SUMMARY
Pseudomonas aeruginosa, a Gram-negative bacterium, is a significant opportunistic pathogen associated with skin and soft tissue infections, nosocomial pneumonia, and sepsis. In addition, it can chronically colonize the lungs of cystic fibrosis (CF) patients. Overproduction of the exopolysaccharide called alginate provides P. aeruginosa with a selective advantage and facilitates survival in the CF lung. The in vitro phenotype of alginate overproduction observed on solid culture media is referred to as mucoid. Expression of the alginate machinery and biosynthetic enzymes are controlled by the extracytoplasmic sigma factor, σ22 (AlgU/T). The key negative regulator of both σ22 activity and the mucoid phenotype is the cognate anti-sigma factor MucA. MucA sequesters σ22 to the inner membrane inhibiting the sigma factor’s transcriptional activity. The well-studied mechanism for transition to the mucoid phenotype is mutation of mucA, leading to loss of MucA function and therefore activation of σ22. Recently, regulated intramembrane proteolysis (RIP) has been recognized as a mechanism whereby proteolysis of the anti-sigma factor MucA leads to active σ22 allowing P. aeruginosa to respond to environmental stress conditions by overproduction of alginate. The goal of this review is to illuminate the pathways leading to RIP that have been identified and proposed.
Keywords: RIP, alginate, σ22, AlgU/T, MucA, AlgW, MucP, Prc, ClpXP, Opr86, KinB
Introduction
The genetic disease cystic fibrosis (CF) affects multiple organ systems of the human body with one of the consequences being increased susceptibility to respiratory infections by opportunistic pathogens (Lyczak et al., 2002). CF is caused by mutation of the CF transmembrane conductance regulator (CFTR) gene (Kerem et al., 1989, Rommens et al., 1989). Mutations in CFTR abrogate lung functions resulting in an impaired ability to eradicate inhaled microorganisms (Welsh et al., 2001). One consequence of CF is the formation of thick mucus in the lungs, creating an environment suitable for microbial growth. With the advances in modern pharmacology, many of the once dominant respiratory pathogens that historically afflicted those with CF can now be controlled, leading to increased life expectancy. Even with such advancements in antibiotic therapies, the Gram-negative bacterium Pseudomonas aeruginosa remains the most common CF pathogen (Govan & Deretic, 1996). Given its inherent antibiotic resistance and plethora of virulence factors, P. aeruginosa is a direct cause of most of the morbidity and mortality in those with CF. The onset of chronic P. aeruginosa infection of the CF respiratory tract is marked by the emergence of the mucoid phenotype. Mucoid P. aeruginosa overproduce and secrete an exopolysaccaride known as alginate (Fig. 1). Alginate forms a capsule that protects P. aeruginosa from various host defenses (Leid et al., 2005), antibiotics (Govan & Fyfe, 1978), and phagocytosis (Schwarzmann & Boring, 1971).
Figure 1. P. aeruginosa mucoid and nonmucoid phenotypes.

Representative strains of the wild-type nonmucoid and mucoid phenotypes are shown. Mucoid strains overproduce the exopolysaccharide known as alginate. Strains were grown on Pseudomonas isolation agar (PIA) for 24 hours at 37°C, and then 24 hours at 25°C.
Several mechanisms exist which lead to the induction of the mucoid phenotype, but most mechanisms converge upon a common pathway through the extracytoplasmic sigma factor, σ22 (AlgU or AlgT). σ22 has 65% similarity to its Escherichia coli homologue σE (Hershberger et al., 1995, Martin et al., 1993a), otherwise known as RpoE. In Table 1, the P. aeruginosa proteins that will be discussed throughout this review are presented, along with their E. coli homologues. P. aeruginosa σ22 directs RNA polymerase to activate expression of the alginate biosynthetic genes, which are encoded in the algD alginate biosynthetic operon at loci PA3540 to PA3551. The functions of the proteins encoded by the alginate biosynthetic genes are well characterized and have recently been expertly reviewed (Franklin et al., 2011, Rehm, 2009).
Table 1.
σ22 and proteins which play roles in regulated intramembrane proteolysis (RIP) of MucA in P. aeruginosa.
| PA Locus # | Protein(s) | Alt. names | Homology (E. coli) | Type of regulator | Localization | Function(s) a | Key Reference(s) b |
|---|---|---|---|---|---|---|---|
| PA0762 | σ22 | AlgU/T | RpoE | Alternative sigma factor | Cytoplasmic | Activates transcription | (DeVries & Ohman, 1994, Martin et al., 1993a) |
| PA0763 | MucA | RseA | Anti-sigma factor | Inner membrane | Sequesters and inhibit sσ22 | (Martin et al., 1993b, Mathee et al., 1997) | |
| PA0764 | MucB | AlgN | RseB | Periplasmic | Protects C-terminus of MucA from proteolysis | (Cezairliyan & Sauer, 2009, Goldberg et al., 1993, Wood & Ohman, 2009) | |
| PA0765 | MucC | AlgM | none | Unknown | Unknown | Unknown | (Boucher et al., 1997) |
| PA0766 | MucD | AlgY | DegP | Chaperone/Protease | Periplasmic | Degrades proteins that activate AlgW and or MucP | (Boucher et al., 1996, Damron & Yu, 2011, Qiu et al., 2007, Wood & Ohman, 2009) |
| PA4446 | AlgW | DegS | Protease | Periplasmc/Inner membrane | Cleaves C-terminus of MucA | (Boucher et al., 1996, Damron & Yu, 2011, Qiu et al., 2007, Wood & Ohman, 2009) | |
| PA3649 | MucP | YaeL | RseP | Protease | Inner membrane | Cleaves MucA near transmembrane domain | (Damron & Yu, 2011, Qiu et al., 2007, Wood & Ohman, 2009) |
| PA4033 | MucE | none | Envelope Protein | Periplasmic | Activates AlgW cleavage of MucA | (Cezairliyan & Sauer, 2009, Qiu et al., 2007) | |
| PA3257 | Prc | Prc | Periplasmic Protease | Periplasmic | Facilitates degradation of mutant MucA | (Reiling et al., 2005) | |
| PA1802 PA1801 |
ClpX/P | ClpX/P | Cytoplasmic Proteases | Cytoplasmic | Facilitates degradation of N-termius of MucA | (Qiu et al., 2008b) | |
| PA3648 | Opr86 | YaeT/BamA | Outer Membrane Biogenesis | Outer membrane | Depletion of Opr86 upregulates MucD | (Tashiro et al., 2009) | |
| PA5484 | KinB | PhoR/NtrB | Sensor Kinase | Inner membrane | Inactivation causes AlgW-cleavage of MucA | (Damron et al., 2009) | |
| PA5483 | AlgB | NtrC | Response Regulator | Cytoplasmic | Required for σ22 activity in kinB null mutant | (Damron et al., 2009) | |
| PA4462 | RpoN | σ54 | RpoN | Alternative Sigma Factor | Cytoplasmic | Required for σ22 activity in kinB null mutant | (Damron et al., 2009) |
Protein may have more functions than indicated. The indicated function is that most relevant to regulated proteolysis of MucA.
Additional references to the function of the proteins listed exist; here only key references relevant to the topic of this review are given.
The principal regulator of σ22 is the cognate anti-sigma factor MucA. σ22 and MucA are encoded in an operon (σ22-mucA-mucB-mucC-mucD) at loci PA0762-PA0766 (Table 1) on the PAO1 genome. MucA localizes to the inner membrane through a single transmembrane domain (Mathee et al., 1997) with the amino- and carboxyl-terminal regions in the cytoplasm and periplasm, respectively. Typically, MucA sequesters σ22 to the inner membrane (Mathee et al., 1997). When MucA is mutated, σ22 may not be sequestered (Martin et al., 1993b). In the absence of MucA repression, σ22 is active and directs transcription with RNA polymerase at σ22-dependent promoters throughout the genome (Firoved et al., 2002). Since alginate genes are regulated by σ22, this loss of MucA-repression results in the constitutive mucoid phenotype. Another negative regulator of σ22 was identified just downstream of mucA (Goldberg et al., 1993, Martin et al., 1993b) and is named mucB (Table 1). It was later shown that MucB is a periplasmic protein (Mathee et al., 1997) that binds MucA (Cezairliyan & Sauer, 2009, Mathee et al., 1997, Wood & Ohman, 2009). mucC is another putative regulatory gene (Table 1) (Boucher et al., 1997). A function of MucC has not been established, but one study has clearly defined the promoter of the downstream gene mucD, to be within the mucC gene (Wood & Ohman, 2006). mucD is the final gene of the σ22 locus which encodes another negative regulator of alginate overproduction. MucD is a periplasmic chaperone protease homologous to E. coli DegP (Table 1) (Boucher et al., 1996).
It is generally accepted the P. aeruginosa strains that initially infect CF patients are obtained directly from the environment, but some studies have documented patient-to-patient spread (Armstrong et al., 2003, Jones et al., 2002, McCallum et al., 2001) and sharing of strains among CF siblings (Renders et al., 1997). The P. aeruginosa strains that initially infect these patients are generally nonmucoid and sensitive to antibiotics (Burns et al., 2001, Doggett et al., 1966). Phenotypically nonmucoid P. aeruginosa strains, when cultured in vitro, produce low levels of alginate (Anastassiou et al., 1987, Pier et al., 1986). CF patients that only have nonmucoid strains infecting their lungs have antibody responses specific for alginate (Pedersen et al., 1990). In fact, the ability to produce alginate is key to establishing chronic colonization of CF mice (Coleman et al., 2003). When low levels of alginate are produced, but the mucoid phenotype is not evident, we will refer to this phenotype as “alginate production”. For clarity throughout this review, we will refer to the mucoid phenotype as “alginate overproduction”.
While the exact conditions to induce the mucoid phenotype are not completely understood, several key studies have recently been performed that show how cell wall stress (Wood et al., 2006) or overexpression of an envelope protein (Qiu et al., 2007) can activate proteolytic degradation of MucA by regulated intramembrane proteolysis (RIP). RIP is a mechanism that is conserved from bacteria to humans (Brown et al., 2000). In the model detailed in this review, proteases respond to environmental conditions and cleave MucA allowing σ22 to initiate transcription of its regulon. It has been proposed that the most opportune time to block chronic infection of P. aeruginosa in CF is before the conversion to constitutively mucoid phenotype (Ramsey & Wozniak, 2005). Building upon this idea, we propose that the RIP proteases responsible for the mucoid phenotype may be potential drug targets for eradication of P. aeruginosa from the lungs of CF patients. The goal of this review is to summarize the key literature that formed the basis for the model of MucA RIP, identify remaining questions, and propose future directions for the field.
Membrane stress leads to expression of the σ22 regulon
Wood et al. hypothesized that the σ22-MucA signal transduction pathways would respond to environmental stresses leading to expression of the σ22 regulon (Wood et al., 2006). To test this hypothesis they utilized a reporter construct to identify stress agents that could induce σ22 activity. Many classes of stress compounds (50 total) were screened for their ability to activate a σ22-dependent promoter fusion of PalgD with a chloramphenicol resistance gene (PalgD-cat). Of the ten compounds that activated PalgD, most were inhibitors of peptidoglycan synthesis. In particular D-cycloserine induced very PalgD high expression (Wood et al., 2006), suggesting that inhibiting peptidoglycan synthesis caused membrane stress and affected the integrity of the cell. These authors also showed that the σ22 gene was required for PalgD activity in the presence of D-cycloserine, suggesting that D-cycloserine induced loss of repression of σ22 by MucA. Transcriptional profiling was performed to determine the genes dysregulated during growth in the presence of D-cycloserine. In addition to the established σ22 regulon (Firoved et al., 2002, Firoved & Deretic, 2003, Firoved et al., 2004a, Firoved et al., 2004b, Tart et al., 2005), D-cycloserine also affected expression of hypothetical, efflux, pyoverdine, peptidoglycan, LPS, and intermediary metabolism genes (Wood et al., 2006).
Activation of AlgW protease leads to degradation of MucA
Activation of σ22 by cell wall inhibitors was a pivotal observation that confirmed the hypothesis that alginate production was a stress response that could be turned on by environmental conditions. Based on this finding, Wood et al. (2006) hypothesized that proteolysis of MucA was a possible mechanism. Genes encoding two envelope proteases, MucD and AlgW were previously implicated as potential regulators of alginate biosynthesis (Table 1) (Boucher et al., 1996). However, both of these proteases were considered negative regulators depending on the strain background. In context of nonmucoid PAO1 background, when the mucD gene was inactivated, alginate was overproduced, but inactivation of algW, did not cause the mucoid phenotype (Boucher et al., 1996). To test the possibility that AlgW could be a positive regulator of alginate overproduction, an algW mutant was subjected to D-cycloserine treatment and, as expected, failed to induce the PalgD activity (Wood et al., 2006). This suggested that without algW, σ22 was not activated and that AlgW was likely a protease of MucA and a positive regulator of σ22 activity (Fig. 2). Furthermore, the overexpression of algW caused alginate overproduction (Wood et al., 2006). From these data, it was concluded AlgW was the functional homologue (42% identical) of E. coli DegS. Figure 3 indicates the protein domains of the various regulatory proteases of P. aeruginosa including AlgW. E. coli DegS acts on the anti-sigma factor RseA (MucA homolog) in response to misfolded or accumulated proteins (such as OmpC) in the periplasm (Ades, 2008). The mechanism of DegS proteolysis of RseA allows stress conditions to activate σE-dependent gene expression in E. coli. Due to the homology of AlgW-DegS and the data that AlgW was required for PalgD expression, Wood et al. proposed a model where membrane stress agents activate AlgW and release σ22 (Wood et al., 2006).
Figure 2. P. aeruginosa wild-type and mutant MucA and associated protease complexes.
Proteolytic activities of proteins are indicated by scissors. A. Full length MucA protein sequestering σ22 is shown with MucB binding the C-termius of MucA. AlgW is indicated as a trimer as previously demonstrated (Cezairliyan & Sauer, 2009). The relative positions where AlgW cleaves are indicated with the major cleavage site. MucP is shown localized to the inner membrane. PDZ domains of each protease are indicated in red and it should be noted that all RIP proteases identified thus far harbor one or two of these domains. Cytoplasmic ClpXP cleaves residual MucA from σ22 in the final step of activation of the σ22 (Qiu et al., 2008b). B. Mutant MucA22 is shown localized to the inner membrane. Prc is a protease that facilitates degradation of mutant MucA proteins. In this review, it is proposed that MucP may play a role in degradation of mutant MucA; however this has not been established. RIP of MucA leads to activation of σ22 and expression of the σ22 regulon. RIP of MucA is accomplished by the proteases AlgW, MucP, ClpXP, and Prc.
Figure 3. Domains of the proteases involved in regulation of alginate overproduction.
The known proteases which are involved in regulating σ22 are shown with their respective domains as indicated by the SMART protein database (Letunic et al., 2009). PDZ domains (red octagons) are protein-protein interaction domains and are found in all of the alginate regulatory envelope proteases. Envelope proteases AlgW and MucD both also have trypsin protease domains (black rectangle). TSPc (purple octagon) indicates the tail-specific protease domain on Prc. Prc also has a C-terminal domain of unknown function, which has yet to be characterized known as DUF3340. The ClpX protein has an AAA (ATPase Associated Activities) domain (gray oval). ClpX and ClpP are both required for the activation of σ22 (Qiu et al., 2008b) which suggests they likely work in concert. ClpX also has a ClpB domain that plays a role in protein recognition and is similar to a PDZ domain. Red blocks in the N-terminus indicate a signal sequence has been identified.
It seemed likely that misfolded proteins in the periplasm may activate degradation of MucA by a mechanism similar to that of E. coli DegS. Since DegS must be activated to degrade RseA, there are likely AlgW activators encoded in the P. aeruginosa genome. In a study by Qiu et al., transposon mutagenesis of nonmucoid strain PAO1 was used to identify new alginate regulatory genes. A mucoid mutant was found with a transposon insertion into the promoter region of the gene encoded at PA4033 (Qiu et al., 2007). Due to the orientation of the transposon and the lack of a transcriptional terminator in the gentamicin resistance gene (aacC1) within the transposon, it was hypothesized that overexpression of PA4033 caused alginate overproduction. To test this, PA4033 was overexpressed in trans from an arabinose inducible PBAD promoter and alginate overproduction was observed (Qiu et al., 2007). Bioinformatics suggested that PA4033 encodes an envelope protein and is now referred to as MucE (Table 1) (Qiu et al., 2007). MucE has a C-terminus that is similar to proteins that activate DegS proteolysis of RseA in E. coli. In P. aeruginosa, when MucE is overexpressed AlgW cleaves MucA, which results in alginate overproduction (Qiu et al., 2007) (Fig. 4). The importance of the C-terminus region of MucE in activating AlgW was investigated by mutating the three C-terminal amino acids (tryptophan, valine, phenylalanine, WVF) of MucE; these studies showed that the WVF motif was required to activate AlgW (Qiu et al., 2007). Furthermore, it was also shown that other triplet C-terminal (YVF, LVF, WIF, and WVW) signatures can also activate AlgW (Qiu et al., 2007). Numerous AlgW-activating sequences are encoded in the P. aeruginosa genome but not all outer membrane or envelope proteins contain these sequences (Qiu et al., 2007). For example, the major outer membrane porin, OprF, lacks an AlgW activating sequence. It has been demonstrated that overexpression of OprF does not cause alginate overproduction, but addition of the AlgW activation signal (WVF) to the C-terminus of OprF caused alginate overproduction (Qiu et al., 2008a).
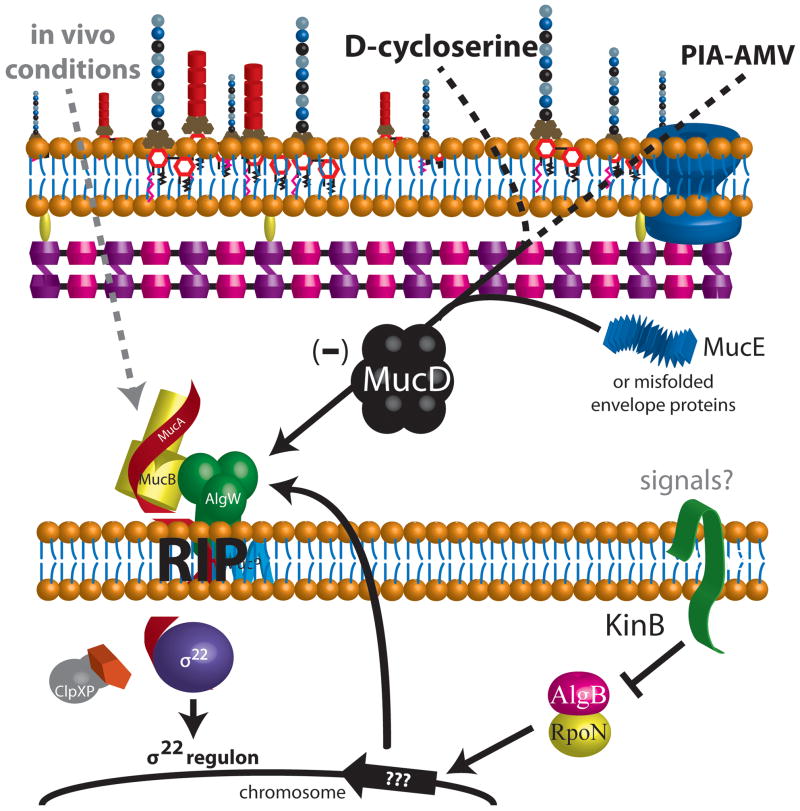
Figure 4. Pathways leading to regulated intramembrane proteolysis (RIP) of MucA.
A composite model of the various pathways in P. aeruginosa, that lead to RIP of MucA and activation of σ22 are shown. Cell wall stress agents such as D-cycloserine can inhibit peptidoglycan synthesis and activate RIP of MucA (Wood et al., 2006, Wood & Ohman, 2009). When the envelope protein MucE is overexpressed, RIP of MucA occurs due to activation of AlgW and MucP proteases (Qiu et al., 2007). Growth on PIA-AMV medium causes RIP of MucA by AlgW and MucP protease, presumably due to misfolded proteins in the envelope (Damron et al., 2011). MucD is a chaperone-protease that is in the periplasm. The protease activity of MucD is required for repression of alginate overproduction. Likely, MucD degrades proteins that accumulate in the periplasm. If the function of MucD is lost then RIP of MucA will occur. MucD overexpression can suppress the MucE signal (Qiu et al., 2007) and is upregulated during growth on PIA-AMV (Damron et al., 2011). However, in the absence of MucD, RIP of MucA and activation of σ22 only requires MucP and not AlgW(Damron & Yu, 2011). When the histidine kinase KinB is inactivated or deleted, AlgW-RIP of MucA occurs and is dependent upon response regulator AlgB and sigma factor RpoN (σ54) (Damron et al., 2009). It is hypothesized that AlgB/RpoN controls expression of genes that influence RIP. It is also not known what signals activate KinB. In vivo conditions are indicated in gray dashed line since a study does indicate alginate production can occur during infection (Bragonzi et al., 2005), but the specifics of this pathway have not been elucidated. Based on the convergence to RIP by multiple pathways, future therapeutics inhibiting the RIP proteases may provide novel treatment options against P. aeruginosa.
Genetic evidence suggested a proteolysis-driven model with AlgW degrading MucA and thus releasing and activating σ22. This model of AlgW proteolysis was confirmed in a separate study by in vitro biochemical analysis using MucE as the activator of AlgW (Cezairliyan & Sauer, 2009). In this study, a major cleavage site as well as three minor cleavage sites were identified in the C-terminus of MucA (Cezairliyan & Sauer, 2009) and are depicted in Figure 2A. The domain structure of AlgW is similar to DegS with a protease domain and one PDZ domain (Fig. 3). PDZ is an acronym derived from the three proteins in which the domain was first identified (Kennedy, 1995). The PDZ domain of E. coli DegS keeps the protease inactive until misfolded proteins bind, inducing a conformational change resulting in proteolytic activity (Hasselblatt et al., 2007, Walsh et al., 2003). Deletion of the PDZ domain of E. coli DegS causes constitutive protease activity (Walsh et al., 2003). However, deletion of the PDZ domain of AlgW renders the protease active but less efficient than wild-type AlgW (Cezairliyan & Sauer, 2009). This data confirmed previous in vivo studies that had shown that the PDZ domain of AlgW was required for proteolysis of MucA (Damron et al., 2009, Qiu et al., 2007). Cezairliyan and Sauer also observed that the “LA loop” of AlgW inhibits MucA binding. LA loops of DegS-like serine proteases have been shown to play roles in hindering the active site (Cezairliyan & Sauer, 2009). Without the LA loop, AlgW could cleave MucA in the absence of MucE, but inefficiently. These experiments show that AlgW is regulated through protein-protein interactions and that the LA loop and the PDZ domain participate in controlling proteolytic activity. In addition to characterizing how AlgW is activated and cleaves MucA, Cezairliyan and Sauer also showed in vitro that MucB binds the C-terminus of MucA, which protects MucA from proteolysis by AlgW (Cezairliyan & Sauer, 2009).
Inner membrane protease MucP acts on MucA
Inspection of the locations that AlgW acts on MucA indicates that after AlgW proteolysis (Cezairliyan & Sauer, 2009) about half of MucA would still remain (Fig. 2A). In E. coli, the MucA homolog RseA requires further degradation by an inner membrane protease named RseP (YaeL) (Alba et al., 2002), which cleaves the transmembrane domain of RseA (Akiyama et al., 2004). In P. aeruginosa PAO1 the homologue of E. coli RseP is PA3649 and referred to as MucP or YaeL (Table 1). Both algW and mucP are required for the mucoid phenotype in a strain overexpressing mucE (Qiu et al., 2007). In later studies, it was established that MucP plays a role in degradation of MucA and activation of σ22 when cells are cultured in the presence of D-cycloserine (Wood & Ohman, 2009). Furthermore, critical residues in the predicted protease domain of MucP are required for activation of σ22 (Damron & Yu, 2011). Unlike AlgW, MucP has two putative PDZ domains presumably for substrate recognition (Fig. 3). After proteolysis of MucA at the major AlgW cleavage site, the remainder of the protein would likely be membrane anchored. Based on this observation, it could be hypothesized that MucP may be required for degrading mutant MucA proteins; however, this has not been investigated.
Chaperone-protease MucD regulates pathways leading to MucA proteolysis
AlgW and MucP are proteases that are positive regulators of alginate production due to their direct action upon MucA. MucD is the only protease identified in P. aeruginosa that is a negative regulator of alginate overproduction (Boucher et al., 1996). Based on its similarity to E. coli DegP and the presence of a leader peptide, MucD is likely localized to the periplasm (Boucher et al., 1996). MucD is encoded in the σ22 regulator operon but unlike the situation in E. coli (Hiratsu et al., 1995), mucD is expressed from both σ22-dependent and -independent promoters (Wood & Ohman, 2006). Inactivation of mucD causes alginate overproduction (Boucher et al., 1996). MucD is an HtrA-family protein and is similar to AlgW because they are both serine proteases (Boucher et al., 1996). The protease domain of MucD is required for its ability to regulate alginate production (Wood & Ohman, 2006, Yorgey et al., 2001). In addition to the protease domain, MucD also has two PDZ domains (Fig. 3). Since the protease domain is required for suppression of alginate overproduction, it is conceivable that MucD may intercept misfolded or accumulated proteins in the periplasm that could activate AlgW. Overexpression of MucD suppresses the MucE-induced mucoid phenotype, which suggests MucD may act on accumulated proteins in the periplasm (Qiu et al., 2007). E. coli DegP recognizes misfolded substrates causing DegP monomers to assemble into a functional oligomer and then the oligomer degrades periplasmic proteins lacking appropriate conformation (Krojer et al., 2008a, Krojer et al., 2008b). It is possible the PDZ domains in MucD are responsible for substrate recognition as they are in E. coli (Ortega et al., 2009). The presumptive chaperone function of MucD (i.e., when it lacks its protease activity) can decrease alginate overproduction (Yorgey et al., 2001). This suggests MucD may not only degrade but may also chaperone misfolded proteins that leads to a slower rate of RIP of MucA and therefore decreased alginate overproduction. It could be suggested that while the direct molecular function of DegP/MucD proteases may be destructive, the goal is to maintain balance, homeostasis, and overall integrity of the cell in stress conditions.
In E. coli, sequential cleavages, first by DegS and then by RseP, are required to degrade RseA and activate σ22 (Ades, 2008). Adapting this model to P. aeruginosa, it would be predicted that AlgW would always be required for activation of σ22. In the absence of MucD, AlgW would likely cleave MucA. However, a mucD/algW double mutant is mucoid on standard laboratory media such as L-agar (unpublished observations) or Pseudomonas isolation agar (PIA) (Damron & Yu, 2011), suggesting that MucA proteolysis can occur even in the absence of AlgW. When mucD and mucP proteases are inactivated, alginate overproduction does not occur and further investigation revealed that MucA degradation is diminished by the inactivation of mucP (Damron & Yu, 2011). These data suggest that while AlgW-MucP sequential digestion of MucA is the normal mechanism, a second pathway, where only MucP proteolysis is required, exists. This discrepancy begs the question: how is MucP activated independent of AlgW cleavage of MucA? Acid stress activates σE in Salmonella enterica serovar Typhimurium (Muller et al., 2009) independent of DegS but dependent upon RseP. In Salmonella, it seems the PDZ domain of RseP is dispensable for induction. From these data, Muller et al. proposed that disruption of the interaction between the RseP PDZ domain and RseA may be a novel signal to activate σ22 (Muller et al., 2009). It would be interesting to see if acid stress can activate RIP of MucA in P. aeruginosa. One study, which was performed prior to the identification of the mechanism of MucA-RIP, does indicate regulated intramembrane proteolysis independent of AlgW; a PAO1 strain lacking algW became mucoid in the presence of paraquat (Boucher et al., 1996). Paraquat is a redox cycling compound that causes superoxide production, which can be detrimental to bacteria. This data suggests that paraquat may activate MucP and that RIP of MucA can occur without AlgW.
ClpXP: positive regulator of σ22 and cytoplasmic protease of MucA
Regulated proteolysis of wild-type MucA begins with degradation by AlgW or MucP, as described above. Membrane stress and overexpression of envelope proteins are at least two mechanisms that can activate this cascade. Presumably, proteolysis of MucA at the transmembrane region (Fig. 2) would leave approximately half of the MucA protein still attached to σ22. This suggests that another protease may participate in the final degradation of MucA from σ22. The muc-25 mutation (Fyfe & Govan, 1983) truncates MucA to 94 amino acids (Qiu et al., 2008b). Due to this short length, it is likely that MucA25 is not localized to the inner membrane, since it lacks a complete transmembrane domain. The MucA25 protein has been used to address the question as to what other proteases in the cytoplasm may be responsible for degradation of MucA and activation of σ22. Through transposon mutagenesis, Qiu et al. identified three proteases, clpP, clpX, and a clpP paralogue that were required for alginate overproduction in the mucA25 strain (Table 1) (Qiu et al., 2008b). ClpXP is a molecular machine that binds unstructured peptides, unfolds, and then degrades the protein (Baker & Sauer, 2011). Since MucA25 is stabilized in absence of the clp genes, it seems that cytoplasmic protease complex ClpXP (Fig. 2) is responsible for removal of residual MucA leading to active σ22 (Qiu et al., 2008b). ClpX contains two PDZ-like domains (Levchenko et al., 1997), which likely recognize unfolded or truncated MucA that is attached to σ22 and then facilitates degradation of MucA by ClpP. The two PDZ-like domains of ClpX are shown in Figure 3 as a single ClpB domain. While ClpXP protease was originally identified for its requirement in a mucA mutant, ClpX is also required for the mucoid phenotype in strains with wild-type mucA (unpublished observations).
Prc: an envelope protease that acts on mutant MucA proteins
MucA22 is a common mutant MucA protein that arises in both CF isolates and laboratory strains (Mathee et al., 1999). The mucA22 mutation is due to loss of a guanine at base 430 resulting in a premature stop codon, truncating the resultant protein to 146 amino acids (Figure 2B). The location of the stop codon of MucA22 suggests that the C-terminus would reside in the periplasm (Figure 2B) and it was also shown that MucA22 remains sequestered σ22 to the inner membrane (Rowen & Deretic, 2000). Thus to release σ22 from a truncated MucA such as MucA22, proteolysis is a plausible mechanism. Using a mucoid mucA22 mutant strain (PAO578), suppressor of mucoidy (SOM) mutants were isolated (Reiling et al., 2005). Complementation with a cosmid library identified several of the SOM mutations were in a gene encoded at locus PA3257. PA3257 was identified as a homologue of the periplasmic protease Prc from E. coli (Table 1) (Reiling et al., 2005). Inactivation of P. aeruginosa prc was shown to only affect strains with mutant and not wild-type MucA proteins, leading the authors to suggest a model where Prc degrades mutated MucA proteins to facilitate activation of σ22; Prc was also shown to promote alginate overproduction in two additional mucoid mucA strains (CF20 and CF25), suggesting that it may have a broad substrate specificity for mucA mutant proteins (Reiling et al., 2005). Based on genetic experiments, it seems clear that Prc has a role in degradation of mutant MucA proteins, but this has yet to be confirmed by Western blot analysis. In terms of structure, Prc has a signal peptide (for periplasmic localization), one PDZ domain, and a tail specific protease domain (TSPc) that cleaves substrates (Fig. 3). Prc also contains an uncharacterized domain that is conserved in other proteases. Since PDZ domains recognize specific sequences and bind the C-termini of proteins, it is possible that the PDZ of Prc interacts with mutant MucA and regulates proteolysis by the tail-specific protease domain.
As a result of degradation by proteases AlgW and MucP, it is possible that truncated MucA proteins exist which would be of similar size to the truncated MucA proteins such as MucA22 (Fig. 2). This begs the question of whether or not Prc has a role in degrading wild-type MucA. Loss of Prc seems to only affect strains with mutant MucA(Reiling et al., 2005) and Prc was not required for the mucoid phenotype of PAO1 when RIP is activated (Damron et al., 2011). Overexpression of Prc does not activate alginate overproduction in PAO1, but PalgD activity suggests Prc may play a slight role in degradation of MucA (Wood et al., 2006). However, stability of MucA was not affected by inactivation of prc (Wood et al., 2006). These conflicting results suggest that Prc likely has a minor role in the degradation of wild-type MucA.
In vitro conditions can induce alginate overproduction
In a recent study, a medium has been formulated which causes wild-type nonmucoid strain PAO1 to overproduce alginate, independent of mutations (Damron et al., 2011). The medium contained the standard components of PIA supplemented with ammonium metavanadate (PIA-AMV). In E. coli, growth on media containing AMV increased σE activity (Tam & Missiakas, 2005). In P. aeruginosa, PIA-AMV medium did not cause mutations in genes such as mucA, but rather resulted in inducible alginate overproduction. It was also shown that triclosan and magnesium chloride components of PIA were necessary for the mucoid phenotype of PAO1 on PIA-AMV (Damron et al., 2011). One result of growing PAO1 on PIA-AMV was the palmitoylation of lipid A (Damron et al., 2011), which has also been observed in chronic CF isolates (Ernst et al., 1999). PhoP is a response regulator that is essential for palmitoylation of lipid A in P. aeruginosa (Ernst et al., 1999) and interestingly, phoP was required for the mucoid phenotype of PAO1 on PIA-AMV. This indicated that growth of PAO1 on PIA-AMV results in remodeling of the outer-leaflet via modification to lipid A but other experiments showed O-antigen LPS chain length was not affected by growth on PIA-AMV (Damron et al., 2011). Predictably, proteases AlgW and MucP were both required for the mucoid phenotype of PAO1 on PIA-AMV; furthermore, degradation of MucA was observed as a result of growth on PIA-AMV. Western blot analysis indicated that chaperone protease MucD was upregulated during growth on PIA-AMV, which suggested that the medium caused membrane stress. From these observations, a model was proposed where PIA-AMV medium affects membrane integrity that may cause misfolding or accumulation of envelope proteins (Damron et al., 2011). Consequently, MucA degradation by AlgW/MucP and σ22 activation were necessary to compensate for the stress of the PIA-AMV medium environment (Damron et al., 2011). However, the question remains: what are the direct molecular targets of AMV? A study has shown that vanadate binds to siderophores inhibiting iron uptake (Baysse et al., 2000) in P. aeruginosa. Also, since vanadate mimics phosphate there are many potential proteins and enzymes that may interact with and/or be inhibited by vanadate. It may be possible to use this PIA-AMV medium to further characterize the RIP pathways leading to induction of alginate overproduction.
Abrogated outer membrane protein processing results in the formation of membrane vesicles
Alginate overproduction on PIA-AMV medium suggested that stress conditions could cause misfolding of proteins in the envelope. Along these same lines, if protein processing is abrogated, then misfolded proteins would accumulate and activate RIP. Upstream of mucP (A3649) there is a gene opr86 (PA3648) that encodes an Omp85/YaeT/BamA family outer membrane protein. In E. coli, YaeT is a part of a complex that directly plays a role in assembly of β-barrel outer membrane proteins (OMPs) (Ruiz et al., 2006). Opr86 localizes to the outer membrane and has been shown to be essential in P. aeruginosa and depletion of Opr86 causes blebbing of the outer membrane (Tashiro et al., 2008). These authors hypothesized that misfolded outer membrane proteins cause membrane vesicle (MV) formation. Since MucD likely acts on misfolded proteins in the periplasm, Tashiro et al., examined the role of mucD on the formation of MV. In the absence of MucD, high amounts of MV were observed (Tashiro et al., 2009). Furthermore, overexpression of mucD or algW caused decreased MV production. Since algW overexpression would activate proteolysis of MucA (Wood et al., 2006) and subsequently σ22 transcriptional activity leading to more MucD expression, these data fit with other previous findings. The authors proposed a three-step model for dealing with accumulated OMPs. In step one, MucD directly acts upon misfolded OMPs. In step two, if misfolded OMP concentrations exceed the capabilities of MucD, then misfolded proteins will be released in MVs. In step three, if release of MV and misfolded proteins cannot lower the level of misfolded proteins to maintain homeostasis, then RIP will activate σ22 (Tashiro et al., 2009). This model fits well into the RIP model and adds an interesting layer to how P. aeruginosa deals with stress conditions by modulating multiple systems.
Unanswered questions
As detailed in this review, AlgW is positive regulator and key protease that acts on MucA to release σ22. However, the study that originally identified algW classified it as a negative regulator of alginate production (Boucher et al., 1996). Expression of plasmid-borne algW could turn off alginate production and PalgD promoter activity in mucoid strain CF31 (Boucher et al., 1996). It was apparent that algW encoded a serine protease due to its homology with HtrA-family proteins; however the mechanism of how AlgW suppressed alginate production was not clear. Here we have proposed that MucP RIP protease may act upon mutant MucA proteins such as MucA22. Previous work in E. coli has indicated that DegS inhibits RseP proteolysis of RseA (Grigorova et al., 2004). Based on algW suppression of the mucoid phenotype in the mucA strain and the inhibition of RseP by DegS in E. coli, we speculate that AlgW may inhibit MucP proteolysis. Furthermore, the degradation profile of epitope-tagged MucA was notably different between the mucoid mucD and mucD/algW strains (Damron & Yu, 2011). Collectively these data suggest AlgW may have an inhibitory effect on MucP but a mechanism may also exist to activate MucP to act directly upon MucA. It is possible AlgW-MucP interactions may occur through one of MucP’s two PDZ domains (Fig. 2 and 3), but this has not been investigated or experimentally determined.
AlgB-KinB is a two-component system (Table 1) that has been shown to regulate alginate overproduction (Goldberg & Ohman, 1984, Ma et al., 1997). AlgB is a response regulator that has been extensively characterized and shown to activate PalgD expression (Goldberg & Dahnke, 1992, Leech et al., 2008, Wozniak & Ohman, 1991). Recently, KinB was characterized as a negative regulator of alginate overproduction in PAO1 (Damron et al., 2009) (Fig. 4). In that study, it was observed that inactivation and deletion of kinB caused AlgW-dependent degradation of MucA. Additionally, the AlgB transcription factor and sigma factor RpoN (σ54) were both required for PalgU activity and complete degradation of MucA (Damron et al., 2009). This suggested novel roles for RpoN and AlgB (Fig. 4) outside of the characterized roles at PalgD. It is not clear why AlgW is degrading MucA in the absence of KinB (Damron et al., 2009) and more studies are required to determine the pathway between KinB and AlgW-RIP. kinB has been shown to be required for virulence in a zebrafish model, biofilm formation and quorum sensing (Chand et al., 2011), which suggested that kinB may control genes in addition to the alginate biosynthetic genes. Microarray analysis comparing a kinB mutant to a kinB/rpoN double mutant has revealed a large number of genes both up- and down-regulated (Damron et al., 2012). Interestingly, it was observed that loss of rpoN decreased algW expression in the kinB mutant. This may explain why rpoN was required for high PalgU and PalgD promoter activities of this strain (Damron et al., 2009). Due to the drastically altered transcriptome and the phenotypic changes that were observed, the kinB mutant and isogenic strains were used to challenge mice in an acute pneumonia model. Mucoid mucA22 mutant (PDO300) was virulent in this model but the mucoid kinB mutant was not virulent (Damron et al., 2012). Collectively these recent studies suggest that sensor kinase KinB, along with RpoN, control many genes related to multiple virulence phenotypes including alginate overproduction. KinB is clearly a pleiotropic regulator, but the environmental conditions controlling KinB and thus influencing gene expression are not known at this time.
MucA degradation by RIP has been observed by two in vitro conditions: membrane stress (growth in presence of D-cycloserine or on PIA-AMV) and mutations in regulators (MucE, MucD, MucB, KinB). How does each of these in vitro conditions relate to in vivo regulation of alginate overproduction? One study has shown that inducible alginate overproduction occurs during P. aeruginosa infection of murine lungs and as a result of anaerobic growth (Bragonzi et al., 2005). This intriguing study suggests that alginate overproduction occurs in strains with wild-type MucA, which can be supported by other in vitro studies described in this review. Another study has indicated that inactivation of σ22 (algU) or algW caused attenuation of PAO1 in a rat chronic respiratory infection model (Potvin et al., 2003). This further suggests MucA proteolysis is a key mechanism for infection. However, it is not clear how AlgW protease is activated in vivo. It can be hypothesized based on in vitro results that AlgW may be activated by misfolded proteins that accumulate during in vivo conditions. It is also possible that overall membrane integrity is compromised which leads to AlgW-mediated RIP. A third possible mechanism is that sensor kinase KinB may respond to in vivo conditions and activate RIP (Damron et al., 2009).
Another currently unanswered question is how MucB is released from MucA to allow RIP via AlgW (Cezairliyan & Sauer, 2009) to proceed. A recent study in E. coli suggests RseB responds to lipid signals and that RseB is a “noise-filtering gatekeeper” which improves the quality of the response (Chaba et al., 2011). Using this as a model, it seems possible in the context of P. aeruginosa that MucB signaling could be affected in situations where the mucoid phenotype is induced. Further research is necessary to understand MucB regulation of MucA and σ22.
Directions towards therapeutics
Now that there is a basic understanding of the regulation of degradation of MucA, research towards impairing the functions of these proteases may be a key to deactivating alginate production. Such treatments may augment already available therapeutics to target this system and eradicate P. aeruginosa before it establishes a chronic infection in CF patients. Two themes can be drawn when thinking about the proteases that act on MucA. The first is that assuming MucP is required for degrading mutant MucA proteins: MucP and ClpXP are the only proteases that are currently recognized as required for degradation of both mutant MucA and wild-type MucA. Since MucP is a zinc metalloprotease, it may be possible to use inhibitors of metalloproteases to block alginate overproduction. Pseudomonas elastase (LasB) is a metalloprotease that plays an important role in virulence. Recently, novel inhibitors of LasB have shown promise as a therapeutic approach to eradicate P. aeruginosa biofilms (Cathcart et al., 2011). Previous studies have shown that overexpression of LasB in nonmucoid strains resulted in alginate overproduction (Kamath et al., 1998). If added to the current model of MucA-RIP, elastase accumulation in the periplasm (Kamath et al., 1998) could cause misfolding in the envelope leading to activation of AlgW and MucP. It would be interesting to determine if zinc metalloprotease inhibitors can block alginate overproduction and increase the efficacy of current therapeutics. The second observation from the MucA-RIP model is that all of the MucA proteases contain PDZ domains (Fig. 3). PDZ domains may be a novel target to arrest RIP and block alginate production. PDZ domains exist in many human proteins, and it is possible that they would not be an advantageous target. However, inhibitors of PDZ domains have been shown to be promising pharmacotherapeutics in neuropathic pain and cocaine addiction (Thorsen et al., 2010).
Multiple pathways in P. aeruginosa converge at RIP of MucA (Fig. 4). From the studies reviewed here, it is clear that RIP occurs in strains with both wild-type and mutant mucA. While an abundance of research on this topic has focused on alginate overproduction, more research is still critical to understanding this stress response system of P. aeruginosa. We are hopeful that blocking this pathway could provide a much needed option for treating P. aeruginosa lung infections in CF.
Acknowledgments
F.H.D. was supported by a postdoctoral fellowship from the Cystic Fibrosis Foundation (DAMRON10F0). J.B.G. was supported grants from the NIH (R01 AI068112) and the Cystic Fibrosis Foundation (GOLDBERG10G0). We would like to thank the anonymous reviewers for their suggestions for improving this manuscript. We would also like to thank all of those who have contributed to the field and apologize if any details have been left out due to space limitations.
References
- Ades SE. Regulation by destruction: design of the sigmaE envelope stress response. Curr Opin Microbiol. 2008;11:535–540. doi: 10.1016/j.mib.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Akiyama Y, Kanehara K, Ito K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 2004;23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 2002;16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiou ED, Mintzas AC, Kounavis C, Dimitracopoulos G. Alginate production by clinical nonmucoid Pseudomonas aeruginosa strains. J Clin Microbiol. 1987;25:656–659. doi: 10.1128/jcm.25.4.656-659.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong D, Bell S, Robinson M, Bye P, Rose B, Harbour C, Lee C, Service H, Nissen M, Syrmis M, Wainwright C. Evidence for spread of a clonal strain of Pseudomonas aeruginosa among cystic fibrosis clinics. J Clin Microbiol. 2003;41:2266–2267. doi: 10.1128/JCM.41.5.2266-2267.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TA, Sauer RT. ClpXP, an ATP-poweredunfolding and protein-degradation machine. Biochim Biophys Acta. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysse C, De Vos D, Naudet Y, Vandermonde A, Ochsner U, Meyer JM, Budzikiewicz H, Schafer M, Fuchs R, Cornelis P. Vanadium interferes with siderophore-mediated iron uptake in Pseudomonas aeruginosa. Microbiology. 2000;146:2425–2434. doi: 10.1099/00221287-146-10-2425. [DOI] [PubMed] [Google Scholar]
- Boucher JC, Martinez-Salazar J, Schurr MJ, Mudd MH, Yu H, Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher JC, Schurr MJ, Yu H, Rowen DW, Deretic V. Pseudomonas aeruginosa in cystic fibrosis: role of mucC in the regulation of alginate production and stress sensitivity. Microbiology. 1997;143:3473–3480. doi: 10.1099/00221287-143-11-3473. [DOI] [PubMed] [Google Scholar]
- Bragonzi A, Worlitzsch D, Pier GB, Timpert P, Ulrich M, Hentzer M, Andersen JB, Givskov M, Conese M, Doring G. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J Infect Dis. 2005;192:410–419. doi: 10.1086/431516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Burns JL, Gibson RL, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith AL, Ramsey BW. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis. 2001;183:444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- Cathcart GR, Quinn D, Greer B, Harriott P, Lynas JF, Gilmore BF, Walker B. Novel inhibitors of the Pseudomonas aeruginosa virulence factor LasB: a potential therapeutic approach for the attenuation of virulence mechanisms in pseudomonal infection. Antimicrob Agents Chemother. 2011;55:2670–2678. doi: 10.1128/AAC.00776-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezairliyan BO, Sauer RT. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol. 2009;72:368–379. doi: 10.1111/j.1365-2958.2009.06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaba R, Alba BM, Guo MS, Sohn J, Ahuja N, Sauer RT, Gross CA. Signal integration by DegS and RseB governs the σE-mediated envelope stress response in Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:2106–2111. doi: 10.1073/pnas.1019277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand NS, Lee JS, Clatworthy AE, Golas AJ, Smith RS, Hung DT. The Sensor Kinase KinB Regulates Virulence in Acute Pseudomonas aeruginosa Infection. J Bacteriol. 2011;193:2989–2999. doi: 10.1128/JB.01546-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman FT, Mueschenborn S, Meluleni G, Ray C, Carey VJ, Vargas SO, Cannon CL, Ausubel FM, Pier GB. Hypersusceptibility of cystic fibrosis mice to chronic Pseudomonas aeruginosa oropharyngeal colonization and lung infection. Proc Natl Acad Sci U S A. 2003;100:1949–1954. doi: 10.1073/pnas.0437901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron FH, Davis MR, Jr, Withers TR, Ernst RK, Goldberg JB, Yu G, Yu HD. Vanadate and triclosan synergistically induce alginate production by Pseudomonas aeruginosa strain PAO1. Mol Microbiol. 2011;81:554–570. doi: 10.1111/j.1365-2958.2011.07715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron FH, Owings JP, Okkotsu Y, Varga JJ, Schurr JR, Goldberg JB, Schurr MJ, Yu HD. Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J Bacteriol. 2012;194:1317–1330. doi: 10.1128/JB.06105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron FH, Qiu D, Yu HD. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J Bacteriol. 2009;191:2285–2295. doi: 10.1128/JB.01490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron FH, Yu HD. Pseudomonas aeruginosa MucD regulates alginate pathway through activation of MucA degradation via MucP proteolytic activity. J Bacteriol. 2011;193:286–291. doi: 10.1128/JB.01132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries CA, Ohman DE. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett RG, Harrison GM, Stillwell RN, Wallis ES. An atypical Pseudomonas aeruginosa associated with cystic fibrosis of the pancreas. J Pediatr. 1966;68:215–221. [Google Scholar]
- Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, Hackett M, Miller SI. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- Firoved AM, Boucher JC, Deretic V. Global genomic analysis of AlgU (sigma(E))-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J Bacteriol. 2002;184:1057–1064. doi: 10.1128/jb.184.4.1057-1064.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoved AM, Deretic V. Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J Bacteriol. 2003;185:1071–1081. doi: 10.1128/JB.185.3.1071-1081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoved AM, Ornatowski W, Deretic V. Microarray analysis reveals induction of lipoprotein genes in mucoid Pseudomonas aeruginosa: implications for inflammation in cystic fibrosis. Infect Immun. 2004a;72:5012–5018. doi: 10.1128/IAI.72.9.5012-5018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoved AM, Wood SR, Ornatowski W, Deretic V, Timmins GS. Microarray analysis and functional characterization of the nitrosative stress response in nonmucoid and mucoid Pseudomonas aeruginosa. J Bacteriol. 2004b;186:4046–4050. doi: 10.1128/JB.186.12.4046-4050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Nivens DE, Weadge JT, Howell PL. Biosynthesis of the Pseudomonas aeruginosa Extracellular Polysaccharides, Alginate, Pel, and Psl. Frontiers in Microbiology. 2011;2 doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe JAM, Govan JRW. Synthesis, regulation and biological function of bacterial alginate. In: Bushell ME, editor. Progress in industrial microbiology. London: Elsevier; 1983. pp. 45–83. [Google Scholar]
- Goldberg JB, Dahnke T. Pseudomonas aeruginosa AlgB, which modulates the expression of alginate, is a member of the NtrC subclass of prokaryotic regulators. Mol Microbiol. 1992;6:59–66. doi: 10.1111/j.1365-2958.1992.tb00837.x. [DOI] [PubMed] [Google Scholar]
- Goldberg JB, Gorman WL, Flynn JL, Ohman DE. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J Bacteriol. 1993;175:1303–1308. doi: 10.1128/jb.175.5.1303-1308.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg JB, Ohman DE. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984;158:1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JR, Fyfe JA. Mucoid Pseudomonas aeruginosa and cystic fibrosis: resistance of the mucoid from to carbenicillin, flucloxacillin and tobramycin and the isolation of mucoid variants in vitro. J Antimicrob Chemother. 1978;4:233–240. doi: 10.1093/jac/4.3.233. [DOI] [PubMed] [Google Scholar]
- Grigorova IL, Chaba R, Zhong HJ, Alba BM, Rhodius V, Herman C, Gross CA. Fine-tuning of the Escherichia coli sigmaE envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev. 2004;18:2686–2697. doi: 10.1101/gad.1238604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselblatt H, Kurzbauer R, Wilken C, Krojer T, Sawa J, Kurt J, Kirk R, Hasenbein S, Ehrmann M, Clausen T. Regulation of the sigmaE stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress. Genes Dev. 2007;21:2659–2670. doi: 10.1101/gad.445307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger CD, Ye RW, Parsek MR, Xie ZD, Chakrabarty AM. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative sigma factor (sigma E) Proc Natl Acad Sci U S A. 1995;92:7941–7945. doi: 10.1073/pnas.92.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Amemura M, Nashimoto H, Shinagawa H, Makino K. The rpoE gene of Escherichia coli, which encodes sigma E, is essential for bacterial growth at high temperature. J Bacteriol. 1995;177:2918–2922. doi: 10.1128/jb.177.10.2918-2922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Webb AK, Govan JR, Hart CA, Walshaw MJ. Pseudomonas aeruginosa cross-infection in cystic fibrosis. Lancet. 2002;359:527–528. doi: 10.1016/S0140-6736(02)07648-1. [DOI] [PubMed] [Google Scholar]
- Kamath S, Kapatral V, Chakrabarty AM. Cellular function of elastase in Pseudomonas aeruginosa: role in the cleavage of nucleoside diphosphate kinase and in alginate synthesis. Mol Microbiol. 1998;30:933–941. doi: 10.1046/j.1365-2958.1998.01121.x. [DOI] [PubMed] [Google Scholar]
- Kennedy MB. Origin of PDZ (DHR, GLGF) domains. Trends Biochem Sci. 1995;20:350. doi: 10.1016/s0968-0004(00)89074-x. [DOI] [PubMed] [Google Scholar]
- Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- Krojer T, Pangerl K, Kurt J, Sawa J, Stingl C, Mechtler K, Huber R, Ehrmann M, Clausen T. Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc Natl Acad Sci U S A. 2008a;105:7702–7707. doi: 10.1073/pnas.0803392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krojer T, Sawa J, Schafer E, Saibil HR, Ehrmann M, Clausen T. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008b;453:885–890. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- Leech AJ, Sprinkle A, Wood L, Wozniak DJ, Ohman DE. The NtrC family regulator AlgB, which controls alginate biosynthesis in mucoid Pseudomonas aeruginosa, binds directly to the algD promoter. J Bacteriol. 2008;190:581–589. doi: 10.1128/JB.01307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J Immunol. 2005;175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko I, Smith CK, Walsh NP, Sauer RT, Baker TA. PDZ-like domains mediate binding specificity in the Clp/Hsp100 family of chaperones and protease regulatory subunits. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Wozniak DJ, Ohman DE. Identification of the histidine protein kinase KinB in Pseudomonas aeruginosa and its phosphorylation of the alginate regulator algB. J Biol Chem. 1997;272:17952–17960. doi: 10.1074/jbc.272.29.17952. [DOI] [PubMed] [Google Scholar]
- Martin DW, Holloway BW, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol. 1993a;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U SA. 1993b;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, Johnsen AH, Givskov M, Ohman DE, Molin S, Hoiby N, Kharazmi A. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- Mathee K, McPherson CJ, Ohman DE. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN) J Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum SJ, Corkill J, Gallagher M, Ledson MJ, Hart CA, Walshaw MJ. Superinfection with a transmissible strain of Pseudomonas aeruginosa in adults with cystic fibrosis chronically colonised by P aeruginosa. Lancet. 2001;358:558–560. doi: 10.1016/s0140-6736(01)05715-4. [DOI] [PubMed] [Google Scholar]
- Muller C, I, Bang S, Velayudhan J, Karlinsey J, Papenfort K, Vogel J, Fang FC. Acid stress activation of the sigma(E) stress response in Salmonellaenterica serovar Typhimurium. Mol Microbiol. 2009;71:1228–1238. doi: 10.1111/j.1365-2958.2009.06597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J, Iwanczyk J, Jomaa A. Escherichia coli DegP: a structure-driven functional model. J Bacteriol. 2009;191:4705–4713. doi: 10.1128/JB.00472-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen SS, Espersen F, Hoiby N, Jensen T. Immunoglobulin A and immunoglobulin G antibody responses to alginates from Pseudomonas aeruginosa in patients with cystic fibrosis. J Clin Microbiol. 1990;28:747–755. doi: 10.1128/jcm.28.4.747-755.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pier GB, Desjardins D, Aguilar T, Barnard M, Speert DP. Polysaccharide surface antigens expressed by nonmucoid isolates of Pseudomonas aeruginosa from cystic fibrosis patients. J Clin Microbiol. 1986;24:189–196. doi: 10.1128/jcm.24.2.189-196.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potvin E, Lehoux DE, Kukavica-Ibrulj I, Richard KL, Sanschagrin F, Lau GW, Levesque RC. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ Microbiol. 2003;5:1294–1308. doi: 10.1046/j.1462-2920.2003.00542.x. [DOI] [PubMed] [Google Scholar]
- Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. PBAD-based shuttle vectors for functional analysis of toxic and highly-regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol. 2008a;74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, V, Eisinger M, Head NE, Pier GB, Yu HD. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology. 2008b;154:2119–2130. doi: 10.1099/mic.0.2008/017368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, V, Eisinger M, Rowen DW, Yu HD. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2007;104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- Rehm B. Alginates: Biologyand Applications. In: Steinbchel A, editor. Microbiology Monographs. New York: Springer; 2009. p. 274. [Google Scholar]
- Reiling SA, Jansen JA, Henley BJ, Singh S, Chattin C, Chandler M, Rowen DW. Prc protease promotes mucoidy in mucA mutants of Pseudomonas aeruginosa. Microbiology. 2005;151:2251–2261. doi: 10.1099/mic.0.27772-0. [DOI] [PubMed] [Google Scholar]
- Renders NH, Sijmons MA, van Belkum A, Overbeek SE, Mouton JW, Verbrugh HA. Exchange of Pseudomonas aeruginosa strains amongcystic fibrosis siblings. Res Microbiol. 1997;148:447–454. doi: 10.1016/s0923-2508(97)83875-2. [DOI] [PubMed] [Google Scholar]
- Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- Rowen DW, Deretic V. Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol Microbiol. 2000;36:314–327. doi: 10.1046/j.1365-2958.2000.01830.x. [DOI] [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- Schwarzmann S, Boring JR. Antiphagocytic effect of slime from a mucoid strain of Pseudomonas aeruginosa. Infect Immun. 1971;3:762–767. doi: 10.1128/iai.3.6.762-767.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam C, Missiakas D. Changes in lipopolysaccharide structure induce the sigma(E)-dependent response of Escherichia coli. Mol Microbiol. 2005;55:1403–1412. doi: 10.1111/j.1365-2958.2005.04497.x. [DOI] [PubMed] [Google Scholar]
- Tart AH, Wolfgang MC, Wozniak DJ. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis byinhibiting expression of fleQ. J Bacteriol. 2005;187:7955–7962. doi: 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y, Nomura N, Nakao R, Senpuku H, Kariyama R, Kumon H, Kosono S, Watanabe H, Nakajima T, Uchiyama H. Opr86 is essential for viability and is a potential candidate for a protective antigen against biofilm formation by Pseudomonas aeruginosa. J Bacteriol. 2008;190:3969–3978. doi: 10.1128/JB.02004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y, Sakai R, Toyofuku M, Sawada I, Nakajima-Kambe T, Uchiyama H, Nomura N. Outer membrane machinery and alginate synthesis regulators control membrane vesicle production in Pseudomonas aeruginosa. J Bacteriol. 2009;191:7509–7519. doi: 10.1128/JB.00722-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen TS, Madsen KL, Rebola N, Rathje M, Anggono V, Bach A, Moreira IS, Stuhr-Hansen N, Dyhring T, Peters D, Beuming T, Huganir R, Weinstein H, Mulle C, Stromgaard K, Ronn LC, Gether U. Identification of a small-molecule inhibitor of the PICK1 PDZ domain that inhibits hippocampal LTP and LTD. Proc Natl Acad Sci U S A. 2010;107:413–418. doi: 10.1073/pnas.0902225107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- Welsh MJ, Ramsey BW, Accurso F, Cutting GR. Cystic fibrosis. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic & molecular bases of inherited disease. New York: McGraw-Hill; 2001. pp. 5121–5188. [Google Scholar]
- Wood LF, Leech AJ, Ohman DE. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: Roles of sigma (AlgT) and the AlgW and Prc proteases. Mol Microbiol. 2006;62:412–426. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- Wood LF, Ohman DE. Independent regulation of MucD, an HtrA-like protease in Pseudomonas aeruginosa, and the role of its proteolytic motif in alginate gene regulation. J Bacteriol. 2006;188:3134–3137. doi: 10.1128/JB.188.8.3134-3137.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LF, Ohman DE. Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol. 2009;72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- Wozniak DJ, Ohman DE. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J Bacteriol. 1991;173:1406–1413. doi: 10.1128/jb.173.4.1406-1413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorgey P, Rahme LG, Tan MW, Ausubel FM. The roles of mucD and alginate in the virulence of Pseudomonas aeruginosa in plants, nematodes and mice. Mol Microbiol. 2001;41:1063–1076. doi: 10.1046/j.1365-2958.2001.02580.x. [DOI] [PubMed] [Google Scholar]