Summary
The importance of miRNAs during development and disease processes is well established. However, most studies have been done in cells or with patient tissues, and therefore the physiological roles of miRNAs are not well understood. To unravel in vivo functions of miRNAs, we have generated conditional, reporter-tagged knockout-first mice for numerous evolutionarily conserved miRNAs. Here we report the generation of 162 miRNA targeting vectors, 64 targeted ES cell lines, and 46 germline-transmitted miRNA knockout mice. In vivo lacZ reporter analysis in 18 lines revealed highly tissue-specific expression patterns and their miRNA expression profiling matched closely with published expression data. Most miRNA knockout mice tested were viable, supporting a mechanism by which miRNAs act redundantly with other miRNAs or other pathways. These data and collection of resources will be of value for the in vivo dissection of miRNA functions in mouse models.
Introduction
With the completion of the mouse and human genome sequencing, attention has shifted to deciphering the function of individual genes systematically to better understand human diseases. In fact, genome-wide gene targeting knockout approaches were taken for protein-coding genes by multiple programs including KOMP, EUCOMM, and NorCOM (Austin et al., 2004; Auwerx et al., 2004; Collins et al., 2007; Friedel et al., 2007). In 2007, these programs were consolidated under the International Knockout Mouse Consortium (IKMC) to facilitate these efforts more effectively (Skarnes et al., 2011).
Unfortunately, only protein-coding genes were included in this global effort because non-coding RNA gene knockouts had not captured the same level of attention at that time. However, the last decade has seen a surge of interest in microRNAs (miRNAs), given their roles in developmental and disease processes. These small RNAs of about 22 nucleotides can bind to target mRNAs based on sequence complementarity and direct post-transcriptional regulation of target gene expression. Computational analyses, microarrays, proteomics approaches, and high-throughput sequencing analyses suggest that one miRNA may regulate hundreds of targets. Given that as many as 500–800 miRNAs may exist in mammalian genomes, these data predict that nearly all genes are likely to be regulated by miRNAs at some level. If so, miRNA knockout mice might be expected to exhibit visible phenotypes, including developmental abnormalities and embryonic lethality.
Several recent studies have described knockout mouse phenotypes for a small number of individual miRNAs in mice and larger numbers of modified ES cells ((Prosser et al., 2011), and for review, see Park et al (Park et al., 2010)), suggesting that noncoding-RNAs deserve increased attention and should be considered for large-scale knockout efforts. Here, we report our ongoing progress on the generation of conditional reporter-tagged knockout-first mouse strains for miRNAs. This study focused on the generation of miRNA knockout mice, and provides a limited set of whole mount lacZ reporter expression analysis in embryos.
Results
Construct design and ES cell targeting
miRNA genes have a great diversity in their genomic organization. About one-half of mammalian miRNAs are expressed from introns of annotated protein-coding genes (intronic miRNAs), whereas the others are found outside the context of an annotated gene (intergenic miRNAs). Intronic miRNAs can be transcribed via the promoter of their host gene, or be transcribed from an intronic promoter; intergenic miRNAs have their own promoter. Regardless of their genomic location, miRNAs are frequently found as polycistronic clusters.
There are at least 450 well-conserved miRNAs between mouse and human residing at more than 166 alleles. In this pipeline we focused on the mouse:human conserved subset of microRNAs, conditionally deleting polycistronic miRNA loci when possible. Given the relatively large numbers of miRKO [microRNA knockout] vectors required for these types of projects, we embraced high-throughput recombineering protocols. For most of the vectors we utilized the knockout-first approach derived by Testa and colleagues (Testa et al., 2004), adapted to permit parallel preparation of many vectors in microtiter dish format (Fu et al., 2010; Nefedov et al., 2011). In this approach, sequential steps of genetic engineering do not require colony isolation and intermediate construct verification. We have generated 162 constructs targeting 194 miRNAs. This includes a second smaller subset of vectors targeting single miRNAs within a cluster (~30 miRNAs). So far we have generated conditional lacZ-reporter targeting vectors covering nearly 50% of the conserved miRNA genes.
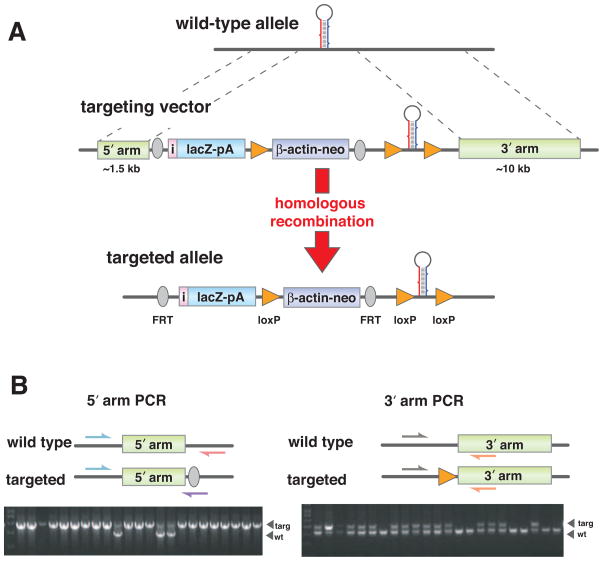
Cre-lox technology was used to generate conditional miRNA knockout mice to ensure that gene function could be studied in a tissue- and temporal-specific manner. LoxP sites approximately 200–250 bases 5′ and 3′ proximal to the miRNA precursor (Figure 1A, middle panel) were placed in the constructs to achieve efficient conditional deletion with a Cre recombinase enzyme. For polycistronic miRNAs, loxP sites were placed outside of the first and the last miRNA precursor in the cluster. The length of polycistronic miRNAs ranged from a few hundred to hundreds of thousands of bases. To maximize efficient Cre recombination, miRNAs were targeted for individual deletion if their resident cluster was comprised of more than 2000 bases. A short 5′-arm (~1–2 kb) to aid PCR genotyping and a long 3′-arm (~10–15 kb) to boost specific targeting of the allele were chosen for homologous recombination.
Figure 1. ES cell targeting and PCR genotyping.
(A) Schematic representation of a miRNA locus and targeting strategy. Two homology arms at the 5′ and 3′ ends of the targeting vector mediate gene-specific targeting by homologous recombination. Targeting leads to insertions of a promoter-less lacZ reporter with an IRES (in black color) and a polyA signal, β-actin-driven neomycin selection marker with a polyA signal, and a miRNA stem-loop flanked by loxP sites into the miRNA locus. (B) PCR genotyping strategy of targeted ES cells. Positions of the PCR primers used for genotyping wild type and targeted alleles are marked with arrows and expected size differences for PCR products are illustrated. To verify homologous recombination on the 5′ arm, PCR was done with 2 gene-specific primers and 1 universal mutant primer. For the longer 3′arm side, PCR was done to verify the insertion of the third loxP site with 2 gene-specific primers. The mutant products were verified by sequencing.
A key addition to the vector design was the inclusion of a promoter-less lacZ reporter and a neomycin selection marker between the 5′ arm and the floxed miRNA precursor sequence, which could be removed with a Flp recombinase (Figure 1A, middle panel). This insertion allowed for the selection of the integrated transgene, and importantly provided the lacZ readout for transcription of the miRNA.
The majority of miRNA expression data have been generated by quantitative RT-PCR (qPCR), microarrays, and sequencing analyses using pools of isolated cells or tissue biopsy. In mammals, in vivo expression data have been limited to the study of a small number of miRNAs, which were mainly studied during early embryo development by in situ hybridization or the use of in vivo miRNA ‘sensors’ (Mansfield et al., 2004; Wienholds et al., 2005). Due to the small size of miRNAs (22–23 nucleotides), the signal to background ratio of miRNA in situ hybridization is very narrow and, therefore, it is difficult to achieve strong and specific signals in whole mount tissues. For this reason, we opted to design a promoter-less lacZ reporter such that it would indicate the activity of the endogenous miRNA promoter. This allowed the temporal and spatial detection of primary miRNA transcriptional activity in mouse tissues or live cells. However, it is important to note that, similar to protein-encoding genes, transcriptional readout will not always be a true readout for the expression of the given gene, since miRNA expression may be regulated post-transcriptionally (Siomi and Siomi, 2010).
Thus far 194 microRNAs (corresponding to 162 constructs) have been electroporated into embryonic stem cells. PCR screening is used to identify the properly targeted ES clones (PCR strategy shown in Figure 1B). For the 5′ short arm, a three-primer PCR protocol was optimized to distinguish between non-targeted and targeted alleles. Given the technical challenge of long-range PCR across the entire ~10–15 kb long arm, the sequence integrity of the 3′-loxP site was verified as a proxy for 3′-end homologous recombination. PCR products from the targeted alleles were sequenced to validate the correct targeting. A diphtheria toxin gene was used as a negative selection marker, and Southern blot analyses were performed to ensure that there were no additional integrations at non-targeted loci (data not shown).
A 129/Ola-derived E14 ES cell line was used for targeting because of its history of highly efficient and reliable germ line transmission (Brenner et al., 2010). We screened 46 ES colonies per construct as a first-pass. This first-pass allowed us to identify loci that were easily targeted and yielded at least 1–2 ES cell lines. This constituted approximately 20% of the targeted loci; those that did not pass the first screen were retargeted until the correctly targeted line(s) were identified.
Generation of knockout mice
To produce reporter-tagged knockouts and conditional knockouts from the same allele, targeting constructs were engineered to contain a combination of FRT and loxP sites (Figure 2A). By crossing a germline-transmitted lacZ-neo-flox mouse with a germline Flp deleter, the reporter and the selection marker can be removed to restore a functional wild type allele (this constitutes a conditional allele, where two loxP sites flank the intact miRNA). Performing this step is critical for intronic miRNAs, as the presence of the lacZ-neo cassette could adversely affect host gene expression, potentially complicating the interpretation of the phenotype. Conditional mice can be crossed either with special Cre strains to generate temporal or tissue-specific knockouts or with a germline Cre-deleter strain to generate constitutive knockouts. Crossing a germline-transmitted lacZ-neo-flox mouse to a Cre deleter line (lacZ-knockout allele) resulted in a knockout line that also reported on the transcription of the miRNA locus. This may be desirable in studies where miRNA expression patterns or the fate of cells harboring a deleted miRNA need to be tracked.
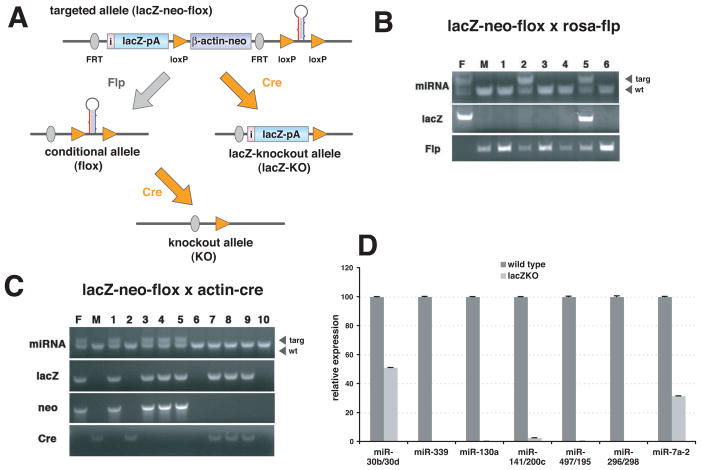
Figure 2. Generation of knockout mice.
(A) Breeding strategy. Combination of FRT and loxP sites allows manipulation of targeted allele in mice. Germline-transmitted mice (lacZ-neo-flox) can be crossed with germline deleter Flp mice, which will remove the reporter and the neomycin cassette to restore the wild type allele (conditional allele/flox). Then conditional mice can be crossed with either germline- or tissue-specific Cre transgenic mice to generate knockouts (knockout allele/KO). Instead of sequential recombination steps, germline-transmitted mice can be crossed with germline deleter Cre mice to produce mice with a reporter-tagged null allele (lacZ-knockout allele/lacZ-KO). (B) Flp-mediated recombination of FRT sites in mice. Germline-transmitted mice were crossed with homozygous Rosa-Flp mice. Parents and offspring were PCR-genotyped to check deletion of the lacZ reporter. Flp excision was not fully penetrant as the lacZ was not deleted in #5 pup, although it was completely excised in #2 pup (F and M indicate father and mother respectively). (C) Cre-mediated recombination of loxP sites in mice. Germline-transmitted mice were crossed with actin-Cre transgenic mice. Parents and offspring were PCR-genotyped to evaluate deletion of the neomycin marker, but not of the lacZ reporter. Excision by actin-Cre was complete among all pups (#7 to #9). (D) qPCR analysis to confirm the loss of miRNA in lacZ-KO mice. Either lung (miR-30b/30d, miR-339, miR-130a, miR-141/200c, miR-479/195 and miR-296/298) or brain (miR-7a-2) of 2 to 6 month-old knockout mice was used to isolate total RNA. RT-PCR was done by using Taqman miRNA assays. PCR were done as triplicates and data were presented as means with standard deviation.
To verify the functionality of the recombination sites in vivo, we performed mating with either Flp or Cre germline deleter mice (Figure 2B and 2C). While Actin-Cre mice were very efficient in transgene excision (Figure 2C), Rosa-Flp mice showed a reduced efficiency (Figure 2B). Actin-Flp also displayed a similar level of recombination activity (data not shown). Incomplete penetrance by Flp recombinases may have been caused by the fact that the distance between FRT sites (~7 kb) is greater than between loxP sites (~3 kb). In any case, we were able to obtain the designed recombination by evaluating a litter or two.
To assess whether miRNAs are essential for normal development in general, we generated knockout-first mouse lines for 11 intergenic miRNA lines (Table 1). Among 11 lacZ-KO lines for intergenic miRNAs tested, we observed embryonic lethality only in the miR-205 lacZ-KO line. To ensure that the surviving homozygous lacZ-KO mice were true knockouts lacking specific miRNAs, we performed qPCRs using lacZ-positive tissues (Figure 2D). We observed near-complete ablations with the exceptions of the miR-30b/30d cluster and miR-7a-2. The remaining signals for miR-30b/30d and miR-7a-2 were most likely generated from related family members, since Southern analysis showed the insertion of the lacZ cassette into specific loci (data not shown). In fact, the miR-30 family is represented by six members (miR-30a, miR-30b, miR-30c-1, miR-30c-2, miR-30d, and miR-30e) and the miR-7 family has three members (miR-7a-1, miR-7a-2 and miR-7b). To confirm the ablation of the targeted miRNAs from related family members, investigators must employ qPCR measuring pre-miRNAs or deep sequencing approaches in the future analysis of miRNA knockouts.
Table 1.
Analysis of embryonic lethality due to loss of miRNAs.
| line | +/+:Cre | −/−:Cre | Total # of pups | lethality |
|---|---|---|---|---|
| miR-7a-2 | 8 (5) | 7 (5) | 41 | no |
| miR-30b/30d | 7 (5) | 6 (5) | 39 | no |
| miR-130a | 6 (5) | 4 (5) | 42 | no |
| miR-141/200c | 4 (5) | 4 (5) | 43 | no |
| miR-146a | 4 (4) | 3 (4) | 33 | no |
| miR-205 | 8 (7) | 0 (7) | 56 | yes |
| miR-210 | 5 (5) | 5 (5) | 40 | no |
| miR-296/298 | 4 (5) | 4 (5) | 39 | no |
| miR-497/195 | 11 (5) | 4 (5) | 41 | no |
| miR-654/376b | 8 (5) | 3 (5) | 43 | no |
| miR-688 | 6 (5) | 6 (5) | 40 | no |
To determine if a miRNA gene (or a cluster) is essential during development, homozygous lacZ-KO mice were generated by intercrossing lacZ-neo-flox/+ and lacZ-KO/+: actin-Cre transgenic mice. About 40 offspring were genotyped and the expected number of animals was indicated inside parenthesis. +/+:Cre denotes wild type/wild type with Cre and −/−:Cre indicates lacZ-KO/lacZ-KO with Cre.
The observed 9% lethality in our screen (1/11) is within the lowest range of estimates of essential genes reported for mice (8~20% based on ENU-mutagenesis screens(Alvarez-Saavedra and Horvitz, 2010). Although the sample size is small, these data may suggest that deletion of most individual murine miRNAs do not lead to embryonic lethality or gross developmental defects. Similar findings have been reported for C. elegans (Miska et al., 2007).
Another important consideration relates to the allele context. For example, miR-301a exists as an intronic miRNA, thus the generation of lacZ-KO mice by Cre recombinase would be expected to introduce an intronic stop cassette, likely altering host gene (fam33a) expression. Since fam33a is a known protein-coding gene with potential roles in cell cycle and cell division processes (Hanisch et al., 2006), it is not possible to distinguish which gene(s) are responsible for embryonic lethality in this case. Thus, for ablation of intronic miRNAs, the appropriate experimental cross will be to employ actin-Flp recombination, followed by Cre-mediated knockout of the miRNA (Figure 2A).
Expression analysis in embryos
As an initial step towards understanding the in vivo function of miRNA genes, we performed expression analyses of the reporter in mouse embryos. The lacZ reporter can provide informative single cell expression patterns in complex tissue types such as the nervous and the immune system. To eliminate the interference of the neomycin cassette with miRNA transcription, we excised the neomycin cassette by crossing lacZ-neo-flox mice with actin-Cre mice (Figure 2A). The resulting lacZ-KO/+ males were crossed with wild type females, and the pregnant females were examined at two time points, 11.5 and 18.5 days post-coitum (E11.5 and E18.5). Embryos at E11.5 were chosen to study early developmental expression patterns, different germ layers, and overall body plan. Embryos at E18.5 were used to collect data on lacZ expression in major organs, including brain, sensory organs in the head, thymus, lung, heart, stomach, spleen, pancreas, intestine, liver, kidney, and bladder.
The lacZ reporter displayed distinctive patterns for different miRNAs and the expression patterns matched well to chicken and fish in situ hybridization patterns on a chicken embryo gene expression database (http://geisha.arizona.edu/geisha), as well as previous moderate- to high-throughput miRNA expression studies (Darnell et al., 2006; Kloosterman et al., 2006; Wienholds et al., 2005).
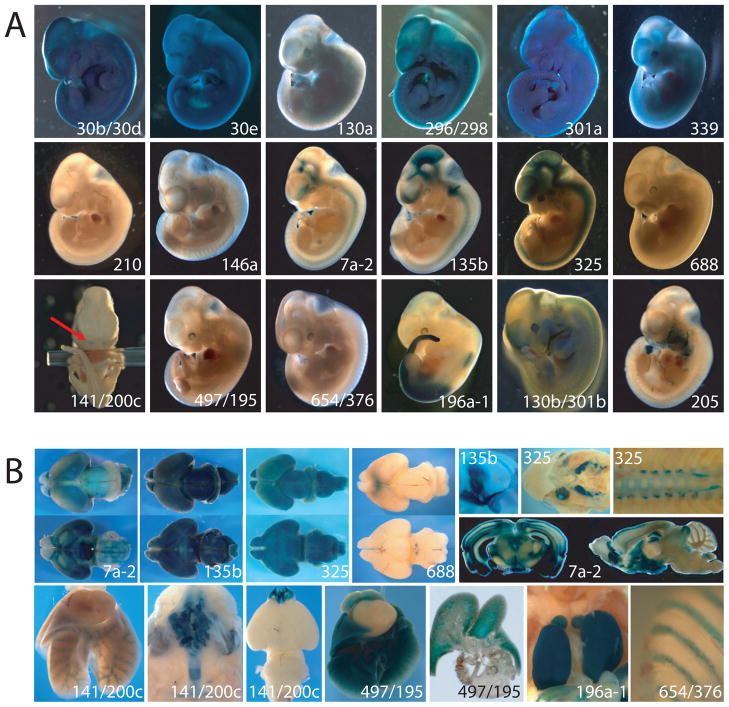
One third (6/18) of embryos displayed ubiquitous expression (for miR-30b/30d, miR-30e, miR-130a, miR-296/298, miR-301a, and miR-339), while approximately two thirds of the embryos showed distinct expression patterns. For miR-210 and miR-146a, we could not detect the lacZ reporter at either E11.5 or E18.5 stages (Figure 3 and data not shown). Expression of miR-210 has been reported in some cancers and hypoxic conditions (Camps et al., 2008; Giannakakis et al., 2008) and miR-146a is mainly observed in a subset of immune and cancer cells (Jazdzewski et al., 2009; Lu et al., 2010; Taganov et al., 2006), suggesting that they may not be expressed during normal development or they are expressed only in a subset of specialized tissues. In fact, the adult mice have shown lacZ activities in a subset of specialized immune cells (Figure S1 and unpublished data).
Figure 3. Expression analysis of miRNA lacZ reporter.
(A) LacZ reporter expression in E11.5 embryos. Embryos for miR-30b/30d, miR-30e, miR-130a, miR-296/298, miR-301a, and miR-339 displayed lacZ expression ubiquitously, whereas embryos for miR-210, miR-146a, mir-688, miR-497/195, and miR-654/376b exhibited no expression. The central nervous system was positive for miR-7a-2, miR-135b, and miR-325. Embryos for miR-141/200c showed staining only in the nostrils, as shown with arrows. Posterior trunk staining was detected for miR-196a-1. Although both expressed in pharyngeal arches and limb buds, expression patterns of miR-130b/301b and miR-205 were distinct. In addition, miR-130b/301b embryos displayed forebrain staining. (B) lacZ reporter expression in E18.5 embryos and adult. Brains of E18.5 embryos for miR-7a-2, miR-135b, and miR-325 displayed broad lacZ expression, whereas miR-688 exhibited very restricted expression domain in the dorsal cortex and ventral mid-brain (for each miRNA, top and bottom panels correspond to dorsal and ventral view respectively). Expression of miR-135b was also found in the inner ears (semicircular canals, vestibule and cochlea), while miR-325 expression was found in the visual sensory system (eyes, optic tracts, and lateral geniculate nucleus). The spinal cord staining was detected for miR-325. The CNS expression pattern of miR-7a-2 was maintained in adults as shown in the vibratome-sectioned brain. Expression of miR-141/200c was found in the airways of the respiratory tract, including in the nasal cavity, trachea, bronchi, and bronchioles, as well as in olfactory bulbs in the brain. Expression of miR-497/195 was found both in the airways and the air sacs of the lungs. Kidneys of miR-196a-1 displayed a striking lacZ staining pattern. For miR-654/376b, the frontal portion of the ribcage (corresponding to cartilage at this developmental stage) stained positive in E18.5 embryos.
We also conducted a limited but focused expression analysis in adult mice, investigating miRNA expression in defined subsets of hematopoietic cells isolated from tail blood using flow cytometry (Figure S1). This single cell-based reporter assay was able to distinguish lacZ activities for the closely related family members such as miR-30 family. The lacZ expression detected in peripheral blood lymphocytes correlated with published expression data in 22 of the 25 mouse strains tested. It is important to note that we unexpectedly did not detect significant lacZ activity for miR-150 in lymphocytes. We suspect that the flow cytometry assay may not be sufficiently robust to detect low levels of lacZ expression, or perhaps the lacZ reporter cassette is silent at this locus.
The expression of miRNAs varies dynamically in the brain both before and after birth in mice (Krichevsky et al., 2003; Miska et al., 2004). As expected, we found several miRNAs that were exclusively expressed in the central nervous system (miR-7a-2, miR-135b, miR-325, and miR-688). In addition to the central nervous system, we observed interesting reporter expression patterns in the respiratory system (miR-141/200c and miR-497/195). We performed limited histological analysis for a handful of lacZ-stained tissues either using vibratome sectioning or traditional paraffin-sectioning and confirmed that the lacZ reporter expression is very specific to a subset of cells in a given tissue and easily tractable with a single cell resolution (data not shown). Detailed expression profiling of tissue-specific miRNAs may shed light on the roles of miRNA genes in that sub-compartment. In addition, the reporter expression pattern will guide the selection of Cre lines for conditional knockouts.
In addition to differential spatial expression patterns, temporal regulation of miRNA expression was observed for several miRNAs in a variety of tissues (miR-688, miR-654/376b, and miR-130b/301b). A more detailed analysis of temporal expression changes may provide additional clues about their function.
Discussion
In this study, we generated a battery of novel miRNA reporter and conditional knockout mouse lines, and collected limited in vivo expression data from a subset of targeted lines. Investigators can use this resource to explore the phenotypic consequences of disruption of miRNAs. Our analysis of a limited (13) set of homozygous miRNA knockouts suggests that ablation of many miRNAs will be tolerated in the mouse. This would be consistent with the systematic ablation of miRNAs in C. elegans, where less than 10% of miRNAs give rise to embryonic developmental or grossly abnormal phenotypes but which may appear in sensitized backgrounds or under stress (Alvarez-Saavedra and Horvitz, 2010; Brenner et al., 2010; Miska et al., 2007). In part, our observations may be partially explained by the functional redundancy of multiple family members. Many miRNAs exist as duplicates or highly similar genes, and therefore deleting one of the members might not reduce expression of target genes below the threshold needed to exhibit phenotypic outcomes. Thus, it may be necessary to generate double, triple, or perhaps more complicated knockouts to totally ablate an entire miRNA family in vivo. In theory, it may be possible to use miRNA inhibitors to ablate all related family members (Krutzfeldt et al., 2005); however, the discrepancy between genetic ablation and the in vivo use of miRNA inhibitors has raised questions about this approach in mice (Patrick et al., 2010; Thum et al., 2008). In any case, genetic ablation is the gold standard for elucidation of gene function and large-scale projects such as this are possible using the pipeline described here.
This miRKO resource should be of value to the community using mouse models to study human diseases. For example, the role of miRNAs in human cancers has been well established during the past decade; there is a veritable need to ablate specific oncomiRs and modifiers of tumorigenesis. Having a lacZ reporter might allow the monitoring of miRNA expression in tumors. In addition, given that these lines are conditional, investigators will have a more powerful tool to perform coincident ablation of multiple genes in tissue-specific contexts. This will allow a more precise dual ablation of genes that would render developmental phenotypes, also aiding miRNA studies in adult tissues and tumor contexts.
This project is continuing to generate additional data. Generated resources including targeting vectors, ES cell lines, and mice will be available to researchers via UC Davis KOMP Repository (https://www.komp.org/index.php) and the Jackson Laboratory (http://jaxmice.org). The active pipeline can be viewed online at the miRKO database (http://rna.keck.ucsf.edu), where each miRNA can be tracked from plasmid production stage to the generation of mice and the deposition of mice to the Jackson Laboratory. Vector maps, ES cell data, high-resolution reporter expression data, and protocols are available for download.
Experimental Procedures
Knockout Vector Construction
Vectors were constructed via a previously described ‘knockout-first’ approach (Testa et al., 2004). Two versions of this approach were implemented to generate knockout vectors from C57BL/6J BACs using a high-throughput recombineering pipeline essentially as described (Fu et al., 2010). For the first ten vectors, the process was started by selecting BAC clones from the CHORI-39 library (http://bacpac.chori.org) by a simultaneous selection and first modification step (Nefedov et al., 2011). The CHORI-39 library has been created in a BAC vector (pBAC-RED) which include the lambda recombineering genes (exo, bet and gam) under control of a thermosensitive cI repressor. All subsequent KO vectors were prepared by the more efficient pipeline developed for the KOMP and EUCOMM projects. In essence, BAC clones were selected from the RPCI-23 and RPCI-24 BAC libraries(Osoegawa et al., 2000) using bio-informatics. All recombineering steps were performed using arabinose-inducible recombineering functions provided by plasmid pSC101-BAD-gbaA-tet(Fu et al.). Although the two approaches provided recombineering functions either in cis or in trans, all other aspects of the process were very similar and made use of the same basic modification cassettes.
Generation of miRNA transgenic mice
E14, a substrain of 129/Ola, was electroporated with targeting constructs and electroporated cells were under G418 selection (250 μg/ml) for one week. Surviving colonies were picked and PCR-genotyped to identify recombined alleles. Correctly targeted ES cells were injected into C57BL/6 blastocysts to generate chimeras. Two to three Chimeras with higher than ~70% coat color chimerism were mated with either wild type or actin-Cre C57BL/6 females to achieve germline transmission. Total of 39 chimeric lines out of 41 tested contributed to colonize germline mice when they have higher than 70% coat color chimerism.
Supplementary Material
Highlights.
Large numbers of miRNAs are conditionally ablated in mouse models
Mouse miRNAs act redundantly with other miRNAs or other pathways
An integrated lacZ reporter gives rich in situ patterns reflecting miRNA expression
Acknowledgments
This work was supported by a grant from the W.M. Keck Foundation to AA, AW, LLL, JAB, and MTM and the NIH 5R24HL092851 to DS, PJD, and MTM. LTJ was supported by a fellowship from the Swiss National Science Foundation PBBS33-118644 and SSMBS PASMP3-124274/1 and a JDRF Scholar Award to JAB. LLL is an American Cancer Society Professor. We thank Dimitri De Kouchkovsky for assistance with mouse work and the Gladstone transgenic core for assistance with blastocyst injection. We thank members of the McManus laboratory and the UCSF Diabetes Center for their help and support throughout the project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, et al. The knockout mouse project. Nat Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P, et al. The European dimension for the mouse genome mutagenesis program. Nat Genet. 2004;36:925–927. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner JL, Jasiewicz KL, Fahley AF, Kemp BJ, Abbott AL. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr Biol. 2010;20:1321–1325. doi: 10.1016/j.cub.2010.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Darnell DK, Kaur S, Stanislaw S, Konieczka JH, Yatskievych TA, Antin PB. MicroRNA expression during chick embryo development. Dev Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- Friedel RH, Seisenberger C, Kaloff C, Wurst W. EUCOMM--the European conditional mouse mutagenesis program. Brief Funct Genomic Proteomic. 2007;6:180–185. doi: 10.1093/bfgp/elm022. [DOI] [PubMed] [Google Scholar]
- Fu J, Teucher M, Anastassiadis K, Skarnes W, Stewart AF. A recombineering pipeline to make conditional targeting constructs. Methods Enzymol. 2010;477:125–144. doi: 10.1016/S0076-6879(10)77008-7. [DOI] [PubMed] [Google Scholar]
- Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, Li C, O’Brien-Jenkins A, Katsaros D, Weber BL, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7:255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. Embo J. 2006;25:5504–5515. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, Jarzab B, de la Chapelle A. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci U S A. 2009;106:1502–1505. doi: 10.1073/pnas.0812591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. Rna. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS genetics. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nefedov M, Carbone L, Field M, Schein J, de Jong PJ. Isolation of specific clones from nonarrayed BAC libraries through homologous recombination. J Biomed Biotechnol. 2011;2011:560124. doi: 10.1155/2011/560124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa K, Tateno M, Woon PY, Frengen E, Mammoser AG, Catanese JJ, Hayashizaki Y, de Jong PJ. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000;10:116–128. [PMC free article] [PubMed] [Google Scholar]
- Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum Mol Genet. 2010;19:R169–175. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest. 2010;120:3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser HM, Koike-Yusa H, Cooper JD, Law FC, Bradley A. A resource of vectors and ES cells for targeted deletion of microRNAs in mice. Nat Biotechnol. 2011 doi: 10.1038/nbt.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional Knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa G, Schaft J, van der Hoeven F, Glaser S, Anastassiadis K, Zhang Y, Hermann T, Stremmel W, Stewart AF. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38:151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
- Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.