Abstract
During the development and adult life of multicellular organisms cells move from one location to another as they assemble into organs, seal a wound or fight pathogens. For navigation, migrating cells follow cues that guide them to their final position. Frequently, a single cue simultaneously guides different cells to different positions. Recent studies of one such cue—the chemokine SDF1—suggest strategies for how the animal achieves this task without causing erroneous migration.
Keywords: CXCR4, CXCR7, SDF1/CXCL12, Cajal-Retzius Cells, Interneurons, Germ cells
1. Introduction
Chemokines are small, mostly secreted proteins of 8–14 kD [1]. They are classified into four groups according to the position of their first two cysteine residues (CXC, CC, C and CX3C) [2]. Humans have at least 46 different chemokines that signal through at least 23 different chemokine receptors [3][4]. All chemokine receptors are G-protein-coupled seven-transmembrane receptors, most of which signal through Gαi and are pertussis toxin sensitive [5]. The predominant function of chemokines is to recruit and activate immune cells [6], but chemokines have also been implicated in human immunodeficiency virus infection [7], autoimmune conditions [8], inflammatory diseases [9] and cancer [10]. Consistent with their essential role in the adaptive immune response, chemokines and their receptors are vertebrate-specific and likely co-evolved with the expansion of lymphocyte function. Jawless fish are the most basal vertebrates known to have chemokine receptors in their genome [4]. The three types of chemokine receptors found in jawless fish correspond to CCR14, CXCR4 and CXCR7, suggesting that these receptors served as the blueprint for the diversification of the mammalian chemokine system.
Intriguingly, one of these ancient chemokine receptors, CXCR4, and its ligand SDF1 provide guidance to many different cell types during development and adult life. SDF1 was initially identified in bone marrow stromal cell lines as a secreted molecule that attracts and stimulates the growth of B-cells [11][12][13] and was shown to signal through the chemokine receptor CXCR4 [14][15][16]. Subsequent studies have demonstrated that this ligand-receptor pair orchestrates additional aspects of the immune system and embryonic development. In the immune system, CXCR4-mediated SDF1 signaling attracts lymphocytes [13], stimulates the proliferation of blood cells [17][18][19] and contributes to the homing of hematopoietic stem cells [20]. These guidance functions are also critical in the developing embryo where SDF1 guides Cxcr4-expressing germ cells [21][22][23], cells of the posterior lateral line primordium [24] and blood vessels [25][26], dentate gyrus granule cells [27][28], trigeminal [29] and dorsal root ganglia sensory neurons [30], gonadotropin-releasing hormone neurons [31] and olfactory neurons [32]. SDF1 also controls the spreading of interneurons [33][34][35][36] and Cajal-Retzius cells across the cortex [37][38], retains cerebellar external granule cells at their intermediate target [39][40][18][19][41] and anchors endoderm cells to the adjacent mesoderm during gastrulation [42][43]. Many of these cell types are guided by SDF1 to different targets at the same time and in close proximity to each other. Remarkably, the embryo orchestrates these concurrent processes with little to no error, suggesting tight control of the spatial and temporal distribution of SDF1. Recent studies have begun to elucidate how the animal achieves such tight control of SDF1 signaling. In this review, we discuss how chemokine clearance and signaling through the second SDF1 receptor CXCR7 and microRNA regulation of chemokine signaling may regulate the spatial and temporal distribution of SDF1 signaling during germ cell migration and cortical interneuron and Cajal-Retzius cell spreading.
2. Primordial germ cell migration
Primordial germ cells (PGCs) are the precursors of the germline stem cells that reside in the gonads and eventually differentiate into either sperm or oocytes. In most animals, PGCs are born far from their final position and need to migrate extensively to reach the gonads [44]. In vertebrates, PGCs express Cxcr4 and are recruited to their final position by the CXCR4 ligand SDF1. The expression domain of SDF1 prefigures the PGC migration route, and loss of Cxcr4 or SDF1 results in mispositioning of PGCs [22][21][23][45][46]. In zebrafish—whose genome contains two copies of SDF1 (SDF1a and SDF1b) [21][22] and Cxcr4 (cxcr4a and cxcr4b) [47]—the SDF1a expression domain closest to the migrating PGCs refines over time towards the future site of the gonads. The PGCs are born onto this domain and remain closely associated with it as it refines (Figure 1A) [22]. It is unclear whether cxcr4b-mediated chemokine signaling is required throughout PGC migration, but the close association of the PGCs with the dynamic chemokine expression domain suggests a continuous requirement for SDF1a in guiding these cells to their final position.
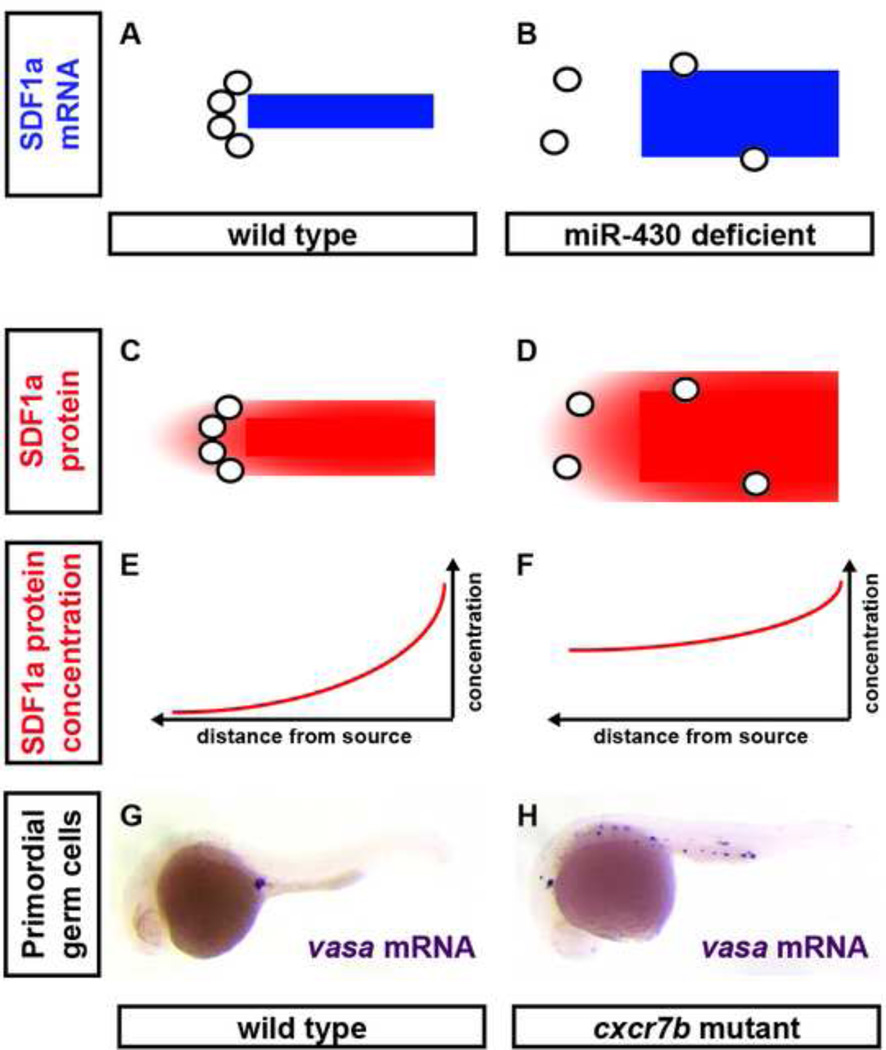
Figure 1. Refinement of SDF1 chemokine signaling during primordial germ cell migration.
The SDF1a expression domain is refined at the level of transcript and protein through miR-430 and Cxcr7b, respectively. Cxcr4b-expressing primordial germ cells (grey circles) are born onto a dynamic SDF1a mRNA expression domain (blue stripe in A and B). This expression domain refines over time towards the future site of the gonads. Primordial germ cells stay closely associated with this shifting expression domain and are thus guided to their final position (A). In the absence of miR-430-mediated regulation of SDF1a transcripts the chemokine expression domain is expanded, and primordial germ cells are misdirected (B). Cxcr7b removes SDF1a protein (red color) from the tissue around the SDF1a expression domain (C) and refines its distribution (E), such that primordial germ cells are guided correctly to the future site of the gonad (G). In the absence of Cxcr7b, excess SDF1a protein is not cleared (D) and spreads further from SDF1a-expressing tissues (F). This results in ectopic primordial germ cells in cxcr7b mutant embryos (H). G and H show wildtype and cxcr7b mutant zebrafish embryos, respectively, stained for vasa mRNA to visualize primordial germ cells at 24 hours post fertilization.
Many tissues in the embryo express SDF1a during PGC migration [22]. PGCs pass near and partly encounter some of these SDF1a expression domains but almost always follow only the domain onto which they were born. In rare cases, the PGCs are attracted to other domains, such as the SDF1a expression domain that guides trigeminal sensory neurons [48][22]. This low error rate may be attributed to the consistent, close association between the PGCs and the dynamic SDF1a expression domain that guides them to their target. SDF1a protein can act at a distance [49] and is thought to form gradients that emanate from the chemokine-producing tissues in the embryo [50][51]. However, PGCs are born onto a domain of chemokine-producing cells and are therefore exposed to high SDF1a protein levels at birth. As the SDF1a expression domain refines, the PGCs are thought to migrate towards the highest levels of the attractant. Repetition of this process would result in maintenance of the close association of PGCs and the SDF1a expression domain onto which they were born [52][51]. Consequently, PGCs would be less likely to be misdirected by more distant sources of SDF1a protein. This model implies that the SDF1a mRNA and protein expression patterns closely reflect the spatial and temporal activity of the SDF1a gene.
Studies in zebrafish have revealed two mechanisms that contribute to the tight correlation of gene activity and SDF1a mRNA and SDF1a protein distribution during PGC migration. First, the ubiquitously expressed microRNA miR-430 targets SDF1a mRNA for degradation. When miR-430 is prevented from binding to its target site in the SDF1a-3’UTR, the mRNA expression domain of SDF1a is enlarged and PGCs are more likely to migrate incorrectly (Figure 1B). This defect can be rescued by lowering SDF1a protein levels, suggesting that in the absence of miR-430-mediated mRNA regulation, SDF1a protein levels are elevated perturbing PGC migration [53]. Second, CXCR7, a recently described second SDF1 receptor [54][55], has been shown to decrease SDF1 protein levels through ligand sequestration (Figure 1C–H) [56][57]. Although CXCR7 seems to act as a signaling receptor in some contexts [54][55][58][59][60], there is compelling evidence for a clearance receptor function for Cxcr7b, one of the two zebrafish Cxcr7 paralogs [32], during PGC migration [56]. First, its function is required not in the PGCs but rather in the tissue through which they migrate. Second, lowering SDF1a protein levels in cxcr7b-depleted embryos partially rescues PGC migration. Third, PGCs behave similarly in cxcr7b-depleted embryos and embryos with elevated SDF1a protein levels. Although the lack of antibodies has precluded direct analysis of SDF1a protein expression, these observations are consistent with a model in which miR-430 and Cxcr7b act together to refine SDF1a protein expression so that it closely mirrors the spatiotemporal dynamics of SDF1a gene transcription (Figure 1).
3. Cortical interneuron migration
Cortical interneurons comprise a diverse set of morphologically and physiologically distinct cells that constitute a major part of the cortex. Locally, they mediate synaptic inhibition, but globally they are thought to shape cortical network oscillations and support many brain functions (reviewed in[61]). In mice, cortical interneurons are born at three different sites in the ventral part of the developing forebrain: the medial and caudal ganglionic eminences [62] and the preoptic area [63][64]. Interneurons migrate from these sites in two streams into the overlying cortex on the dorsal side of the forebrain. One stream of interneurons traverses the cortex along the marginal zone (MZ), just underneath the meninges of the dorsal forebrain. The other, more prominent stream of interneurons migrates into the cortex through the intermediate zone/subventricular zone (IZ/SVZ). These two streams travel in parallel to each other and are separated by the developing cortical plate. Because the interneurons migrate in parallel to the meningeal membranes and the ventricular surface, this phase of their migration is referred to as tangential migration. Initially, there is very little mixing between the two streams [65] Tanaka:2003hn, [34][35][66][67]. However, once the interneurons have emigrated from the ventral forebrain and spread across the cortex in the MZ and IZ/SVZ, they change their direction of migration; interneurons from the MZ cell stream turn ventrally while interneurons from the IZ/SVZ cell stream turn dorsally, initiating cortical plate entry of neurons from both streams. Thus, while interneurons initially migrate tangentially to spread across the cortex in two distinct streams, they later migrate radially to populate the cortical plate (reviewed in [68]).
In vivo and in vitro live imaging studies have shown that interneurons migrate in all directions within the planes of the MZ and IZ/SVZ but exhibit a slight rostro-caudal preference. This type of cell movement is referred to as multidimensional tangential migration [69][66][70][71]. Intriguingly, based on the direction of cellular protrusions, the interneurons at the leading edge of the streams are oriented dorsally towards areas not yet occupied by interneurons while interneurons in densely populated regions are oriented in various directions [67]. This suggests that interneurons at the edge of the streams sense directional information while interneurons within the streams sense little or no directional information.
Tangential and radial migration both depend on SDF1 chemokine signaling for directional information. In Cxcr4 and SDF1 mutant mice, interneurons are not restricted to the two streams in the MZ and IZ/SVZ. Instead, they are found throughout the developing cortex [34][72][35][33]. Additionally, removing Cxcr4 activity after interneurons have spread across the cortex results in premature radial migration and invasion of the cortical plate [35]. Consistent with the requirement for SDF1 signaling in tangential and radial migration, interneurons express Cxcr4 and migrate from the ganglionic eminences into the cortex along two stripes of SDF1 expression (Figure 2A–B) [73][35][74][34][33][75][76]. These two stripes of SDF1 expression delineate the migratory routes along the MZ and IZ/SVZ. While SDF1 is expressed along the MZ throughout tangential and radial migration, its expression ceases in the IZ/SVZ as interneurons change from tangential to radial migration. In contrast to primordial germ cells, which follow a refining SDF1a expression domain, interneurons migrate across two seemingly uniform stripes of SDF1 expression. Although interneurons are attracted by SDF1 both in vitro and in vivo [73][35] and display increased motility in response to SDF1 [74][34], it is puzzling how uniform chemokine expression can provide directionality to the migrating interneurons. Recent analysis of the role of the second SDF1 receptor, CXCR7, in interneuron migration provides insights into how this may be achieved. Interneurons in the MZ and IZ/SVZ express Cxcr7 during tangential migration, and cells in the cortical plate express Cxcr7 for a short period at the onset of migration [77][73][74]. Intriguingly, Cxcr7 function is required in both of these cell populations for correct interneuron migration (Figure 2K–L) [73][74]. This dual requirement for Cxcr7 is reflected in the two ways Cxcr7 functions during interneuron migration. First, in interneurons, binding of SDF1 to CXCR7 activates the MAP kinase pathway [74][73] independently of Cxcr4 [73]. Consistent with this, both CXCR7 and CXCR4 are essential to recruit interneurons to sites of ectopic SDF1 expression in vivo [73]. Second, in addition to signaling through the MAP kinase pathway, CXCR7 also binds, internalizes and degrades SDF1 in interneurons [74]. In contrast to zebrafish, where—based on a qualitative assay—Cxcr7b but not Cxcr4b seems to sequester SDF1a protein [56], both receptors contribute equally to SDF1 protein uptake by mouse interneurons [74]. In interneurons, this chemokine clearance function of CXCR7 leads to lower SDF1 protein levels around the cells. In the absence of Cxcr7, interneurons do not clear SDF1 protein and are therefore exposed to increased local chemokine levels. This increase in SDF1 protein levels leads to excessive activation of CXCR4 resulting in complete internalization and degradation [74]. Importantly, CXCR4 degradation in Cxcr7 mutant interneurons is blocked when SDF1 is removed from the media of cultured interneuron explants [74]. Therefore, the membranes of Cxcr7 mutant interneurons are devoid of CXCR4 receptors [74], and, thus, the interneurons are probably unable to sense and respond to SDF1. These two distinct functions of CXCR7—intracellular signaling and chemokine clearance—are reflected at the level of cellular behavior. If the interneuron migration defect in Cxcr7 mutants were solely due to elevated SDF1 protein levels and resultant CXCR4 internalization and degradation, then one would expect Cxcr4 and Cxcr7 mutant interneurons to behave similarly. However, they do not: Cxcr4 mutant interneurons transition more readily from tangential to radial migration and move tangentially at a greater rate than Cxcr7 mutant interneurons [73], suggesting that the contribution of CXCR7 to the speed and direction of migration is at least partially independent of CXCR4.
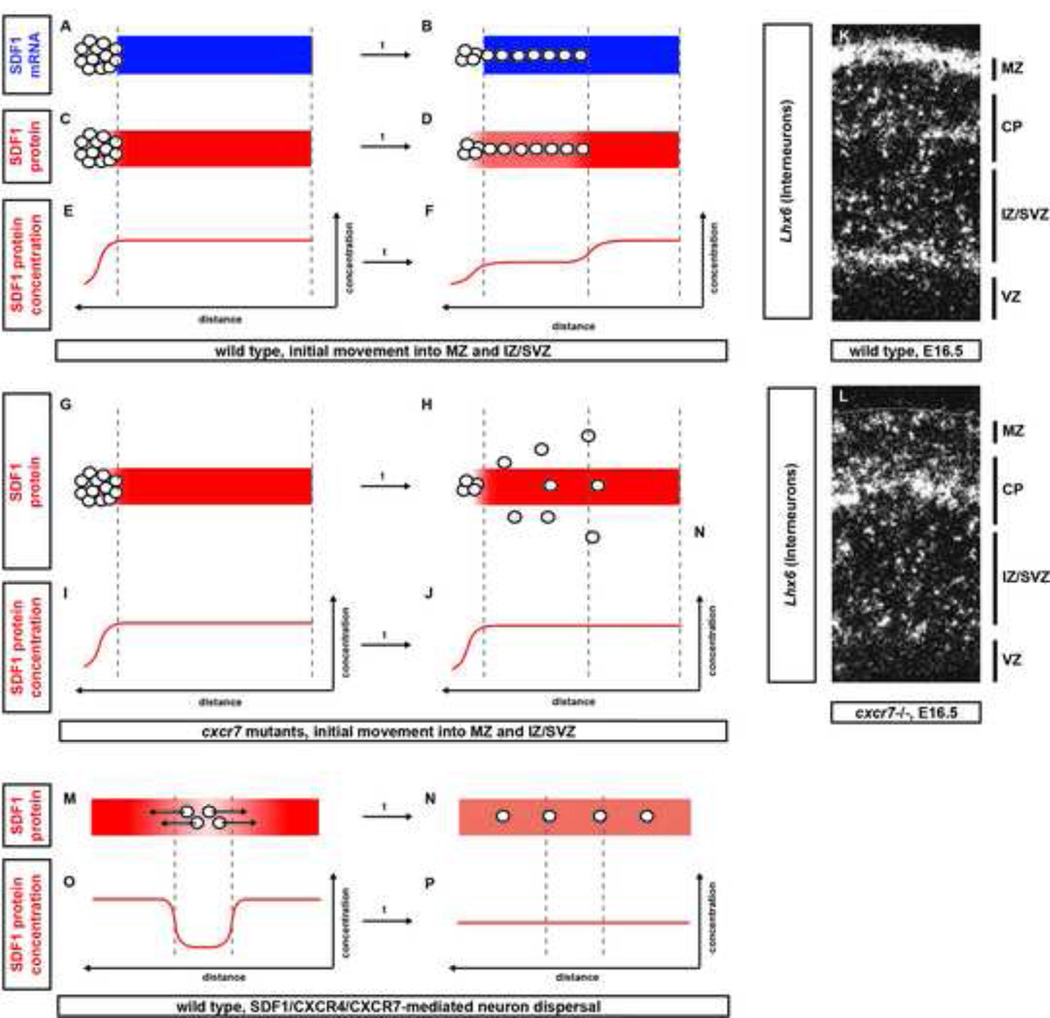
Figure 2. Attractive dispersion model for SDF1 chemokine signaling in interneuron migration.
The attractive dispersion model postulates that interneurons modulate the SDF1 protein distribution to facilitate their even dispersal across the cortex. Cxcr4 and Cxcr7 expressing interneurons (grey circles) are born next to one of two stripes of uniform SDF1 mRNA expression in the MZ and the IZ/SVZ (A). For simplicity, only one stripe of SDF1 mRNA expression is shown (blue stripe in A and B). Upon recruitment onto this expression domain (B), interneurons remove SDF1 protein through CXCR7-mediated chemokine clearance (compare C with D). This creates a graded distribution of SDF1 protein with less attractant behind than in front of them (compare E with F). In the absence of CXCR7, interneurons cannot refine the SDF1 protein expression pattern [74] (H and J), do not respond to SDF1 protein [73] and do not disperse evenly along the SDF1 expression stripe (H). In the wild-type cortex, interneurons are restricted to the MZ and IZ/SVZ at E16.5 (K). In Cxcr7 mutants, they prematurely enter the CP (L). Panels in K and L are adapted with permission from [74]. Panels M-P describe how, according to the attractive dispersion model, the interaction of CXCR7 and CXCR4 with SDF1 could facilitate the redistribution of neurons from an over-populated region of the cortex to a region populated by less neurons. MZ, marginal zone; CP, cortical plate; IZ/SVZ, intermediate zone/subventricular zone; VZ, ventricular zone.
Taken together, these observations suggest an “attractive dispersion model.” According to this model, newly born interneurons are initially attracted to the tip of one of the two uniform stripes of SDF1 expression in the cortex (Figure 2A). When they the first encounter the SDF1 expressing tissue, the interneurons start to lower the SDF1 protein concentration around them through CXCR7-mediated SDF1 protein clearance (Figure 2C). This creates a local dip in the otherwise uniform SDF1 protein distribution along the stripe of chemokine expression (Figure 2E). The interneurons are therefore exposed to more SDF1 protein in front of them than underneath them. As a consequence of this locally generated gradient, the interneurons migrate further along the stripe of chemokine expression toward higher levels of SDF1 protein (Figure 2D). As additional interneurons move onto the stripe of chemokine expression and others advance further along the stripe, the interneurons continue to create local dips in the SDF1 protein concentration (Figure 2F) resulting in further spreading of interneurons across the chemokine expression domain. Therefore, interneurons within the stream migrate with little to no directional bias while interneurons at the edge migrate preferentially towards areas not yet occupied by interneurons, a prediction that is consistent with live imaging experiments [69][66][70][71][67]. Intriguingly, such a mechanism also ensures even distribution of interneurons throughout the stripes of SDF1 expression in the MZ and IZ/SVZ. According to the model, clustering of interneurons within the chemokine expression domain results in increased local CXCR7-mediated SDF1 protein clearance. This in turn generates a local SDF1 protein dip (Figure 2M). Consequently, interneurons within a local cluster move away from one another toward areas with higher concentrations of SDF1 protein (Figure 2O). Repetition of this process ultimately results in even spreading of interneurons across the uniform stripes of SDF1 expression in the cortex (Figure 2N and P). Moreover, SDF1 protein clearance through the initial expression of Cxcr7 in the cortical plate might help restrict the chemokine distribution to the overlying MZ and the underlying IZ/SVZ and, thus, prevent interneurons from entering the cortical plate before they are evenly dispersed.
Once the interneurons have spread across the cortex, they change from tangential to radial migration and start to infiltrate the cortical plate. Interneurons in the MZ stream descend ventrally and interneurons in the IZ/SVZ stream ascend dorsally into the cortical plate. Intriguingly, co-culture experiments have shown that interneurons invade the cortical plate only when they are confronted with older cortical slices [34]. Although this suggests that the early cortical plate constitutes a non-permissive environment for interneurons, this does not seem to be the case; interneurons placed next to cortical plate isolates of the same age readily infiltrate them [34]. Similarly, live imaging studies of cortical slice cultures have demonstrated that interneurons occasionally pass through the cortical plate from the MZ to the IZ/SVZ and vice versa but do not settle in the cortical plate until they have dispersed across the cortex [69][66][70][71][67][34][73]. Thus, it is likely that there is an activity in the early but not in the mature cortex that prevents interneurons from settling in the cortical plate. On a molecular level, this cortical maturation process correlates with the dynamic expression pattern of Cxcr7 in the interneurons. Concomitant with entry into the cortical plate, interneurons cease to express Cxcr7 but continue to express Cxcr4 [73][74][77]. Since both chemokine receptors are essential for the attraction of interneurons by SDF1 in vivo [73,74][35], this is consistent with the idea that downregulation of Cxcr7 renders interneurons less sensitive to SDF1, allowing them to change their direction of migration and enter the cortical plate. Additionally, downregulation of SDF1 expression in the IZ/SVZ coincides with downegulation of Cxcr7 in the interneurons and cortical plate entry [73][35][74][34][33][78][75][76]. Therefore, interneurons in the IZ/SVZ stream not only become less sensitive to SDF1 but also lose the source of the attractant itself. This change in responsiveness to SDF1 could be the molecular correlate of the maturation process that interneurons need to undergo before they change from tangential to radial migration and enter the cortical plate. One prediction from this model is that premature loss of Cxcr7 expression in interneurons should shorten the maturation period and result in earlier entry of interneurons into the cortical plate. Although this has not been tested directly, it has been shown that conditional inactivation of Cxcr4 after interneurons have spread across the cortex accelerates interneuron entry into the cortical plate [35], suggesting that desensitization to SDF1 is important for cortical plate entry.
Although the attractive dispersion model is an appealingly simple model, live imaging studies suggest that loss of responsiveness to SDF1 is probably not the only trigger for cortical plate entry. Before entry, MZ interneurons extend long protrusions into the cortical plate but frequently choose not to enter [67], suggesting that the cortical plate is composed of both favorable and non-favorable environments for interneuron entry. Although this could be due to physical or molecular barriers that hinder interneuron entry, cells in the cortical plate may also secrete attractants that recruit interneurons once they are no longer retained by SDF1 in the MZ and IZ/SVZ. Alternatively, the meninges and/or the IZ/SVZ may secrete repellents from which interneurons migrate away once they are no longer attracted by SDF1. Together, these observations suggest that SDF1 may facilitate both the tangential migration and even distribution of interneurons throughout the MZ and IZ/SVZ and the initiation of radial migration into the cortical plate, possibly in conjunction with other attractants and repellents in the cortical plate.
4. Cajal-Retzius cell migration
Cajal-Retzius (CR) cells are a transient population of neurons at the surface of the cerebral cortex. They regulate the radial migration and laminar arrangement of neurons in the dorsal forebrain (reviewed in [79][80]). For a long time, it was thought that CR cells are born in the ventricular zone of the dorsal forebrain, a conclusion supported by fate mapping and loss of function and expression analyses [81][82]. More recently, though, experiments that combined fate mapping with tissue ablation demonstrated that CR cells are also born in the cortical hem [83], the septum and the pallial-subpallial boundary [64]. From these three sites, CR cells migrate tangentially into the cortex just beneath the meninges and disperse across the cortex. At the sites in the marginal zone where the CR cells from different origins meet, the cells intermingle but do not disperse into areas that are already occupied by other CR cells. However, if pallial-subpallial boundary-derived or hem-derived CR cells are genetically ablated, CR cells from other sources spread into the unoccupied area and—at least transiently—compensate for the genetically ablated population [64][83]. These observations suggest that, rather than being restricted from certain areas in the MZ by a repellent, CR cells instead sense the density of their sibling cells, a property that limits their dispersal.
Although it is not clear how CR cells born in the septum and the pallial-subpallial boundary navigate into the cortex, surgical removal of the meninges locally blocks hem-derived CR cell migration [38]. Conversely, removal of all cortical structures except the meninges does not perturb hem-derived CR cell migration [38]. These findings suggest that the meninges are both necessary and sufficient for the migration of hem-derived CR cells. Consistent with this idea, CR cells are attracted to meningeal tissue but not to neocortical tissue in co-cultures [38]. Intriguingly, meningeal tissues from different locations in the cortex attract hem-derived CR cells to the same degree [38]. Moreover, transplanted hem-derived CR cells disperse in all directions along the meninges with no apparent directional bias regardless of the cortical region in which they are placed [38]. Together, these observations suggest that the meninges provide a permissive environment for the hem-derived CR cells to disperse within the MZ. In addition to permitting dispersal across the cortex, the meninges are also required for retention of CR cells in the MZ. Chemical disruption of the meninges after CR cells have dispersed across the cortex results in CR cell redistribution to deeper cortical layers, a defect that can be rescued by a secreted factor from the meninges [37].
SDF1 seems to provide the molecular basis for the attraction and retention of CR cells in the MZ adjacent to the meninges. In the absence of CXCR4-mediated SDF1 signaling, hem-derived CR cells still enter the cortex but are not confined to a stream of cells beneath the meninges in the MZ [38][37]. Instead, they infiltrate all layers of the cortex. Moreover, resupplying SDF1 protein to cortical slices in which CR cells have redistributed to deeper cortical layers due to chemical ablation of the meninges restores CR cell positioning in the MZ [37]. The expression of Cxcr4 and SDF1 mRNAs are consistent with these observations: Hem-derived CR cells express Cxcr4 and the meninges express SDF1 [38][37][72]. Since the expression of SDF1 in the meninges also guides the interneurons in the MZ stream [34][72][35][33], CR cells and MZ interneurons likely use the same SDF1 source to enter the cortex. Intriguingly, though, these two cell populations migrate beneath the meninges in opposite directions. While the CR cells migrate from the hem outwards towards more lateral positions, the interneurons migrate from the ganglionic eminences inwards towards more medial positions. Thus, Cxcr4-expressing CR cells and interneurons migrate from opposing ends of a uniform stripe of SDF1 expression. When CR cells and interneurons meet in the MZ, the CR cells remain directly beneath the meninges, and the interneurons populate the space beneath the CR cells [67]. This topography might reflect the qualitatively different response to SDF1 observed in explants: While CR cells are attracted towards co-cultured SDF1 expressing cells [38], interneurons gain motility in response to SDF1-expressing cells but do not show a clear directional bias [34][74]. Importantly, after CR cells and interneurons have arranged into two layers, both cell types exhibit similar polarity [67], suggesting that they perceive the same guidance cue.
The attractive dispersion model as described above (Section 3 and Figure 2), provides an appealing explanation for how migrating interneurons might themselves sculpt the uniform expression of SDF1 into a gradient that recruits and disperses them across the MZ and IZ/SVZ in the cortex. Although the role of CXCR7 in CR cell migration is not clear, it is tempting to speculate that CR cells might use the strategy described by the attractive dispersion model for their dispersal beneath the meninges. However, while CR cells also express Cxcr7 [77][78][73], their distribution and positioning does not seem to be affected when Cxcr7 function is selectively removed from the cortical glutamatergic lineages using the Emx1-Cre driver [73]. It is unclear, though, whether Emx1 drives Cre expression in all CR cells and how efficiently Cre mediates recombination in these cells [84]. These caveats in combination with the observation that hem-derived and pallial-subpallial boundary-derived CR cells can compensate for each other [83][64] suggest that the consequences of incomplete loss of Cxcr7 function in CR cells could be mild and difficult to detect. Additionally, both CXCR7 and CXCR4 sequester SDF1 in interneurons [74], an observation also made in cell culture experiments [57]. Therefore, it is conceivable that CXCR4 contributes to the clearance of SDF1 by CR cells, partially compensating for the loss of CXCR7-mediated SDF1 protein clearance.
The attractive dispersion model could also provide a molecular explanation for the behavior of CR cells in three other experimental situations. First, CR cells from the hem and the pallial-subpallial boundary form a diffuse boundary where cells of both origins mix [83][64], suggesting that the CR cells do not repel one another. Second, when CR cells from the pallial-subpallial boundary are ablated, CR cells—likely hemderived—disperse into the area normally covered by the pallial-subpallial boundary-derived CR cells [64]. This observation suggests that CR cells from different sources may limit each other’s dispersal. Third, a modest reduction in hemderived CR cells reduces the final cell number but does not prevent dispersal of CR cells across the cortex [83], suggesting that there is a mechanism that ensures even distribution of the cells. In the first scenario, CR cells from the hem and the pallial-subpallial boundary stream would modulate the uniform SDF1 expression through CXCR7-mediated SDF1 protein clearance to generate local gradients that attracts cells to areas that are not yet occupied. Once the two CR cell streams encounter each other, cells at the leading edge would infiltrate the opposite stream until uniform cell density results in loss of local SDF1 gradients. If one stream is absent—as in the second scenario—CR cells from the remaining population would continue to spread into unoccupied areas and, thus, compensate for ablated CR cells. In the third scenario, the mechanism is conceptually identical: The CR cells would disperse across the cortex under the influence of local SDF1 gradients until a uniform cell density—although lower than in wild-type—results in the dissipation of local SDF1 gradients.
5. Conclusion
Guiding migrating cells through the body is a difficult task. Animals have developed many layers of control to ensure that cells navigate to their final position with little or no error. Recent work on SDF1-guided cell migration has uncovered two new layers of control: The distribution of SDF1 is controlled at the transcript level through the microRNA miR-430 and at the protein level through the chemokine clearance receptor CXCR7. Moreover, CXCR7 not only removes the chemokine but also mediates SDF1 activation of the MAPK pathway. Together, these two aspects of CXCR7’s function modulate the cell’s response to SDF1.
Refinement of the SDF1 expression domain through miR-430 during germ cell migration is the first example of microRNA-mediated regulation of chemokine signaling. It is conceivable that similar refinement of the SDF1 expression domain is required in other chemokine-guided migration events. Rapid changes in SDF1 expression as observed during germ cell migration or the establishment of sharp expression borders as observed in cortical interneuron migration could rely on such a mechanism. In the first case, clearance of SDF1 transcripts would ensure that promoter shutdown is reinforced by transcript degradation in tissues that no longer express SDF1. In the second case, transcript degradation in domains with leaky SDF1 expression could sharpen the borders between tissues that should or should not express SDF1 at any given time. Moreover, graded microRNA activity could generate precise SDF1 transcript gradients across tissues, as is seen along the migratory route of gonadotropin-releasing hormone neurons [31].
The discovery of the role of CXCR7 in SDF1 protein clearance during primordial germ cell and interneuron migration suggests that other SDF1-guided processes may also rely on this function. For example, cerebellar external granule cells are born in the rhombic lip of the hindbrain and migrate tangentially beneath the meninges to cover the surface of the cerebellum. Once they have covered the cerebellum, they change direction and migrate through the molecular and Purkinje cell layers to form the internal granule cell layer. Analogous to interneuron and CR cell migration in the cortex, external granule cells express and rely on SDF1 to spread across the cerebellar surface [41][18][40]. Although it is unclear whether the external granule cells express Cxcr7, it is tempting to speculate that they might use the same strategy as interneurons for dispersal. Additionally, the juxtaposition of the expression domains of SDF1 and Cxcr7 at multiple sites in the brain [75][77][78][76] suggests that SDF1 protein clearance through CXCR7 could sharpen the borders of SDF1 protein expression domains and restrict the domains through which chemokine responsive cells migrate.
In addition to clearing SDF1 protein, CXCR7 also activates the MAP kinase pathway [85][74][73][60]. This signaling activity has been suggested to be Cxcr4-independent [73][60][86]. However, in some cases, CXCR4 seems to modulate CXCR7 signaling, possibly through CXCR4-CXCR7 heterodimerization [87]. Therefore, it will be interesting to see how these non-scavenging functions of CXCR7 contribute to SDF1-guided cell migration.
Highlights.
-
>
Review of SDF1-guided cell migration
-
>
Discussion of the control of SDF1 through microRNAs and CXCR7
-
>
New model for dispersion of cells across a uniform SDF1 expression domain
Acknowledgements
The authors wish to thank members of the Knaut and Torres-Vázquez labs for discussion and comments. We apologize to any colleague whose work was not mentioned due to space restrictions. This work is supported by the National Institutes of Health grant NS069839 to H.K. S.L. was supported by a National Institutes of Health training grant HD007520.
Abbreviations
- PGC
primordial germ cell
- MZ
marginal zone
- IZ
intermediate zone
- SVZ
subventricular zone
- CR cells
Cajal-Retzius cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Stephen W. Lewellis, Email: stephen.lewellis@med.nyu.edu.
Holger Knaut, Email: holger.knaut@med.nyu.edu.
References
- 1.Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu Rev Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez EJ, Lolis E. Structure, function, and inhibition of chemokines. Annu. Rev. Pharmacol. Toxicol. 2002;42:469–499. doi: 10.1146/annurev.pharmtox.42.091901.115838. [DOI] [PubMed] [Google Scholar]
- 3.Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006;7:243. doi: 10.1186/gb-2006-7-12-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nomiyama H, Osada N, Yoshie O. A family tree of vertebrate chemokine receptors for a unified nomenclature. Developmental & Comparative Immunology. 2011;35:705–715. doi: 10.1016/j.dci.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mantovani A, Bonecchi R, Locati M. Tuning inflammation and immunity by chemokine sequestration: decoys and more. Nat Rev Immunol. 2006;6:907–918. doi: 10.1038/nri1964. [DOI] [PubMed] [Google Scholar]
- 7.Kuhmann SE, Hartley O. Targeting Chemokine Receptors in HIV: A Status Report. Annu. Rev. Pharmacol. Toxicol. 2008;48:425–461. doi: 10.1146/annurev.pharmtox.48.113006.094847. [DOI] [PubMed] [Google Scholar]
- 8.Szekanecz Z, Kim J, Koch AE. Chemokines and chemokine receptors in rheumatoid arthritis. Semin. Immunol. 2003;15:15–21. doi: 10.1016/s1044-5323(02)00124-0. [DOI] [PubMed] [Google Scholar]
- 9.Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2:108–115. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 10.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 11.Tashiro K, Tada H, Heilker R, Shirozu M, Nakano T, Honjo T. Signal sequence trap: a cloning strategy for secreted proteins and type I membrane proteins. Science. 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 12.Nagasawa T, Kikutani H, Kishimoto T. Molecular cloning and structure of a pre-B-cell growth-stimulating factor. Proc Natl Acad Sci USA. 1994;91:2305–2309. doi: 10.1073/pnas.91.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 15.Bleul CC, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, et al. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 16.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier JL, Arenzana-Seisdedos F, et al. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 17.Lataillade J-J, Clay D, Bourin P, Hérodin F, Dupuy C, Jasmin C, et al. Stromal cell-derived factor 1 regulates primitive hematopoiesis by suppressing apoptosis and by promoting G(0)/G(1) transition in CD34(+) cells: evidence for an autocrine/paracrine mechanism. Blood. 2002;99:1117–1129. doi: 10.1182/blood.v99.4.1117. [DOI] [PubMed] [Google Scholar]
- 18.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 19.Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, et al. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci USA. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Q, Jones D, Springer TA. The chemokine receptor CXCR4 is required for the retention of B lineage and granulocytic precursors within the bone marrow microenvironment. Immunity. 1999;10:463–471. doi: 10.1016/s1074-7613(00)80046-1. [DOI] [PubMed] [Google Scholar]
- 21.Knaut H, Werz C, Geisler R, Nüsslein-Volhard C. Tübingen 2000 Screen Consortium, A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature. 2003;421:279–282. doi: 10.1038/nature01338. [DOI] [PubMed] [Google Scholar]
- 22.Doitsidou M, Reichman-Fried M, Stebler J, Köprunner M, Dörries J, Meyer D, et al. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647–659. doi: 10.1016/s0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- 23.Molyneaux KA, Zinszner H, Kunwar PS, Schaible K, Stebler J, Sunshine MJ, et al. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development. 2003;130:4279–4286. doi: 10.1242/dev.00640. [DOI] [PubMed] [Google Scholar]
- 24.David NB, Sapède D, Saint-Etienne L, Thisse C, Thisse B, Dambly-Chaudière C, et al. Molecular basis of cell migration in the fish lateral line: role of the chemokine receptor CXCR4 and of its ligand, SDF1. Proc Natl Acad Sci USA. 2002;99:16297–16302. doi: 10.1073/pnas.252339399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bussmann J, Wolfe SA, Siekmann AF. Arterial-venous network formation during brain vascularization involves hemodynamic regulation of chemokine signaling. Development. 2011;138:1717–1726. doi: 10.1242/dev.059881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siekmann AF, Standley C, Fogarty KE, Wolfe SA, Lawson ND. Chemokine signaling guides regional patterning of the first embryonic artery. Genes Dev. 2009;23:2272–2277. doi: 10.1101/gad.1813509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu M, Grove EA, Miller RJ. Abnormal development of the hippocampal dentate gyrus in mice lacking the CXCR4 chemokine receptor. Proc Natl Acad Sci USA. 2002;99:7090–7095. doi: 10.1073/pnas.092013799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bagri A, Gurney T, He X, Zou Y-R, Littman DR, Tessier-Lavigne M, et al. The chemokine SDF1 regulates migration of dentate granule cells. Development. 2002;129:4249–4260. doi: 10.1242/dev.129.18.4249. [DOI] [PubMed] [Google Scholar]
- 29.Knaut H, Blader P, Strähle U, Schier AF. Assembly of trigeminal sensory ganglia by chemokine signaling. Neuron. 2005;47:653–666. doi: 10.1016/j.neuron.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Belmadani A, Tran PB, Ren D, Assimacopoulos S, Grove EA, Miller RJ. The chemokine stromal cell-derived factor-1 regulates the migration of sensory neuron progenitors. J Neurosci. 2005;25:3995–4003. doi: 10.1523/JNEUROSCI.4631-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarting GA, Henion TR, Nugent JD, Caplan B, Tobet S. Stromal cell-derived factor-1 (chemokine C-X-C motif ligand 12) and chemokine C-X-C motif receptor 4 are required for migration of gonadotropin-releasing hormone neurons to the forebrain. J Neurosci. 2006;26:6834–6840. doi: 10.1523/JNEUROSCI.1728-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyasaka N, Knaut H, Yoshihara Y. Cxcl12/Cxcr4 chemokine signaling is required for placode assembly and sensory axon pathfinding in the zebrafish olfactory system. Development. 2007;134:2459–2468. doi: 10.1242/dev.001958. [DOI] [PubMed] [Google Scholar]
- 33.Tiveron M-C, Rossel M, Moepps B, Zhang YL, Seidenfaden R, Favor J, et al. Molecular interaction between projection neuron precursors and invading interneurons via stromal-derived factor 1 (CXCL12)/CXCR4 signaling in the cortical subventricular zone/intermediate zone. J Neurosci. 2006;26:13273–13278. doi: 10.1523/JNEUROSCI.4162-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.López-Bendito G, Sánchez-Alcañiz JA, Pla R, Borrell V, Picó E, Valdeolmillos M, et al. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J Neurosci. 2008;28:1613–1624. doi: 10.1523/JNEUROSCI.4651-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G, Adesnik H, Li J, Long J, Nicoll RA, Rubenstein JLR, et al. Regional distribution of cortical interneurons and development of inhibitory tone are regulated by Cxcl12/Cxcr4 signaling. J Neurosci. 2008;28:1085–1098. doi: 10.1523/JNEUROSCI.4602-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stumm RK, Zhou C, Ara T, Lazarini F, Dubois-Dalcq M, Nagasawa T, et al. CXCR4 regulates interneuron migration in the developing neocortex. Journal of Neuroscience. 2003;23:5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paredes MF, Li G, Berger O, Baraban SC, Pleasure SJ. Stromal-derived factor-1 (CXCL12) regulates laminar position of Cajal-Retzius cells in normal and dysplastic brains. Journal of Neuroscience. 2006;26:9404–9412. doi: 10.1523/JNEUROSCI.2575-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borrell V, Marín O. Meninges control tangential migration of hem-derived Cajal-Retzius cells via CXCL12/CXCR4 signaling. Nat Neurosci. 2006;9:1284–1293. doi: 10.1038/nn1764. [DOI] [PubMed] [Google Scholar]
- 39.Klein RS, Rubin JB, Gibson HD, DeHaan EN, Alvarez-Hernandez X, Segal RA, et al. SDF-1 alpha induces chemotaxis and enhances Sonic hedgehog-induced proliferation of cerebellar granule cells. Development. 2001;128:1971–1981. doi: 10.1242/dev.128.11.1971. [DOI] [PubMed] [Google Scholar]
- 40.Reiss K, Mentlein R, Sievers J, Hartmann D. Stromal cell-derived factor 1 is secreted by meningeal cells and acts as chemotactic factor on neuronal stem cells of the cerebellar external granular layer. Neuroscience. 2002;115:295–305. doi: 10.1016/s0306-4522(02)00307-x. [DOI] [PubMed] [Google Scholar]
- 41.Zhu Y, Yu T, Zhang X-C, Nagasawa T, Wu JY, Rao Y. Role of the chemokine SDF-1 as the meningeal attractant for embryonic cerebellar neurons. Nat Neurosci. 2002;5:719–720. doi: 10.1038/nn881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizoguchi T, Verkade H, Heath JK, Kuroiwa A, Kikuchi Y. Sdf1/Cxcr4 signaling controls the dorsal migration of endodermal cells during zebrafish gastrulation. Development. 2008;135:2521–2529. doi: 10.1242/dev.020107. [DOI] [PubMed] [Google Scholar]
- 43.Nair S, Schilling TF. Chemokine signaling controls endodermal migration during zebrafish gastrulation. Science. 2008;322:89–92. doi: 10.1126/science.1160038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson BE, Lehmann R. Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat Rev Mol Cell Biol. 2010;11:37–49. doi: 10.1038/nrm2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stebler J, Spieler D, Slanchev K, Molyneaux KA, Richter U, Cojocaru V, et al. Primordial germ cell migration in the chick and mouse embryo: the role of the chemokine SDF-1/CXCL12. Dev Biol. 2004;272:351–361. doi: 10.1016/j.ydbio.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 46.Ara T, Nakamura Y, Egawa T, Sugiyama T, Abe K, Kishimoto T, et al. Impaired colonization of the gonads by primordial germ cells in mice lacking a chemokine, stromal cell-derived factor-1 (SDF-1) Proc Natl Acad Sci USA. 2003;100:5319–5323. doi: 10.1073/pnas.0730719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chong SW, Emelyanov A, Gong Z, Korzh V. Expression pattern of two zebrafish genes, cxcr4a and cxcr4b. Mech Dev. 2001;109:347–354. doi: 10.1016/s0925-4773(01)00520-2. [DOI] [PubMed] [Google Scholar]
- 48.Weidinger G, Wolke U, Köprunner M, Klinger M, Raz E. Identification of tissues and patterning events required for distinct steps in early migration of zebrafish primordial germ cells. Development. 1999;126:5295–5307. doi: 10.1242/dev.126.23.5295. [DOI] [PubMed] [Google Scholar]
- 49.Blaser H. Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J Cell Sci. 2005;118:4027–4038. doi: 10.1242/jcs.02522. [DOI] [PubMed] [Google Scholar]
- 50.Raz E, Mahabaleshwar H. Chemokine signaling in embryonic cell migration: a fisheye view. Development. 2009;136:1223–1229. doi: 10.1242/dev.022418. [DOI] [PubMed] [Google Scholar]
- 51.Knaut H, Schier AF. Clearing the path for germ cells. Cell. 2008;132:337–339. doi: 10.1016/j.cell.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 52.Schier AF. Chemokine signaling: rules of attraction. Curr Biol. 2003;13:R192–R194. doi: 10.1016/s0960-9822(03)00122-2. [DOI] [PubMed] [Google Scholar]
- 53.Staton AA, Knaut H, Giraldez AJ. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat Genet. 2011;43:204–211. doi: 10.1038/ng.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balabanian K, Lagane B, Infantino S, Chow KYC, Harriague J, Moepps B, et al. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005;280:35760–35766. doi: 10.1074/jbc.M508234200. [DOI] [PubMed] [Google Scholar]
- 55.Burns JM, Summers BC, Wang Y, Melikian A, Berahovich R, Miao Z, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boldajipour B, Mahabaleshwar H, Kardash E, Reichmanfried M, Blaser H, Minina S, et al. Control of Chemokine-Guided Cell Migration by Ligand Sequestration. Cell. 2008;132:463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 57.Naumann U, Cameroni E, Pruenster M, Mahabaleshwar H, Raz E, Zerwes H-G, et al. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS ONE. 2010;5:e9175. doi: 10.1371/journal.pone.0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valentin G, Haas P, Gilmour D. The Chemokine SDF1a Coordinates Tissue Migration through the Spatially Restricted Activation of Cxcr7 and Cxcr4b. Current Biology. 2007;17:1026–1031. doi: 10.1016/j.cub.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, Shiozawa Y, Wang J, Wang Y, Jung Y, Pienta KJ, et al. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J Biol Chem. 2008;283:4283–4294. doi: 10.1074/jbc.M707465200. [DOI] [PubMed] [Google Scholar]
- 60.Odemis V, Boosmann K, Heinen A, Kury P, Engele J. CXCR7 is an active component of SDF-1 signalling in astrocytes and Schwann cells. J Cell Sci. 2010:1–8. doi: 10.1242/jcs.062810. [DOI] [PubMed] [Google Scholar]
- 61.Cossart R. The maturation of cortical interneuron diversity: how multiple developmental journeys shape the emergence of proper network function. Current Opinion in Neurobiology. 2011;21:160–168. doi: 10.1016/j.conb.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 62.Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. Journal of Neuroscience. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gelman DM, Martini FJ, Nobrega-Pereira S, Pierani A, Kessaris N, Marin O. The Embryonic Preoptic Area Is a Novel Source of Cortical GABAergic Interneurons. Journal of Neuroscience. 2009;29:9380–9389. doi: 10.1523/JNEUROSCI.0604-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bielle F, Griveau A, Narboux-Nême N, Vigneau S, Sigrist M, Arber S, et al. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat Neurosci. 2005;8:1002–1012. doi: 10.1038/nn1511. [DOI] [PubMed] [Google Scholar]
- 65.Polleux F, Whitford KL, Dijkhuizen PA, Vitalis T, Ghosh A. Control of cortical interneuron migration by neurotrophins and PI3-kinase signaling. Development. 2002;129:3147–3160. doi: 10.1242/dev.129.13.3147. [DOI] [PubMed] [Google Scholar]
- 66.Yokota Y, Gashghaei HT, Han C, Watson H, Campbell KJ, Anton ES. Radial glial dependent and independent dynamics of interneuronal migration in the developing cerebral cortex. PLoS ONE. 2007;2:e794. doi: 10.1371/journal.pone.0000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ang ESBC, Haydar TF, Gluncic V, Rakic P. Four-dimensional migratory coordinates of GABAergic interneurons in the developing mouse cortex. Journal of Neuroscience. 2003;23:5805–5815. doi: 10.1523/JNEUROSCI.23-13-05805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marín O, Rubenstein JLR. Cell migration in the forebrain. Annu. Rev. Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 69.Tanaka DH, Maekawa K, Yanagawa Y, Obata K, Murakami F. Multidirectional and multizonal tangential migration of GABAergic interneurons in the developing cerebral cortex. Development. 2006;133:2167–2176. doi: 10.1242/dev.02382. [DOI] [PubMed] [Google Scholar]
- 70.Nadarajah B, Alifragis P, Wong ROL, Parnavelas JG. Neuronal migration in the developing cerebral cortex: observations based on real-time imaging. Cereb. Cortex. 2003;13:607–611. doi: 10.1093/cercor/13.6.607. [DOI] [PubMed] [Google Scholar]
- 71.Tanaka D, Nakaya Y, Yanagawa Y, Obata K, Murakami F. Multimodal tangential migration of neocortical GABAergic neurons independent of GPI-anchored proteins. Development. 2003;130:5803–5813. doi: 10.1242/dev.00825. [DOI] [PubMed] [Google Scholar]
- 72.Stumm RK, Zhou C, Ara T, Lazarini F, Dubois-Dalcq M, Nagasawa T, et al. CXCR4 regulates interneuron migration in the developing neocortex. Journal of Neuroscience. 2003;23:5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, Li G, Stanco A, Long JE, Crawford D, Potter GB, et al. CXCR4 and CXCR7 Have Distinct Functions in Regulating Interneuron Migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sánchez-Alcañiz JA, Haege S, Mueller W, Pla R, Mackay F, Schulz S, et al. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69:77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 75.Daniel D, Rossel M, Seki T, König N. Stromal cell-derived factor-1 (SDF-1) expression in embryonic mouse cerebral cortex starts in the intermediate zone close to the pallial-subpallial boundary and extends progressively towards the cortical hem. Gene Expr Patterns. 2005;5:317–322. doi: 10.1016/j.modgep.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 76.Stumm R, Kolodziej A, Schulz S, Kohtz JD, Höllt V. Patterns of SDF-1alpha and SDF-1gamma mRNAs, migration pathways, and phenotypes of CXCR4-expressing neurons in the developing rat telencephalon. J Comp Neurol. 2007;502:382–399. doi: 10.1002/cne.21336. [DOI] [PubMed] [Google Scholar]
- 77.Schönemeier B, Kolodziej A, Schulz S, Jacobs S, Hoellt V, Stumm R. Regional and cellular localization of the CXCl12/SDF-1 chemokine receptor CXCR7 in the developing and adult rat brain. J Comp Neurol. 2008;510:207–220. doi: 10.1002/cne.21780. [DOI] [PubMed] [Google Scholar]
- 78.Tiveron M-C, Boutin C, Daou P, Moepps B, Cremer H. Expression and function of CXCR7 in the mouse forebrain. Journal of Neuroimmunology. 2010;224:72–79. [PubMed] [Google Scholar]
- 79.Medina L, Abellán A. Development and evolution of the pallium. Semin Cell Dev Biol. 2009;20:698–711. doi: 10.1016/j.semcdb.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 80.Soriano E, Del Río JA. The cells of cajal-retzius: still a mystery one century after. Neuron. 2005;46:389–394. doi: 10.1016/j.neuron.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 81.Hevner RF, Neogi T, Englund C, Daza RAM, Fink A. Cajal-Retzius cells in the mouse: transcription factors, neurotransmitters, and birthdays suggest a pallial origin. Brain Res Dev Brain Res. 2003;141:39–53. doi: 10.1016/s0165-3806(02)00641-7. [DOI] [PubMed] [Google Scholar]
- 82.Shinozaki K, Miyagi T, Yoshida M, Miyata T, Ogawa M, Aizawa S, et al. Absence of Cajal-Retzius cells and subplate neurons associated with defects of tangential cell migration from ganglionic eminence in Emx1/2 double mutant cerebral cortex. Development. 2002;129:3479–3492. doi: 10.1242/dev.129.14.3479. [DOI] [PubMed] [Google Scholar]
- 83.Yoshida M, Assimacopoulos S, Jones KR, Grove EA. Massive loss of Cajal-Retzius cells does not disrupt neocortical layer order. Development. 2006;133:537–545. doi: 10.1242/dev.02209. [DOI] [PubMed] [Google Scholar]
- 84.Guo H, Hong S, Jin XL, Chen RS, Avasthi PP, Tu YT, et al. Specificity and efficiency of Cre-mediated recombination in Emx1-Cre knock-in mice. Biochemical and Biophysical Research Communications. 2000;273:661–665. doi: 10.1006/bbrc.2000.2870. [DOI] [PubMed] [Google Scholar]
- 85.Rajagopal S, Kim J, Ahn S, Craig S, Lam CM, Gerard NP, et al. -arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proceedings of the National Academy of Sciences. 2009:1–5. doi: 10.1073/pnas.0912852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hartmann TN, Grabovsky V, Pasvolsky R, Shulman Z, Buss EC, Spiegel A, et al. A crosstalk between intracellular CXCR7 and CXCR4 involved in rapid CXCL12-triggered integrin activation but not in chemokine-triggered motility of human T lymphocytes and CD34+ cells. Journal of Leukocyte Biology. 2008;84:1130–1140. doi: 10.1189/jlb.0208088. [DOI] [PubMed] [Google Scholar]
- 87.Sierro F, Biben C, Martínez-Muñoz L, Mellado M, Ransohoff RM, Li M, et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci USA. 2007;104:14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]