Abstract
Background
Cardiomyopathy is a heterogeneous disease with a strong genetic component. A research-based pediatric cardiomyopathy registry (PCMR) identified familial, syndromic, or metabolic causes in 30% of children. However, these results pre-dated clinical genetic testing.
Methods and Results
We determined the prevalence of familial, syndromic, or metabolic causes in eighty-three consecutive unrelated patients referred for genetic evaluation of cardiomyopathy from 2006–2009. Seventy-six percent of probands (n=63) were categorized as familial, syndromic, or metabolic. Forty-three percent (n=18) of hypertrophic cardiomyopathy (HCM) patients had mutations in sarcomeric genes, with MYH7 and MYBPC3 mutations predominating. Syndromic (17%, n=7) and metabolic (26%, n=11) causes were frequently identified in HCM patients. The metabolic subgroup was differentiated by decreased endocardial shortening fraction on echocardiography. Dilated cardiomyopathy (DCM) patients had similar rates of syndromic (20%, n=5) and metabolic (16%, n=4) causes, but fewer familial cases (24%, n=6) than HCM patients.
Conclusions
The cause of cardiomyopathy is identifiable in a majority of affected children. An underlying metabolic or syndromic cause is identified in greater than 35% of children with HCM or DCM. Identification of etiology is important for management, family based risk assessment, and screening.
Keywords: cardiomyopathy, heart failure, genetics, mutation, genetic testing
Introduction
Cardiomyopathy is a genetically and clinically heterogeneous disease of the myocardium that causes systolic and/or diastolic dysfunction. In affected individuals, pediatric cardiomyopathy has severe consequences with up to 40% of children progressing to death or transplant within five years of diagnosis (1–4).
Cardiomyopathy can be classified into five clinical phenotypes: hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), restrictive cardiomyopathy (RCM), left ventricular non-compaction cardiomyopathy (LVNC), and arrhythmogenic right ventricular cardiomyopathy (ARVC). Phenotypic classification is based on the clinical taxonomy and informs management decisions, but provides little insight into the underlying etiology. In 1994, the Pediatric Cardiomyopathy Registry (PCMR) initiated studies on the epidemiologic features of cardiomyopathy in children with diagnostic categories that included myocarditis, inborn errors of metabolism, malformation syndromes, neuromuscular disease, familial, and unknown (idiopathic) causes (5). These data pre-dated many clinically available genetic tests, and along with other epidemiologic studies highlighted the uncertain etiologic basis of cardiomyopathy: the majority of HCM and DCM patients were identified as idiopathic (1, 3, 4, 6). Recently, the development of molecular testing for cardiomyopathy has led to new recommendations for diagnostic evaluation and family screening incorporating genetic testing (7–9), but the yield of such testing in children is unclear.
HCM is commonly considered a disease of the sarcomere, with mutations in genes encoding sarcomeric proteins identified in 40–70% of adults with HCM (8, 10). More recently, mutations in these genes have also been found to cause DCM, RCM, and LVNC (11–18). While recent publications have demonstrated a prevalence of sarcomeric mutations in children with HCM similar to rates identified in adults (19,20,21) these studies excluded potential syndromic or metabolic cases. As a result, the impact of genetic and metabolic testing as a part of routine clinical care in pediatric cardiomyopathy is largely unknown.
The objectives of this study were (1) to characterize the phenotypic groups of cardiomyopathy patients presenting to a single center multidisciplinary clinic, (2) to test the hypothesis that clinical genetic testing improves diagnostic rates across clinical phenotypes, and (3) to evaluate these groups for potential genotype/phenotype correlations that may inform future management.
Methods
Patient Cohort and Clinical Evaluation
A retrospective review of patients diagnosed with cardiomyopathy at Cincinnati Children’s Hospital Medical Center (CCHMC) from October, 2006 – October, 2009 was performed with approval of the CCHMC Institutional Review Board. In families with multiple affected members, only the proband was included in order to accurately determine diagnostic yield. This study focuses on pediatric patients with unexplained cardiomyopathy at the time of presentation and therefore excluded patients with previously diagnosed neuromuscular disease (e.g. Duchenne muscular dystrophy and Friedreich ataxia). Additional exclusion criteria included cardiomyopathy secondary to congenital heart disease, arrhythmia, chemotherapy, myocarditis, or environmental toxins.
Definition of Diagnostic Categories
Patients were grouped using categories set forth by the PCMR with modifications to incorporate new clinical genetic testing capabilities (3–5). The familial classification was updated to include isolated, heritable cardiomyopathy detectable by clinically available molecular testing. Therefore, familial cardiomyopathy was defined as an affected proband with 1) documented cardiomyopathy or sudden cardiac death (SCD) in a first degree relative and/or 2) a disease-causing mutation identified by clinical genetic testing for HCM or DCM or ARVC. This is consistent with the existing classification scheme of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases (11). The metabolic disease group includes patients with inborn errors of metabolism (e.g. storage diseases) or patients fulfilling modified Walker criteria for mitochondrial disease (Supp. Table 1) (22–24). The syndromic group includes patients meeting clinical criteria and dysmorphology for well characterized genetic syndromes and/or patients with genetic testing identifying a syndromic cause. Patients for whom family history, clinical criteria, and/or molecular testing was unable to establish a classification were classified as idiopathic This included patients with genetic syndromic diagnoses not previously associated with cardiomyopathy.
Clinical Evaluation
All patients were evaluated by a cardiologist and clinical geneticist. The age of presentation was defined by clinical symptoms and/or positive cardiac imaging. All patients had a three generation pedigree developed from their reported family history. For this study, a positive family history was defined as cardiomyopathy or SCD reported at presentation, and confirmed by obtaining clinical records and/or autopsy records. New York Heart Association (NYHA) symptoms scores were assigned with modifications made for the pediatric age group (25,26).
Laboratory testing was ordered by a clinical geneticist based on clinical presentation. Metabolic testing included some or all of the following: serum amino acids, urine organic acids, acylcarnitine profile, lactate, pyruvate, coenzyme Q10 panel, creatinine kinase levels, electron transport chain analysis of fibroblasts or muscle, muscle histology and electron microscopy, fibroblast enzyme assays, and fibroblast beta oxidation analysis for fatty acid oxidation disorders. Modified Walker criteria were utilized to diagnose mitochondrial disease (22–24). These criteria combine clinical, laboratory, and molecular findings and are a rigorous and standardized clinical diagnostic tool (Supp. Table 1). Genetic testing was ordered based on clinical evaluation and family history. HCM or DCM gene panel testing was ordered from commercial laboratories. All patients undergoing HCM gene panel testing were tested for MYH7, MYBPC3, MYL2, MYL3, and TNNT2 genes. In addition, those undergoing HCM gene panel testing in later years of the study also may have had testing for TNNI3, TPM1, ACTC, LAMP2, PRKAG2, GLA, CAV3, TNNC1, TTR, MTTG, MTTI, MTTK. Patients undergoing DCM panel testing were tested for MYH7, MYBPC3, TNNT2, TNNI3, TPM1, ACTC, LDB3, LMNA, PLN, and TAZ. For this study, mutations interpreted by the commercial laboratory as disease causing were considered positive. Genetic test results interpreted by the commercial laboratory as inconclusive or variants of uncertain significance were considered negative and are provided in Supp. Table 2. Patients with a suspected genetic syndrome underwent appropriate molecular or cytogenetic testing. Molecular testing included Noonan spectrum gene panel (PTPN11, SOS1, RAF1, MEK1, MEK2, HRAS, KRAS, BRAF), Pompe disease testing (GAA), mitochondrial DNA sequencing, Alström syndrome testing (ALMS1), and CHARGE testing (CHD7).
Echocardiographic Data
Transthoracic two-dimensional, color Doppler and M-mode echocardiography was performed at rest using standard methods (27, 28). All M-mode measurements were indexed to age and body-surface area and corresponding z-scores were derived. Protocols are described in supplemental methods.
Statistical Analyses
Descriptive statistics for continuous variables are presented as means and ranges; categorical variables are presented as frequencies and percentages. A two-sample test of proportion was used to determine whether current testing significantly improved diagnostic rates compared to the PCMR. A Chi-square test of homogeneity was applied to the distribution of phenotypes in the current study and the PCMR population. The criterion for statistical significance was set at the nominal α = 0.05 level. Analyses were conducted using SAS v9.
Results
Study Population
The demographics of 83 probands are summarized in Table 1. The phenotype distribution included 51% HCM, 30% DCM, 5% RCM, 13% LVNC and 1% ARVC with an even gender distribution. The ethnic profile of the study group was 67% Caucasian, 22% African-American, 3% Hispanic and 8% other.
Table 1.
Characteristics of 83 Probands According to Cardiomyopathy Phenotype
| Characteristic | All | HCM | DCM | RCM | LVNC | ARVC |
|---|---|---|---|---|---|---|
| Gender (n) | ||||||
| Male | 38 | 18 | 10 | 3 | 6 | 1 |
| Female | 45 | 24 | 15 | 1 | 5 | 0 |
| Total | 83 | 42 | 25 | 4 | 11 | 1 |
| Age at presentation (%) | ||||||
| <1 y | 41 | 45 | 48 | 0 | 27 | 0 |
| 1 to <6 y | 11 | 5 | 20 | 25 | 9 | 0 |
| 6 to <13 y | 19 | 19 | 12 | 75 | 18 | 0 |
| 13 to <18 y | 25 | 26 | 20 | 0 | 36 | 100 |
| 18 to 21y | 4 | 5 | 0 | 0 | 9 | 0 |
| Family history (%) | 36 | 40 | 12 | 75 | 27 | 100 |
| Family history of sudden death (%) | 16 | 17 | 4 | 75 | 18 | 100 |
The HCM patients had a bi-modal age distribution; the largest group was diagnosed prior to one year of age with a second cluster after late childhood. A positive family history of cardiomyopathy was present in 40% of subjects at their initial evaluation, consistent with reports from other childhood HCM populations (4, 6, 19–21). Seven patients (17%) had a family history of SCD.
The DCM population had the youngest mean age of presentation, with 48% presenting before one year of age. DCM subjects had a positive family history of cardiomyopathy in 12% and of SCD in 4%. Eleven patients had isolated LVNC without hypertrophy, dilation or congenital heart disease. Interestingly, in the 27% that had a positive family history, two subjects had a family history of HCM and one had a family history of “mixed” phenotype with DCM and LVNC.
Diagnostic Evaluation
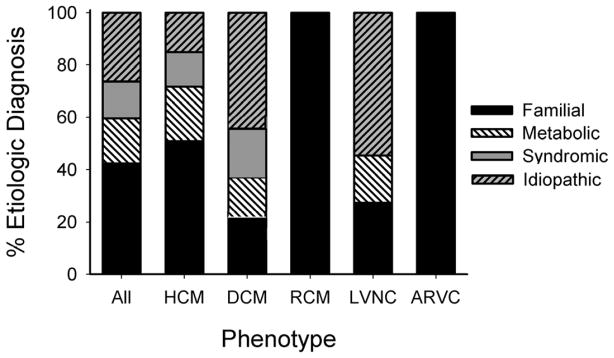
By combining clinical assessment with testing, 76% (63/83) of patients were placed in a diagnostic category, while 24% (20/83) were idiopathic (Figure 1). This rate is significantly higher than the PCMR findings prior to clinically available genetic testing, in which approximately 30% of patients were given an etiologic diagnosis (p<0.01) (3, 4). This increased yield does not result from intrinsic differences in patient population or improvement within a single category (Chi square test of homogeneity p=0.43).
Figure 1.
Cause of cardiomyopathy by phenotype. For each cardiomyopathy subtype, the relative contribution of each diagnostic category is shown as a percent of total cases.
The HCM subgroup had the highest rate (88%) of categorical classifications, and LVNC had the lowest (45%). In each phenotypic subgroup, familial disease was the most commonly identified etiology, occurring in 42% of all patients. This is likely a minimum estimate since not all patients in the DCM or LVNC categories underwent gene panel testing. Metabolic disease was identified in 17/83 patients (20.5%) of which 11 had HCM, 4 DCM and 2 LVNC. Twelve children (14.5%) were categorized as syndromic, 7 with HCM and 5 with DCM.
The results for patients with positive molecular testing are summarized in Table 2. In the familial classification, 23 mutations were identified in 22 probands. All HCM patients without syndromic or metabolic disease underwent HCM gene panel testing (n=25). In the DCM and LVNC groups, only a minority of patients underwent gene panel testing (n=9 and n=3, respectively). Only one missense mutation, MYBPC3 R502W, was identified in more than one proband, occurring in two unrelated infants that presented with severe HCM. In our cohort, 30/83 (36%) patients had a positive family history at the time of initial evaluation, of which 17 (57%), were found to carry mutations. The testing yield was highest in the HCM group where 82% of patients with a positive family history had a mutation identified. In this group, 56% of mutations were in MYH7 and 39% in MYBPC3. There was no difference between mutation-negative and mutation-positive patients with respect to age of presentation, clinical features, or findings on echocardiography. The DCM population consisted of 25 patients, 6 of whom were categorized as familial (24%). Two probands in the DCM group had sarcomeric mutations, one in MYH7 and the other in TNNT2. The yield for sarcomeric mutation testing was lowest in the LVNC group, in which no patients with a positive family history had positive genetic testing (n = 3). Clinical genetic testing can identify variants of uncertain significance (VUS) (Supp. Table 2). Patients with VUS were classified as idiopathic.
Table 2.
Positive Genetic Testing Results
| Etiology | Phenotype | Age of Cardiac Presentation | Age of Genetic Diagnosis | Gene | Mutation | Positive family history: CM/SCD |
|---|---|---|---|---|---|---|
| Familial | ARVC | 14 | 14 | DSP | R425X | +/+ |
| DCM | 3 | 8 | MYH7 | Q1346X | 0/0 | |
| DCM | 15 | 15 | TNNT2 | K210del | +/0 | |
| HCM | 15 | 15 | ACTC | L10M | 0/0 | |
| HCM | 8 | 14 | MYH7 | R663H | +/+ | |
| HCM | 40 | 40 | MYH7 | L908V | +/0 | |
| HCM | 12 | 12 | MYH7 | R403W | +/0 | |
| HCM | 12 | 12 | MYH7 | G741R | +/0 | |
| HCM | 10 | 14 | MYH7 | G768R | +/+ | |
| HCM | 16 | 16 | MYH7 | V606M | +/0 | |
| HCM | >19 | >19 | MYH7 | R1475C | 0/0 | |
| HCM | 19 | 19 | MYH7 | P710H | +/+ | |
| HCM | 6 | 6 | MYH7 | R403Q | +/+ | |
| HCM | 16 | 16 | MYH7 | V606M | +/0 | |
| HCM | <1 | <1 | MYBPC3 | R502W | +/0 | |
| HCM | 35 | 35 | MYBPC3 | F412fsX1+ R177C | +/+ | |
| HCM | 11 | 11 | MYBPC3 | E258K | +/+ | |
| HCM | <1 | <1 | MYBPC3 | R502W | 0/0 | |
| HCM | 3 | 3 | MYBPC3 | F448S | 0/0 | |
| HCM | 11 | 11 | MYBPC3 | D1064GfsX38 | +/0 | |
| HCM | 8 | 8 | MYBPC3 | D770N | +/0 | |
| RCM | 2 | 3 | MYH7 | G768R | 0/0 | |
| Metabolic | HCM | <1 | 3 |
MTATP6 MTATP8 |
m.8528T>C M1T; W55R |
0/0 |
| HCM | <1 | <1 | GAA | c.1437(+1)G>A+ c.1437(+1)G>A | 0/0 | |
| HCM | <1 | <1 | GAA | R660H+ R854X | 0/0 | |
| HCM | <1 | <1 | GAA | E176RfsX44+ T737N | 0/0 | |
| HCM | <1 | <1 | GAA | E176RfsX44 + E176RfsX44 | 0/0 | |
| Syndromic | HCM | 17 | 9 | Multiple | Deletion 1p36 | 0/0 |
| HCM | <1 | <1 | CHD7 | I1097A | 0/0 | |
| HCM | <1 | <1 | RAF-1 | S257L | 0/0 | |
| HCM | <1 | 15 | PTPN11 | T468M | 0/0 | |
| HCM | 16 | 16 | SOS1 | E846K | 0/0 | |
| HCM | 1 | 3 | PTPN11 | T468M | 0/0 | |
| DCM | <1 | 10 | ALMS1 | Q3495KfsX14 + R2828S | 0/0 | |
| DCM | 15 | 19 | ALMS1 | D2457X + D2459X | +/0 |
Mutations are listed with protein designations except where indicated as cDNA (c.) or mitochondrial DNA (m.) CM = Cardiomyopathy; SCD = Sudden Cardiac Death;
The metabolic category had two major diagnoses: Pompe disease and mitochondrial disorders. Molecular results for 4/7 patients with Pompe disease are shown in Table 2. Half of the Pompe patients presented with acute cardiopulmonary disease while the other half presented for evaluation as part of a larger systemic work up for non-cardiac symptoms. Patients with mitochondrial disorders presented with diverse cardiomyopathy phenotypes - 4 HCM, 4 DCM, and 2 LVNC. The syndromic group consisted of 12 patients (7 HCM, 5 DCM), including 3 HCM patients with Noonan syndrome and 2 with LEOPARD syndrome. Other syndromes identified included CHARGE syndrome (CHD7 mutation), 1p36 deletion syndrome, and two unrelated patients with Alström syndrome (ALMS1 mutations).
Clinical and Echocardiographic Findings in HCM Probands
Table 3 summarizes data for the HCM group. Eighty-five percent of the familial and idiopathic patients and 71% of syndromic patients were asymptomatic at presentation, as is common in the pediatric age range. In contrast, 55% (Pompe disease n=3; mitochondrial disease n=3) of the metabolic group presented with NYHA class IV symptoms, early feeding problems, and respiratory distress in the first year of life. Two patients with Pompe and two with mitochondrial disease died from heart failure before 18 months.
Table 3.
Clinical Characteristics and Echocardiographic Findings in HCM Patients
| Familial n = 20* | Metabolic n = 11* | Syndromic n = 7 | Idiopathic n = 5 | |
|---|---|---|---|---|
| NYHA | ||||
| Class I | 17* | 4 | 5 | 5 |
| Class II | 3 | 0 | 0 | 0 |
| Class III | 0 | 0 | 0 | 1 |
| Class IV | 0 | 6 | 1 | 0 |
| Unknown | 0 | 1 | 1 | 0 |
| Echocardiography | ||||
| LVMWT z-score (Range) | 4.7 (2.7 to 6.9) | 4.2 (2.3 to 6.6) | 5.4 (3.0 to 7.2) | 6.3 (4.8 to 8.7) |
| LVEDD z-score (Range) | 1.6 (−3.3 to 0.8) | 0.4 (−5.3 to 4.1) | −2.3 (−3.7 to −1.3) | −2.9 (−4.5 to −1.4) |
| FS z-score (Range) | 5.4 (−0.1 to 16) | −2.9 (−14 to 9.2) | 2.6 (−2.6 to 8.4) | 7.1 (1.9 to 15) |
| Mean LA size, z-score(Range) | 0.8 (−1.6 to 3.3) | 0 (−0.9 to 1.1) | 1.6 (−0.2 to 3.1) | 0.2 (−1.4 to 2.7) |
| Arrhythmia (%) | 6 (22%) | 0 | 0 | 0 |
| SCD (%) | 1 (4%) | 0 | 0 | 0 |
| ICD (%) | 5 (19%) | 0 | 0 | 0 |
| Transplant (%) | 0 | 0 | 0 | 0 |
| Deceased (%) | 1 (4%) | 4 (36%) | 0 | 0 |
NYHA = New York Heart Association functional class; LVMWT = Left Ventricular Maximal Wall Thickness; LVEDD = Left Ventricular End Diastolic Dimension; FS=Fracional Shortening; LA = Left Atrium; SCD = Sudden Cardiac Death; ICD = Implantable Cardiac Defibrillator.
Includes one patient with both familial and mitochondrial disease.
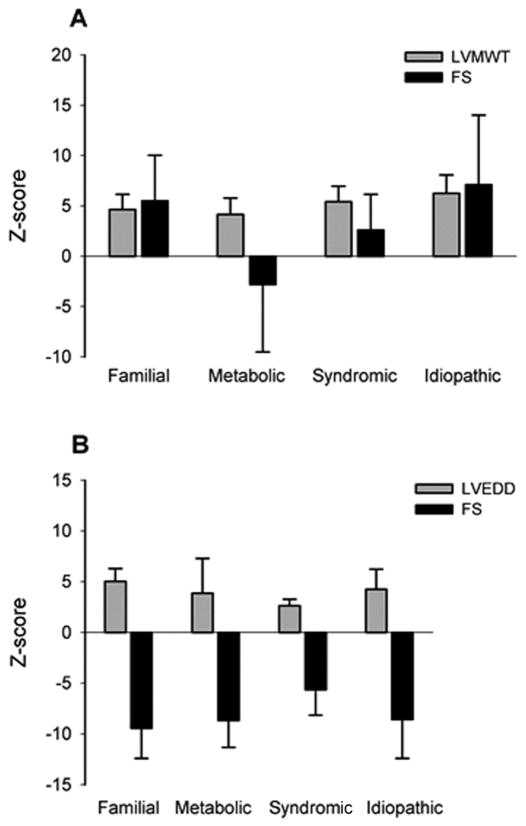
The idiopathic and syndromic groups showed a trend toward larger left ventricular maximal wall thickness (LVMWT) measurements despite the small size of our sub-populations. Interestingly, these thicker hearts did not correlate to more severe symptoms or worse outcomes as these groups had no mortalities. The familial, syndromic and idiopathic subgroups had increased endocardial shortening fraction (FS) as is expected in HCM (Figure 2A). The metabolic group, however, had 6/11 patients with left ventricular dysfunction and an average FS z-score of −2.9. The LVMWT in the metabolic group was similar despite differences in underlying disease pathogenesis. Children with Pompe disease demonstrated similar average LVMWT z-scores compared to children with mitochondrial disease (4.6 vs 3.6), but had worse ventricular function with a decreased average FS percentage z-score (−3.3 vs −1.5).
Figure 2.
Comparison of echocardiographic findings by diagnostic categories. (A) Mean z-score left ventricular maximal wall thickness (LVMWT) and endocardial shortening fraction (FS) in patients with HCM of distinct etiologies. Error bars represent standard deviation. (B) Mean z-score left ventricular end diastolic dimension (LVEDD) and endocardial shortening fraction (FS) in patients with DCM of distinct etiologies. Error bars represent standard deviation.
Clinical and Echocardiographic Findings in DCM Patients
The majority of DCM patients, regardless of etiology, presented with NYHA class IV symptoms (76%) (Table 4). Four patients presented without heart failure symptoms, 2 each in the syndromic and idiopathic groups. Arrhythmias were also common in the DCM patients, with 3 patients presenting with symptoms including palpitations, supraventricular tachycardia, and syncope with pre-excitation on electrocardiogram, and another 4 in whom rhythm disturbances complicated their clinical course. The phenotypic severity, as measured by left ventricular end-diastolic dimension (LVEDD) and FS, was similar for all groups except the syndromic group. In this group, there was a trend towards smaller LVEDD z-scores and less depressed shortening fraction (Figure 2B). Despite these findings, the syndromic group had poor outcomes with 2/5 patients dying during follow-up, one with SCD and one from heart failure.
Table 4.
Clinical Characteristics and Echocardiographic Findings in DCM Patients
| Familial n = 6 | Metabolic n = 4 | Syndromic n = 5 | Idiopathic n = 10 | |
|---|---|---|---|---|
| NYHA | ||||
| Class I | 0 | 0 | 2 | 2 |
| Class II | 0 | 0 | 0 | 0 |
| Class III | 1 | 0 | 1 | 0 |
| Class IV | 5 | 4 | 2 | 8 |
| Unknown | 0 | 0 | 0 | 0 |
| Echocardiography | ||||
| LVMWT z-score (Range) | 1.1 (−0.1 to 2.7) | 1.0 (0.2 to 2.6) | 0.8 (−0.2 to 2.2) | 1.7 (−0.4 to 3.9) |
| LVEDD z-score (Range) | 5.0 (4.1 to 6.8) | 3.9 (−0.7 to 8.0) | 2.7 (2.2 to 3.7) | 4.1 (2.4 to 7.8) |
| FS z-score (Range) | −9.5 (−13 to −5) | −8.7 (−11 to −5) | −5.3 (−7.8 to −1.5) | −8.4 (−12.7 to −0.6) |
| LA size z-score (Range) | 3.7 (1.6 to 5) | 2.8 (0.4 to 4.1) | 2.8 (1.9 to 3.9) | 1.9 (1.4 to 3.5) |
| Arrhythmia (%) | 1 (16.7%) | 2 (50%) | 2 (50%) | 2 (15.3%) |
| SCD (%) | 1 (16.7%) | 0 | 1 (25%) | 0 |
| ICD (%) | 0 | 0 | 1 (25%) | 1 (7.6%) |
| Transplant (%) | 0 | 2 (50%) | 0 | 1 (7.6%) |
| Deceased (%) | 1 (4%) | 0 | 2 (50%) | 0 |
NYHA = New York Heart Association functional class; LVMWT = Left Ventricular Maximal Wall Thickness; LVEDD = Left Ventricular End Diastolic Dimension; FS=Fractional Shortening; LA = Left Atrium; SCD = Sudden Cardiac Death; ICD = Implantable Cardiac Defibrillator.
Discussion
Predicting long-term prognoses and outcomes in children with cardiomyopathy remains a clinical challenge in part due to limited understanding of underlying etiology of disease in many patients. Improved diagnostic rates will allow the development of more disease-specific management. Using rigorous diagnostic criteria, approximately 75% of children with cardiomyopathy were assigned to a diagnostic group using clinical evaluation and improved clinical molecular genetic testing.
Familial Disease
Familial disease, defined as presence of an affected first degree family member and/or positive HCM or DCM gene panel mutation testing by a commercial laboratory, was identified in 42% of cardiomyopathy patients. To our knowledge, this is the first prevalence estimate for pediatric patients with cardiomyopathy across phenotypes since molecular genetic testing became clinically available. Given the increasing number of genes now tested on HCM or DCM gene panels, this likely represents a minimum estimate within our population. The high yield supports the utility of genetic testing in children with cardiomyopathy. Only 2 of 20 familial HCM cases were classified based on family history alone due to normal HCM gene panel testing. In contrast, 4 of 6 familial DCM cases were classified based on documented positive family history with either no genetic testing or normal results.
Two previous studies have addressed the yield of molecular testing specifically in pediatric HCM cases (19, 20) with discrepant findings regarding outcome in mutation positive versus mutation negative probands. In our study, mutation positive patients did not differ from mutation negative patients with respect to echocardiographic findings for any phenotypic group. As clinical testing identifies mutations in an increasing number of children, both pre-symptomatic and long term follow-up studies are necessary to better define natural history. Our positive testing rates in the HCM subset, in which greater than 80% of patients with an affected first degree relative and 43% of all HCM patients had an identifiable mutation, agrees with published studies using selected populations (65% and 55%(19); 61% and 53%(20)). In addition, 4/18 infantile HCM cases were familial, two of whom had identifiable sarcomeric gene mutations. This finding parallels those by Kaski et al. in which 16% of infants with HCM were positive for a sarcomeric gene mutation (20). These findings reinforce that infantile HCM can result from sarcomeric gene mutations, albeit potentially at lower rates than identified in older children.
Although it has been suggested that digenic inheritance or multiple sarcomeric mutations could explain early onset and severe disease in pediatric patients, only one cohort patient was identified with two mutations, a lower yield than the 6–14% rate quoted previously despite testing the same number of sarcomeric genes. A recent report on 4 patients with HCM, each with 3 distinct sarcomeric mutations, indicated that they presented as adults (29). Thus, further studies will be required to delineate the effect of multiple sarcomeric mutations on disease severity and progression. It is interesting to note that the most severely affected HCM infants in our study inherited their mutation from a mildly affected parent, indicating a role for environmental or genetic effects not testable using current molecular panels.
A sarcomeric or cytoskeletal gene mutation (13, 30) causes DCM in up to 20–35% of adults but a similar role for these mutations has not been defined yet in children. Our data demonstrate that pediatric DCM can be caused by sarcomeric gene mutations. The rate of positive molecular testing within the DCM familial group (33%) is consistent with recent studies in adult DCM patients(31). Taken together, 24% of all DCM patients were categorized as familial. This is higher than that seen in a registry review from Australia (15%) and from the PCMR (5%) (2, 3). Strikingly, we identified 100% familial disease in our RCM cohort, although our number of probands was small, consistent with the rare nature of this disease. In one patient, an MYH7 mutation previously shown to be disease causing in HCM (16) was identified by clinical testing. Two recent publications have also reported sarcomeric gene mutations in a small number of LVNC patients (17, 18). Our LVNC study group, which represents a population of isolated LVNC without accompanying dilation, hypertrophy or structural heart disease, had no mutations identified by HCM gene panel testing. However, 27% of these patients had pedigrees demonstrating autosomal dominant inheritance, suggesting that alternate genetic pathways are important for the development of this phenotype.
Genetic Syndromes and Metabolic Disease
This study identified significant rates of metabolic (19%) and syndromic disease (14.5%) in children with cardiomyopathy, illustrating the importance of a broad differential for clinical evaluation of children. This result was particularly striking for infants less than one year of age with HCM where 9/19 (47%) had a metabolic diagnosis and 4/19 (21%) had syndromic disease. These diagnoses allow for disease specific treatment, education on the natural history of disease, management of extracardiac manifestations and reproductive counseling for parents in a subgroup that historically has very poor outcomes.
Limitations of phenotypic classification
A number of patients in this study presented with “mixed” cardiomyopathy phenotypes. These patients present a difficult quandary in clinical practice due to the challenge of assigning them to a specific phenotypic subgroup and management pathway. Within our HCM group, 10% had abnormally elevated LVEDD measurements. Interestingly, all of these patients presented in infancy and had an underlying metabolic etiology. The majority of these patients presented in overt heart failure, potentially indicating patients of mixed phenotype as a high risk group that warrants further study. In our DCM group, 5 subjects (20%) had LVNC and 5 additional patients (20%) had LVMWT z-scores > 2. These overlapping phenotypes emphasize the complex presentations of cardiomyopathy and demonstrate the limitations of current clinical taxonomy.
Implications for Clinical Evaluation
Our data support the usefulness of clinical genetic testing. However, its improved availability did not entirely account for the higher rate of etiologic diagnoses in this study. Approximately 33% of HCM patients, 70% of DCM patients, and 27% of LVNC patients were identified as familial cases by eliciting a pedigree and obtaining recommended cardiac screening of first degree relatives. Our data support the recently published screening guidelines that recommend a 3 generation pedigree for new HCM, DCM, and RCM patients with the use of appropriate imaging and genetic screening in at risk family members (8).
In some cases, infants with syndromic or metabolic disease may not develop full manifestations of disease until childhood or later, making diagnoses more challenging. The inherent difficulties in correctly identifying an etiology in very young children further argue for a multidisciplinary approach to optimize early and accurate diagnosis. For pediatric patients with cardiomyopathy, an initial evaluation for syndromic or metabolic features and extra-cardiac manifestations is indicated, and periodic follow-up is recommended. Multidisciplinary clinics are useful to broaden the clinical evaluation, direct appropriate clinical testing, provide interpretation of results, and manage screening of at risk family members.
In summary, this study presents the first analysis of diagnostic rates in a diverse group of pediatric cardiomyopathy patients in an outpatient setting, and provides etiologic prevalence estimates. Current clinical genetic testing and a multidisciplinary approach can significantly improve diagnostic rates for children with cardiomyopathy. Cardiomyopathy is more diverse in children than adults, and proper identification of causes is necessary for improved cardiac treatment, extra-cardiac management, and family based risk assessment and screening. Improved diagnostic precision may allow future studies to address risk stratification, management, and long term outcome based on cause.
Supplementary Material
Acknowledgments
J.A.T. and S.M.W. are supported in part by NIH R01 HL087000-01A1 funding for the Pediatric Cardiomyopathy Specimen Repository. This work was supported by a grant from the Children’s Cardiomyopathy Foundation (S.M.W.).
Footnotes
Disclosures
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arola A, Jokinen E, Ruuskanen O, Saraste M, Pesonen E, Kuusela AL, et al. Epidemiology of idiopathic cardiomyopathies in children and adolescents. A nationwide study in Finland. Am J Epidemiol. 1997 Sep 1;146(5):385–93. doi: 10.1093/oxfordjournals.aje.a009291. [DOI] [PubMed] [Google Scholar]
- 2.Daubeney PE, Nugent AW, Chondros P, Carlin JB, Colan SD, Cheung M, et al. Clinical features and outcomes of childhood dilated cardiomyopathy: results from a national population-based study. Circulation. 2006 Dec 12;114(24):2671–8. doi: 10.1161/CIRCULATIONAHA.106.635128. [DOI] [PubMed] [Google Scholar]
- 3.Towbin JA, Lowe AM, Colan SD, Sleeper LA, Orav EJ, Clunie S, et al. Incidence, causes, and outcomes of dilated cardiomyopathy in children. JAMA. 2006 Oct 18;296(15):1867–76. doi: 10.1001/jama.296.15.1867. [DOI] [PubMed] [Google Scholar]
- 4.Colan SD, Lipshultz SE, Lowe AM, Sleeper LA, Messere J, Cox GF, et al. Epidemiology and cause-specific outcome of hypertrophic cardiomyopathy in children: findings from the Pediatric Cardiomyopathy Registry. Circulation. 2007 Feb 13;115(6):773–81. doi: 10.1161/CIRCULATIONAHA.106.621185. [DOI] [PubMed] [Google Scholar]
- 5.Grenier MA, Osganian SK, Cox GF, Towbin JA, Colan SD, Lurie PR, et al. Design and implementation of the North American Pediatric Cardiomyopathy Registry. Am Heart J. 2000 Feb;139(2 Pt 3):S86–95. doi: 10.1067/mhj.2000.103933. [DOI] [PubMed] [Google Scholar]
- 6.Nugent AW, Daubeney PE, Chondros P, Carlin JB, Cheung M, Wilkinson LC, et al. The epidemiology of childhood cardiomyopathy in Australia. N Engl J Med. 2003 Apr 24;348(17):1639–46. doi: 10.1056/NEJMoa021737. [DOI] [PubMed] [Google Scholar]
- 7.Judge DP. Use of genetics in the clinical evaluation of cardiomyopathy. JAMA. 2009 Dec 9;302(22):2471–6. doi: 10.1001/jama.2009.1787. [DOI] [PubMed] [Google Scholar]
- 8.Hershberger RE, Lindenfeld J, Mestroni L, Seidman CE, Taylor MRG, Towbin JA. Genetic Evaluation of Cardiomyopathy-A Heart Failure Society of America Practice Guideline. Journal of Cardiac Failure. 2009 Mar;15(2):83–97. doi: 10.1016/j.cardfail.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Bos JM, Towbin JA, Ackerman MJ. Diagnostic, prognostic, and therapeutic implications of genetic testing for hypertrophic cardiomyopathy. J Am Coll Cardiol. 2009 Jul 14;54(3):201–11. doi: 10.1016/j.jacc.2009.02.075. [DOI] [PubMed] [Google Scholar]
- 10.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006 Apr 11;113(14):1807–16. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 11.Marian AJ. Phenotypic plasticity of sarcomeric protein mutations. J Am Coll Cardiol. 2007;49:2427–9. doi: 10.1016/j.jacc.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morimoto S. Sarcomeric proteins and inherited cardiomyopathies. Cardiovasc Res. 2008;77:659–66. doi: 10.1093/cvr/cvm084. [DOI] [PubMed] [Google Scholar]
- 13.Moller DV, Andersen PS, Hedley P, et al. The role of sarcomere gene mutations in patients with idiopathic dilated cardiomyopathy. Eur J Hum Genet. 2009;17:1241–9. doi: 10.1038/ejhg.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaski JP, Syrris P, Burch M, et al. Idiopathic restrictive cardiomyopathy in children is caused by mutations in cardiac sarcomere protein genes. Heart. 2008;94:1478–84. doi: 10.1136/hrt.2007.134684. [DOI] [PubMed] [Google Scholar]
- 15.Mogensen J, Kubo T, Duque M, et al. Idiopathic restrictive cardiomyopathy is part of the clinical expression of cardiac troponin I mutations. J Clin Invest. 2003;111:209–16. doi: 10.1172/JCI16336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ware SM, Quinn ME, Ballard ET, Miller E, Uzark K, Spicer RL. Pediatric restrictive cardiomyopathy associated with a mutation in beta-myosin heavy chain. Clin Genet. 2008;73:165–70. doi: 10.1111/j.1399-0004.2007.00939.x. [DOI] [PubMed] [Google Scholar]
- 17.Xing Y, Ichida F, Matsuoka T, et al. Genetic analysis in patients with left ventricular noncompaction and evidence for genetic heterogeneity. Mol Genet Metab. 2006;88:71–7. doi: 10.1016/j.ymgme.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Dellefave LM, Pytel P, Mewborn S, et al. Sarcomere mutations in cardiomyopathy with left ventricular hypertrabeculation. Circ Cardiovasc Genet. 2009;2:442–9. doi: 10.1161/CIRCGENETICS.109.861955. [DOI] [PubMed] [Google Scholar]
- 19.Morita H, Rehm HL, Menesses A, et al. Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med. 2008;358:1899–908. doi: 10.1056/NEJMoa075463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaski JP, Syrris P, Esteban MT, et al. Prevalence of sarcomere protein gene mutations in preadolescent children with hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2009;2:436–41. doi: 10.1161/CIRCGENETICS.108.821314. [DOI] [PubMed] [Google Scholar]
- 21.Frisso G, Limongelli G, Pacileo G, et al. A child cohort study from southern Italy enlarges the genetic spectrum of hypertrophic cardiomyopathy. Clin Genet. 2009;76:91–101. doi: 10.1111/j.1399-0004.2009.01190.x. [DOI] [PubMed] [Google Scholar]
- 22.Bernier FP, Boneh A, Dennett X, Chow CW, Cleary MA, Thorburn DR. Diagnostic criteria for respiratory chain disorders in adults and children. Neurology. 2002;59:1406–11. doi: 10.1212/01.wnl.0000033795.17156.00. [DOI] [PubMed] [Google Scholar]
- 23.Munnich A, Rustin P. Clinical spectrum and diagnosis of mitochondrial disorders. Am J Med Genet. 2001;106:4–17. doi: 10.1002/ajmg.1391. [DOI] [PubMed] [Google Scholar]
- 24.Walker UA, Collins S, Byrne E. Respiratory chain encephalomyopathies: a diagnostic classification. Eur Neurol. 1996;36:260–7. doi: 10.1159/000117269. [DOI] [PubMed] [Google Scholar]
- 25.Ross RD, Bollinger RO, Pinsky WW. Grading the severity of congestive heart failure in infants. Pediatr Cardiol. 1992;13:72–5. doi: 10.1007/BF00798207. [DOI] [PubMed] [Google Scholar]
- 26.Rosenthal D, Chrisant MR, Edens E, et al. International Society for Heart and Lung Transplantation: Practice guidelines for management of heart failure in children. J Heart Lung Transplant. 2004;23(12):1313–33. doi: 10.1016/j.healun.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Lang RM, Bierig M, Devereux RB, et al. Recommendations for chamber quantification: a report from the American Society of Echocardiography’s Guidelines and Standards Committee and the Chamber Quantification Writing Group, developed in conjunction with the European Association of Echocardiography, a branch of the European Society of Cardiology. J Am Soc Echocardiogr. 2005;18:1440–63. doi: 10.1016/j.echo.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Cheitlin MD, Armstrong WF, Aurigemma GP, et al. ACC/AHA/ASE 2003 Guideline Update for the Clinical Application of Echocardiography: summary article. A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (ACC/AHA/ASE Committee to Update the 1997 Guidelines for the Clinical Application of Echocardiography) J Am Soc Echocardiogr. 2003;16:1091–110. doi: 10.1016/S0894-7317(03)00685-0. [DOI] [PubMed] [Google Scholar]
- 29.Girolami F, Ho CY, Semsarian C, Baldi M, Will ML, Baldini K, et al. Clinical features and outcome of hypertrophic cardiomyopathy associated with triple sarcomere protein gene mutations. J Am Coll Cardiol. 2010;55(14):1444–53. doi: 10.1016/j.jacc.2009.11.062. [DOI] [PubMed] [Google Scholar]
- 30.Hershberger RE, Norton N, Morales A, Li D, Siegfried JD, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3(2):155–61. doi: 10.1161/CIRCGENETICS.109.912345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hershberger RE, Morales A, Siegfried JD. Clinical and genetic issues in dilated cardiomyopathy: a review for genetics professionals. Genet Med. 2010 Nov;12(11):655–67. doi: 10.1097/GIM.0b013e3181f2481f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.