to that in ribosomes, of “specialized” spliceosomes with different versions of the core components. However, it seems likely that the cell should use less radical strategies to generate different spliceosome variants that could be the basis for some alternative splicing. Some spliceosomal components are encoded by several genes (see [79], for example) and they may assemble into spliceosomes with different properties. Similarly, other components are not essential, or their properties could be post-translationally modified. There is much to be learned as to how homogeneous spliceosomes are, and how this relates to the many cellular factors that control splicing (SR proteins and hnRNPs). Even in yeast, with a simplified splicing network, how the core spliceosome elicits specific splicing outcomes (like the nutrient response in yeast [67]) remains to be explained. Furthermore, many metazoan transcripts encoding regulatory splicing factors undergo regulated splicing themselves. Thus, the splicing code involving regulatory factors and pre-mRNA features [80] may need to be expanded to include the inherent properties of the spliceosome. This may seem a daunting task, but analyses of the increasing amount of transcriptome data are improving our ability to predict which will be the final mRNA synthesized from a given pre-mRNA and set of conditions.
In yeast, with just a relative handful of introns, splicing has a central role in the control of gene expression. Moreover, the possibility that its streamlined spliceosome can be subjected to the same regulatory strategies, including those involving co-transcriptional splicing through a chromatin template and cross exon interactions, is particularly exciting. Thus, future studies that take advantage of the yeast system are likely to yield more fundamental insights into complex aspects of splicing regulation.
Highlights.
- Two simple and consecutive chemical reactions are catalyzed cotrancriptionally by a large macromolecular complex
-The spliceosome is essential for generating mature mRNA.
- The cell takes advantage of this to generate different amounts and isoforms of mRNAs, according to its needs.
- In yeast, control of pre-mRNA splicing of critical genes allows the cell to affect all the pathways in gene expression
-The spliceosome can be both the sensor and controller to fine tune genetic output
Acknowledgments
We sincerely appreciate the constructive critical evaluation of the manuscript by three anonymous reviewers. We thank Charles Query for discussions. This work was supported by grants to TLJ from the National Science Foundation (NSF CAREER Award to T.L.J. MCB-0448010 and MCB-1051921), and to JV by grants from the Spanish Ministry of Science (BIO2008-01091 and 363) and CSIC (200920I195). We apologize to anyone's whose work was not cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 2.Will CLL. R. . Spliceosome structure and function. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Third Edition Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]
- 3.Du H, Rosbash M. The U1 snRNP protein U1C recognizes the 5' splice site in the absence of base pairing. Nature. 2002;419:86–90. doi: 10.1038/nature00947. [DOI] [PubMed] [Google Scholar]
- 4.Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 5.Lallena MJ, Chalmers KJ, Llamazares S, Lamond AI, Valcarcel J. Splicing regulation at the second catalytic step by Sex-lethal involves 3' splice site recognition by SPF45. Cell. 2002;109:285–296. doi: 10.1016/s0092-8674(02)00730-4. [DOI] [PubMed] [Google Scholar]
- 6.Meyer M, Plass M, Perez-Valle J, Eyras E, Vilardell J. Deciphering 3'ss Selection in the Yeast Genome Reveals an RNA Thermosensor that Mediates Alternative Splicing. Mol Cell. 2011;43:1033–1039. doi: 10.1016/j.molcel.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 7.Gahura O, Hammann C, Valentova A, Puta F, Folk P. Secondary structure is required for 3' splice site recognition in yeast. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 9.Osheim YN, Miller OL, Jr., Beyer AL. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell. 1985;43:143–151. doi: 10.1016/0092-8674(85)90019-4. [DOI] [PubMed] [Google Scholar]
- 10.Kotovic KM, Lockshon D, Boric L, Neugebauer KM. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol. 2003;23:5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5'ss base pairing in yeast. Mol Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Lacadie SA, Tardiff DF, Kadener S, Rosbash M. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 2006;20:2055–2066. doi: 10.1101/gad.1434706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 17.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Howe KJ, Kane CM, Ares M., Jr. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011 doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Munoz MJ, de la Mata M, Kornblihtt AR. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem Sci. 2010;35:497–504. doi: 10.1016/j.tibs.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Oesterreich FC, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends Cell Biol. 2011;21:328–335. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi JK, Bae JB, Lyu J, Kim TY, Kim YJ. Nucleosome deposition and DNA methylation at coding region boundaries. Genome Biol. 2009;10:R89. doi: 10.1186/gb-2009-10-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nahkuri S, Taft RJ, Mattick JS. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle. 2009;8:3420–3424. doi: 10.4161/cc.8.20.9916. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 28.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 30.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hnilicova J, Stanek D. Where splicing joins chromatin. Nucleus. 2011;2:182–188. doi: 10.4161/nucl.2.3.15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5:e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunderson FQ, Merkhofer EC, Johnson TL. Dynamic histone acetylation is critical for cotranscriptional spliceosome assembly and spliceosomal rearrangements. Proc Natl Acad Sci U S A. 2011;108:2004–2009. doi: 10.1073/pnas.1011982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, Andrau JC, Ferrier P, Carmo-Fonseca M. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 35.Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci U S A. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallwitz D, Sures I. Structure of a split yeast gene: complete nucleotide sequence of the actin gene in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980;77:2546–2550. doi: 10.1073/pnas.77.5.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 38.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 39.Ares M, Jr., Grate L, Pauling MH. A handful of intron-containing genes produces the lion's share of yeast mRNA. RNA. 1999;5:1138–1139. doi: 10.1017/s1355838299991379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parenteau J, Durand M, Veronneau S, Lacombe AA, Morin G, Guerin V, Cecez B, Gervais-Bird J, Koh CS, Brunelle D, Wellinger RJ, Chabot B, Abou Elela S. Deletion of many yeast introns reveals a minority of genes that require splicing for function. Mol Biol Cell. 2008;19:1932–1941. doi: 10.1091/mbc.E07-12-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parenteau J, Durand M, Morin G, Gagnon J, Lucier JF, Wellinger RJ, Chabot B, Abou Elela S. Introns within Ribosomal Protein Genes Regulate the Production and Function of Yeast Ribosomes. Cell. 2011;147:320–331. doi: 10.1016/j.cell.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 42.Juneau K, Palm C, Miranda M, Davis RW. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc Natl Acad Sci U S A. 2007;104:1522–1527. doi: 10.1073/pnas.0610354104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vilardell J, Warner JR. Regulation of splicing at an intermediate step in the formation of the spliceosome. Genes Dev. 1994;8:211–220. doi: 10.1101/gad.8.2.211. [DOI] [PubMed] [Google Scholar]
- 44.Vilardell J, Warner JR. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol Cell Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemieux C, Marguerat S, Lafontaine J, Barbezier N, Bahler J, Bachand F. A Pre-mRNA Degradation Pathway that Selectively Targets Intron-Containing Genes Requires the Nuclear Poly(A)-Binding Protein. Mol Cell. 2011;44:108–119. doi: 10.1016/j.molcel.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 46.Fewell SW, Woolford JL., Jr. Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18S rRNA. Mol Cell Biol. 1999;19:826–834. doi: 10.1128/mcb.19.1.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, Paulovich AG, Woolford JL., Jr. Feedback inhibition of the yeast ribosomal protein gene CRY2 is mediated by the nucleotide sequence and secondary structure of CRY2 pre-mRNA. Mol Cell Biol. 1995;15:6454–6464. doi: 10.1128/mcb.15.11.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilbert WV. Functional specialization of ribosomes? Trends Biochem Sci. 2011;36:127–132. doi: 10.1016/j.tibs.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Macías S, Bragulat M, Tardiff DF, Vilardell J. L30 binds the nascent RPL30 transcript to repress U2 snRNP recruitment. Mol Cell. 2008;30:732–742. doi: 10.1016/j.molcel.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Vilardell J, Chartrand P, Singer RH, Warner JR. The odyssey of a regulated transcript. Rna. 2000;6:1773–1780. doi: 10.1017/s135583820000145x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vilardell J, Yu SJ, Warner JR. Multiple functions of an evolutionarily conserved RNA binding domain. Mol Cell. 2000;5:761–766. doi: 10.1016/s1097-2765(00)80255-5. [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 54.Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cuenca-Bono B, Garcia-Molinero V, Pascual-Garcia P, Dopazo H, Llopis A, Vilardell J, Rodriguez-Navarro S. SUS1 introns are required for efficient mRNA nuclear export in yeast. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hossain MA, Rodriguez CM, Johnson TL. Key features of the two-intron Saccharomyces cerevisiae gene SUS1 contribute to its alternative splicing. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Preker PJ, Guthrie C. Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA. RNA. 2006;12:994–1006. doi: 10.1261/rna.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Preker PJ, Kim KS, Guthrie C. Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA. 2002;8:969–980. doi: 10.1017/s1355838202020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong S, Li C, Zenklusen D, Singer RH, Jacobson A, He F. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol Cell. 2007;25:559–573. doi: 10.1016/j.molcel.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 62.Fededa JP, Petrillo E, Gelfand MS, Neverov AD, Kadener S, Nogues G, Pelisch F, Baralle FE, Muro AF, Kornblihtt AR. A polar mechanism coordinates different regions of alternative splicing within a single gene. Mol Cell. 2005;19:393–404. doi: 10.1016/j.molcel.2005.06.035. [DOI] [PubMed] [Google Scholar]
- 63.Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 64.Munding EM, Igel AH, Shiue L, Dorighi KM, Trevino LR, Ares M., Jr. Integration of a splicing regulatory network within the meiotic gene expression program of Saccharomyces cerevisiae. Genes Dev. 2010;24:2693–2704. doi: 10.1101/gad.1977410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiu ZR, Schwer B, Shuman S. Defining the Mer1 and Nam8 meiotic splicing regulons by cDNA rescue. RNA. 2011;17:1648–1654. doi: 10.1261/rna.2792011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spingola M, Ares M., Jr. A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol Cell. 2000;6:329–338. doi: 10.1016/s1097-2765(00)00033-2. [DOI] [PubMed] [Google Scholar]
- 67.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McKay SL, Johnson TL. A bird's-eye view of post-translational modifications in the spliceosome and their roles in spliceosome dynamics. Mol Biosyst. 2010;6:2093–2102. doi: 10.1039/c002828b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clark TA, Sugnet CW, Ares M., Jr. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 71.Moldon A, Malapeira J, Gabrielli N, Gogol M, Gomez-Escoda B, Ivanova T, Seidel C, Ayte J. Promoter-driven splicing regulation in fission yeast. Nature. 2008;455:997–1000. doi: 10.1038/nature07325. [DOI] [PubMed] [Google Scholar]
- 72.Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell. 2008;31:360–370. doi: 10.1016/j.molcel.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 2011;3:a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell. 2011;147:1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Mol Biosyst. 2010;6:481–493. doi: 10.1039/b919670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vorlova S, Rocco G, Lefave CV, Jodelka FM, Hess K, Hastings ML, Henke E, Cartegni L. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol Cell. 2011;43:927–939. doi: 10.1016/j.molcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, Krainer AR. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shen H, Zheng X, Luecke S, Green MR. The U2AF35-related protein Urp contacts the 3' splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev. 2010;24:2389–2394. doi: 10.1101/gad.1974810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 81.Hossain MA, Claggett JM, Nguyen T, Johnson TL. The cap binding complex influences H2B ubiquitination by facilitating splicing of the SUS1 pre-mRNA. RNA. 2009;15:1515–1527. doi: 10.1261/rna.1540409. [DOI] [PMC free article] [PubMed] [Google Scholar]