Summary
Marine life forms are an important source of structurally-diverse and biologically-active secondary metabolites, several of which have inspired the development of new classes of therapeutic agents. These success stories have had to overcome difficulties inherent to natural products-derived drugs, such as adequate sourcing of the agent and issues related to structural complexity. Nevertheless, several marine-derived agents are now approved, most as `first-in-class' drugs, with 5 of 7 appearing in the past few years. Additionally, there is a rich pipeline of clinical and pre-clinical marine compounds to suggest their continued application in human medicine. Understanding of how these agents are biosynthetically assembled has accelerated in recent years, especially through interdisciplinary approaches, and innovative manipulations and re-engineering of some of these gene clusters are yielding novel agents of enhanced pharmaceutical properties compared with the natural product.
Introduction
Familiar to all, the coastal margin of the world's oceans are extraordinarily rich in species diversity, especially in tropical environments. The last 40 years has seen a remarkable exploration of these unique organisms for their structurally diverse natural products. Early on, this was motivated by a need to detect and understand the chemical basis of various toxins with which people came into contact, such as saxitoxin, tetrodotoxin and the brevetoxins (Grindberg et al., 2008), and subsequently from a fundamental interest to describe the unique adaptations that marine life possess in this regard. For example, a distinctive and pervasive feature of these is the incorporation of halogen atoms, bromide or chloride, covalently attached to organic compounds (Molinski et al, 2009; Villa and Gerwick, 2010; Sashidhara et al, 2009). However, as the field has progressed, efforts have focused more on biomedical screening programs, and now several drugs have been developed from leads generated from marine natural products (Newman and Cragg, 2011). More recently, the enzymatic and genetic basis by which these unique secondary metabolites arise has become a focal point with an ultimate vision of capturing and harnessing these unique biochemical catalysts so as to create molecular diversity of biomedical and agrochemical utility (Gulder and Moore, 2009). In this perspective review, we trace these developments in the field of marine natural products drug discovery and chemical biology, describe some of the success stories of these efforts, and then provide some vision of the future of this area of investigation.
Tracing the history of marine natural products drug discovery
The early days of marine natural products discovery efforts focused on those organisms most conspicuous and easily collected. Indeed, before the advent of SCUBA, exploration of the marine world languished behind that of the terrestrial world for exactly this cause; an inability to access marine life beyond the intertidal. Thus, the large and showy creatures of the sea, such as red algae, sponges, and soft corals, were early on shown to produce a multitude of quite unique molecular species, such as highly halogenated terpenes and acetogenins from Rhodophytes such as Laurencia (Suzuki and Vairappan, 2005), the highly toxic polyketide `tedanolide' from the fire sponge Tedania ignis (Taylor, 2008), and prostaglandins from the gorgonian coral Plexaura homomalla (Weinheimer, 1974). With continued exploration, other groups of organisms, some more cryptic in space and time, were studied, and the repertoire of unique molecular species grew. By the turn of the century, marine natural products had become an established sub-discipline of natural products chemistry, and several thousand compounds had been described. Some groups of marine organisms were fairly well characterized for their major metabolites, and more often known compounds or their close relatives were being re-encountered (Faulkner, 2002).
Thus, attention turned to smaller and smaller creatures that had previously escaped collection and examination, such as marine cyanobacteria, marine fungi, and diverse other groups of marine eubacteria. This emphasis on microorganisms has been rewarded with a wealth of new natural products chemistry, as well as the realization that many compounds previously isolated from macroorganisms, such as sponges and tunicates, are actually metabolic products of associated microbes (Piel, 2009). As a result, a number of groups around the world have sought to culture marine bacteria from various sources, including shallow and deep water sediments, animate as well as inanimate surfaces, and from within the tissues of other macroorganisms (Williams, 2008). Productive as this has been, it has still only begun to sample the phylogenetic richness present within microbial groups in the sea. From seawater alone, it is estimated that only 1% of bacteria present have been cultured, and from culture independent methodologies, many major lineages remain without any or only a few cultured members. From another perspective, while Actinomycetes and Myxobacteria are the richest terrestrial groups of bacteria in terms of number and diversity of natural product structures, these microbes were largely unrecognized in the world's oceans until recently, and exploration of the extent and diversity of these two groups in the sea is ongoing (Fenical and Jensen, 2006; Schaeberle et al., 2010).
Combined with the exploration of marine microbial life forms, largely by fermentation methods, have been improvements in various approaches and strategies for interfacing natural products with modern biology and screening platforms. Automated extraction protocols coupled to prefractionation strategies have enhanced the quality and number of materials being evaluated in biological assays (Johnson et al., 2010; Bugni et al, 2008). In particular, the pre-fractionation of extracts to single compounds or reduced complexity mixtures enhances hit rates, due to the concentration of low abundance actives, and accelerates the identification process as the active compound is already pure or nearly so. However, there are two long standing schools of thought on natural products discovery: `isolate and then test' versus `test and then isolate', and both have a historical track record of success. Thus, a fusion approach is commonly employed wherein extracts or fractions are tested for bioactive compounds, and if a “strong” activity is detected, then bioassay-guided approaches are used. On the other hand, profiling of the material by 1H NMR and/or LC-ESIMS for unique chemical constituents can be followed by their isolation and broad evaluation in diverse biological assays. The strategies in biological assays have similarly shifted, from at one time in vivo screens to in vitro cell assays to isolated protein biochemical screens, and most recently, to high content phenotypic assays (Swinney and Anthony, 2011). The great gains achieved in throughput have resulted in large part from advances in automation, however, some of these approaches have emerged to be overly costly and lacking in flexibility; it is predicted that future development in HTS will focus more on quality of assays and their physiological relevance, rather than further miniaturization (Mayr and Bojanic, 2009). Thus, the newest approaches accentuate flexibility and cost effectiveness matched with quality test compounds being evaluated in new and intriguing areas of disease biology. Final analytical HPLC scale purifications on small samples can occur very rapidly to yield sufficient compound for structure elucidation by very sensitive MS and NMR instrumentation (dubbed `nanoscale structure elucidation') (Molinski, 2010), such that leads are generated from materials available on only the microgram scale. These data, coupled with informative databases of natural products that contain analytical information (e.g. SciFinder, AntiMarine, MarinLit, ChemSpider) allows for the rapid identification of compounds as being previously known, the so-called process of “dereplication”, or representing new chemical entities.
However, an obvious consequence and issue that has emerged from finding and determining active molecules on an ever smaller scale is that there is not sufficient compound produced to adequately explore their biological properties. Oftentimes, samples are obtained in this small scale because the producing organisms are present in small natural abundance and are difficult or impossible to recollect. Thus, supply problems emerge at an even earlier stage in the drug discovery process, and underscore the critical importance of synthetic and biosynthetic methods to re-supply compounds and analogs. Moreover, structures determined at the nanoscale, just as for compounds elucidated when more plentiful, should be confirmed for structural and stereochemical accuracy by independent methodologies, such as total chemical synthesis (Nicolaou and Snyder, 2005). Finally, while chemoinformatics and in silico screening of natural products to protein targets has occurred to some extent, this is a relatively little developed aspect of the field and thus represents an exciting frontier, as described further below.
Origins and development of marine chemical biology
The molecular revolution has forever changed the face of marine natural products chemistry (Lane and Moore, 2011). With the explosion of numerous “omic” approaches – genomics, proteomics, metabolomics, transcriptomics – now employed by marine natural product practitioners, new areas of science and inquiry have emerged over the past decade that begin to answer long-term questions in the field and provide fresh ideas for new research directions. The bridging of natural products chemistry with modern molecular biology has empowered researchers with the ability to address fundamental questions about the biosynthetic capacity of marine organisms, the synthetic role of microbes in marine invertebrate natural product chemistry, and the bioengineering potential of marine drugs. In this section, we highlight recent trends in marine natural product research that have been impacted by modern omic approaches with a focus on small molecule biosynthesis.
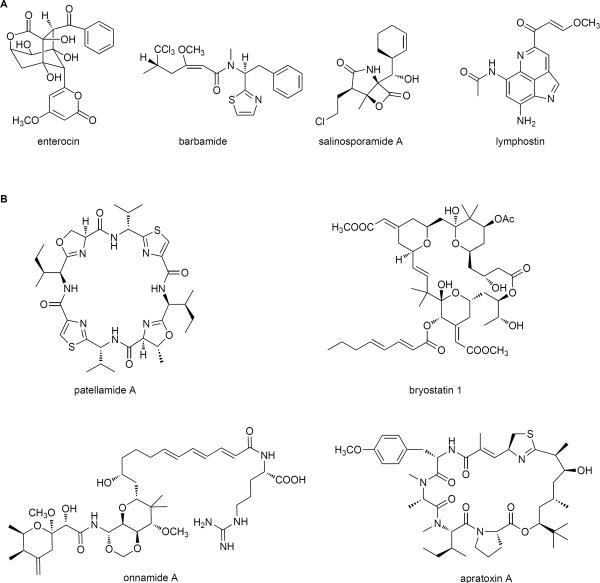
Without a doubt, marine organisms synthesize a plethora of small molecules with fascinating chemical structures and potent biological properties. Early biosynthetic knowledge developed primarily from isotope tracer experiments involving a wide range of marine microbes, algae, and invertebrates (Moore, 2005; Moore, 2006). These studies often revealed biosynthetic strategies in marine organisms distinct from their terrestrial counterparts. At the turn of the 21st century, the molecular basis of marine microbial natural product biosynthesis was first established for the streptomycete antibiotic enterocin (Piel et al, 2000) and the cyanobacterial agent barbamide (Chang et al, 2002) based on the targeted cloning and sequencing of their respective biosynthetic gene clusters (Figure 4). This targeted gene sequencing approach to the identification of secondary metabolic pathways relied upon the ability to generate specific gene probes that in some instances were time-consuming to develop and at other times misleading to implement. Nonetheless, over the past 10 years numerous discoveries were established of natural product biosynthetic gene sets from laboratory-cultured marine isolates to field-collected samples (Lane and Moore, 2011).
Figure 4.
Examples of marine natural products from (A) laboratory cultured and (B) environmental uncultured marine microbes whose biosynthetic pathways have been established by a variety of omic approaches (includes ecteinascidin-743 shown in Figure 3).
With the emergence of faster and cheaper DNA sequencing protocols, it is now much more efficient to sequence entire genomes rather than specific gene sets, especially when searching for biosynthetic pathways with little biochemical precedence or exploring organisms with multiple pathways of interest. Not only does genome sequencing provide key molecular information about how, when and sometimes why a natural product is assembled, it also bestows additional information about precursor supply networks, competing or complementary metabolic pathways, and basic genetic traits of the producing organism, inter alia. The first completed marine actinomycete genome sequence was for the 5,183,331-bp Salinispora tropica in 2007 and revealed the molecular basis of at least 17 natural product biosynthetic pathways spanning approximately 10% of the genome (Udwary et al., 2007). Over the past few years, knowledge of the benthic S. tropica genome facilitated the molecular cloning of the salinosporamide (Eustaquio et al., 2009), sporolide (McGlinchey et al, 2008), and lymphostin (Miyanaga et al, 2011) pathways, the discovery of the novel macrolactam salinilactam A (Udwary et al., 2007), a new salinosporamide analog (Eustaquio et al., 2011) and numerous biosynthetic enzymes, and the linking of natural products to functional adaptation concepts in the Salinispora genus (Penn et al, 2009). This example highlights the far-reaching translational opportunities of having access to a completed genome in being able to speed up the rate of scientific discovery.
More recently, the draft genome of the benthic filamentous cyanobacterium Moorea producta 3L (formally Lyngbya majuscula; Engene et al., 2011), a well-known producer of complex natural products such as barbamide (Chang et al., 2002) and curacin A (Chang et al., 2004), was found to harbor several surprises, including a relatively limited number of natural product pathways (Jones et al., 2011). Its 8.5 Mbp draft genome supports approximately 293 kbp of DNA sequence in secondary metabolism, far too low to encode the full suite of reported M. producta metabolites of ca. 200 different compounds. This maiden genome sequencing project suggests that the rich complement of natural products in M. producta will ultimately be strain specific.
Genome sequencing of organisms not normally associated for their biosynthetic prowess, such as the pelagic cyanobacteria Trichodesmium erythraeum ISM101 and Prochlorococcus MIT9313, has enabled through sequence gazing, or “genome mining”, the hypothesis-driven discovery of the postranslationally-modified ribosomal peptides trichamide (Sudek et al., 2006) and the prochlorosins (Li et al., 2010), respectively. This genomics-driven discovery of new chemical entities is a key, first step in understanding the specialized chemical biology of these important planktonic organisms. The prochlorisins represent a fascinating finding from a biosynthetic perspective in which a diverse collection of polycyclic, conformationally constrained lantipeptides are efficiently synthesized from up to 29 different ribosomally synthesized peptide precursors by the action of a single promiscuous processing enzyme (Li et al., 2010). In this way, although the genome size of P. MIT9313 is a modest 2,410,873 bp (Rocap et al., 2003), it still has the capacity to synthesize a collection of specialized chemicals more often associated with antibiotic-producing streptomycetes whose genomes are several times larger (Nett et al., 2009).
Marine macroorganisms, such as sponges, tunicates and bryozoans, have long been known to harbor natural products of biomedical significance that structurally resemble compounds produced by terrestrial microorganisms (Piel, 2004). This structural resemblance for decades begged the question of the biosynthetic source of invertebrate natural product chemistry, yet went unanswered due to difficulties in the isolation and culturing of community-based, natural product-producing microbes. Early experimental work involving cell sorting and in situ hybridization studies helped establish the symbiosis hypothesis (Haygood et al., 1999), yet unequivocal proof was not ultimately achieved until metagenomic approaches confirmed the central importance of bacteria in marine natural product biosynthesis (Piel, 2009).
The success of the metagenomic method is based on the relatively unbiased sequence analysis of total environmental DNA, which in the examples discussed here involves the marine organism and its associated microflora. Large clonal libraries often numbering in the millions are prepared and screened by phenotype or genotype to identify biosynthetic genes or pathways of interest (Piel, 2011). However, due to the large size of natural product biosynthetic gene clusters that make them difficult to heterologously express, genotype screens are often preferred over phenotype screens. In this way, the bacterial origin and molecular basis were established for the pederin family of polyketide antitumor agents [onnamide (Piel et al., 2004) and psymberin (Fisch et al, 2009)] from sponges, the bryostatin macrolide anticancer agents from bryozoans (Sudek et al., 2007), and the cyanobactin family of posttranslationally modified ribosomal peptides from ascidians (Schmidt et al., 2005). Onnamide, psymberin and bryostatin all share a common biosynthetic strategy involving large, trans-AT PKSs that to date have precluded their heterologous expression; this is in contrast to the patellamide and other cyanobactin ribosomal peptides whose biosynthetic gene clusters are considerably smaller allowing not only for functional expression but also for their genetic remodeling (Donia et al., 2008). These seminal studies confirm the power of metagenomics in capturing the genetic blueprint of natural product biosynthesis in marine invertebrates and point toward the future biotechnological approaches in helping solve the supply problem that often limits marine natural products from clinical development.
Some marine drug discovery success stories
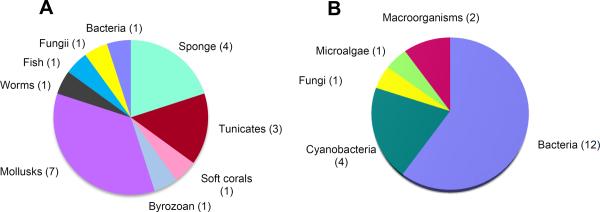
To date, there are seven therapeutic agents (four anticancer, one antiviral, one pain control, and one for hypertriglyceridemia) which derive from the marine environment in some sense (Mayer et al., 2010). Of these, two are the actual chemical structure as isolated, and five others are synthetic agents which capture their chemical idea from a marine product. In addition, a further 13 agents are in phase I, II or III clinical trials (Table 1). Figure 1a summarizes the collected sources of organisms that have yielded these agents, and reveals that mollusks, sponges, and tunicates are the richest collected source of these most valuable to substances. However, as noted above, the collected source has oftentimes been shown or is strongly suspected of harboring or feeding upon microorganisms that are the actual producers of the bioactive agent. Figure 1b displays the actual or suspected metabolic source of the most important agents and reveals that heterotrophic bacteria and cyanobacteria are the real metabolic jewels of the world's oceans, accounting for fully 80% of these clinical trial and approved pharmaceutical agents.
Table 1.
FDA-approved and Clinical Trial Agents Originating from (6 Agents) or Inspired by (14 Agents) Marine Natural Products with Details on Collected Source, Predicted Biosynthetic Source, Molecular Target and Disease Treated (modified from Mayer et al., 2010 and Newman and Cragg, 2011).
| Clinical Status | Compound Name | Natural Product or Derivative | Collected Source Organism | Predicted Biosynthetic Source | Biosynthetic Class of Agent | Molecular Target | Disease Area |
|---|---|---|---|---|---|---|---|
| FDA-Approved | Cytarabine (Ara-C) | Derivative | Sponge | Bacterium | Nucleoside | DNA Polymerase | Cancer |
| FDA-Approved | Vidarabine (Ara-A) | Derivative | Sponge | Bacterium | Nucleoside | Viral DNA polymerase I | Antiviral |
| FDA-Approved | Ziconotide | Natural Product | Cone Snail | Mollusk | Cysteine Knot Peptide | N-type Ca channel | Pain |
| FDA-Approved | Eribulin Mesylate (E7389) | Derivative | Sponge | Bacterium | Complex Polyketide | Microtubules | Cancer |
| FDA-Approved | Omega-3-acid ethyl esters | Derivative | Fish | Microalgae | Omega-3 fatty acids | Triglyceride-synthesizing enzymes | Hypertriglyceridemia |
| FDA-Approved | Trabectedin (ET-743) (EU registered only) | Natural Product | Tunicate | Bacterium | NRPS-derived Alkaloid | Minor groove of DNA | Cancer |
| FDA-Approved | Brentuximab vedotin (SGN-35) | Derivative | Mollusk | Cyanobacterium | Antibody drug conjugate (MM auristatin E) – Linear NRPS/PKS | CD30 & microtubules | Cancer |
| Phase III | Plitidepsin (Aplidine) | Natural Product | Tunicate | Bacterium | Cyclic Depsipeptide | Rac1 & JNK activation | Cancer |
| Phase II | DMXBA (GTS-21) | Derivative | Worm | Worm? | Alkaloid | α7 nicotinic acetylcholine receptor | Cognition, Schizophrenia |
| Phase II | Plinabulin (NPI 2358) | Derivative | Fungus | Fungus | Diketopiperazine | Microtubules & JNK stress protein | Cancer |
| Phase II | Elisidepsin | Derivative | Mollusk | Bacterium | Cyclic Depsipeptide | Plasma membrane fluidity | Cancer |
| Phase II | Zalypsis (PM00104) | Derivative | Nudibranch | Bacterium | Alkaloid | DNA binding | Cancer |
| Phase II | Glembatumumab vedotin (CDX-011) | Derivative | Mollusk | Cyanobacterium | Antibody drug conjugate (MM auristatin E) – Linear NRPS/PKS | Glycoprotein NMB & microtubules | Cancer |
| Phase I | Marizomib (Salinosporamide A; NPI-0052) | Natural Product | Bacterium | Bacterium | NRPS with β-lactone & γ-lactam | 20S proteasome | Cancer |
| Phase I | Trabectedin analog (PM01183) | Derivative | Tunicate | Bacterium | NRPS Alkaloid | Minor groove of DNA, Nucleotide Excision Repair | Cancer |
| Phase I | SGN-75 | Derivative | Mollusk | Cyanobacterium | Antibody drug conjugate (MM auristatin F) – Linear NRPS/PKS | CD70 & microtubules | Cancer |
| Phase I | ASG-5ME | Derivative | Mollusk | Cyanobacterium | Antibody drug conjugate (MM auristatin E) – Linear NRPS/PKS | ASG-5 & microtubules | Cancer |
| Phase I | Hemiasterlin derivative (E7974) | Derivative | Sponge | Bacterium | Modified linear tripeptide (NRPS-PKS) | Microtubules | Cancer |
| Phase I | Bryostatin 1 | Natural Product | Bryozoan | Bacterium | Polyketide | Protein kinase C | Cancer, Alzheimer's |
| Phase I | Pseuclopterosins | Natural Product | Soft Coral | Bacterium? | Diterpene glycoside | Eicosanoid metabolism | Wound healing |
Figure 1.
A) Pie chart illustrating the original collected sources of marine natural product derived or inspired agents currently as approved drugs or in clinical trials (20 total). B) Pie chart of the marine-derived drugs and clinical trial agents divided by their subsequently shown or predicted source organisms (20 total).
An early finding of the marine drug discovery field was that the Caribbean sponge Cryptotethia crypta possessed metabolites with a very interesting but relatively simple modification of a nucleoside; the normal 2-deoxyribose ring of deoxythymine and uracil are replaced by β-D-arabinofuranose (Bergman et al., 1957). Ensuing biological and medicinal chemistry exploration of the impacts of such a subtle alteration revealed that cytosine arabinoside was a potent disrupter of DNA replication and led to cellular toxicity (Brunton et al., 2011), while the arabinoside derivative of adenosine had antiviral effects (Sagar et al., 2010). Metabolic activation of Ara-C to the corresponding triphosphate yields a substrate mimic of deoxycytidine 5'-triphosphate, and following incorporation into the DNA backbone, inhibits the DNA polymerase as well as DNA repair enzymes. While Ara-C has found greatest utility in inducing remissions of acute myelocytic leukemia, more significantly these discoveries from nature helped illuminate nucleoside chemistry as a viable therapeutic strategy that later gained favor in antiviral chemotherapy.
Soon after the discovery of unusual nucleoside sponge-based chemistry, potent anticancer activity was detected in extracts of the tunicate Ecteinascidia turbinata. However it would be nearly 30 years until the structure of the active compound, ecteinascidin 743 (ET-743, = trabectedin), was finally elucidated (Rinehart et al., 1990; Wright et al., 1990). Commercialized by Pharmamar as Yondelis®, its development was severely hampered by lack of compound supply. Ultimately, this need was met by semi-synthesis from the microbial natural product, cyanosafracin B, a fermentation product of the bacterium Pseudomonas fluorescens (Cuevas et al., 2000). On cells, the mechanism of action of ET-743 interacts with DNA, and results in a G2/M block and ensuing p53-independent apoptosis. The subcellular target has been identified as the minor groove of DNA; once bound, it interferes with functioning of the Nucleotide Excision Repair system so as to bring about a cytotoxic effect (Soares et al., 2010). The utility of Yondelis® so far has been in the treatment of soft tissue sarcomas and relapsed ovarian cancer, although clinical trials are ongoing with other tumor types.
A third marine anticancer agent success story results from the 1986 discovery of the polyether metabolite halichondrin A from the sponge Halichondria okadai (Hirata and Uemura, 1986). The natural product was subsequently shown to possess exquisite cancer cell toxicity through an antitubulin mechanism, and perhaps more importantly, have a new mechanism of action by binding near the vinca site on β-tubulin but having rather different biochemical effects, including microtubule dynamics, as compared with other agents (Jordan et al., 2005). During the course of synthetic efforts in this area, it was found that the macrocyclic portion of the molecule was responsible for the potent biological activity, and this could be enhanced through modification of the macrolactone functionality to a macrocarbocyclic ketone, giving rise to eribulin (Yeung, 2011). While simplified in structure compared to the natural product, its commercial chemical synthesis is nothing short of heroic with an overall molecular size of 731 daltons and 19 chiral centers. Eisai Pharmaceuticals in the US recently launched this agent under the product name Halaven®, and it is quickly finding utility in the treatment of drug refractory breast cancer (Menis and Twelves, 2011).
The most recent addition to the arsenal of approved anticancer agents from the marine environment is brentuximab vedotin, a chimeric antibody attached through a protease-cleavable linker to a derivative of the potent antitubulin agent dolastatin 10 (Katz et al., 2011). Dolastatin 10, first reported in 1987 by the Pettit group from the Indian Ocean sea hare Dolabella auricularia (Pettit et al, 1987), was later found to originate from the gastropod's cyanobacterial (Symploca sp.) diet (Luesch et al., 2001). Phase I and II clinical trials of dolastatin 10 and the water soluble analog auristatin PE were unsuccessful due to lack of efficacy and induced peripheral neuropathy. However, linking of another analog of dolastatin 10, namely monomethyl auristatin E, to an antibody which targets CD30, a cell membrane protein present on the surface of Hodgkin's lymphoma cells, has resulted in a highly effective and well tolerated agent, brentuximab vendotin (Katz et al., 2011). An accelerated FDA approval of this agent for use in Hodgkin's lymphoma and anaplastic large cell lymphoma was granted in August 2011 and is marketed as Adcetris® by Seattle Genetics.
A peptide toxin from the fish-hunting marine mollusk, Conus magus, has found utility as a therapeutic for severe pain (Teichart and Olivera, 2010). The ω-conotoxin peptide known as ziconotide (Prialt®) functions through binding as an antagonist to N-type voltage-gated calcium channels. Because of its peptide nature, it must be administered as an infusion directly into the cerebrospinal fluid much like it is injected via a harpoon-like structure in nature to immobilize its prey. For patients with severe chronic pain, this is a useful agent and has the additional benefit of not inducing tolerance. Additional conotoxins are in clinical development with potential applications in pain management and are widely employed tool compounds in neurotoxin research (Daly and Craik, 2011).
While the benefits of fish oil in the diet are widely known and appreciated, including lowered coronary arteriosclerosis risk, a product (Lovaza®) has recently appeared on the market with the specific therapeutic goal of reducing serum triglycerides (Dayspring, 2011). This product is composed of the ethyl esters of various ω-3 fatty acids obtained from fish, mainly comprised of EPA (20:5n3) and DHA (22:6n3), administered as 400 mg/day. The fish used in making this material are reported as coming from the South Pacific and to include Anchovies, Herring, Salmon, Mackerel, Smelts and Jacks (http://www.gsksource.com/gskprm/en/US/adirect/gskprm?cmd=ProductDetailPage&product_id=1250802208236&featureKey=601474). Combined with diet and exercise, this agent has been shown to significantly lower serum triglycerides in treated patients, principally triglyceride and VLDL-cholesterol levels, and may therefore have health benefits such as lowered risk of cardiovascular disease in patients with dyslipdemia (Barter and Ginsberg, 2008).
As noted above, some 13 agents with a marine origin are in current clinical trials, 11 with an indication for cancer, one for cognition and schizophrenia, one for Alzheimer's (along with cancer), and one for wound healing. Interestingly, 3 of these agents (one in phase II and two in phase I trials), are antibody conjugates of dolastatin 10 analogs, and are closely related to brentuximab vedotin as described above. Thus, the pipeline of promising marine derived or inspired agents is very strong, and we will certainly see several of these agents enter the marketplace and clinic in the coming years (Mayer et al., 2010).
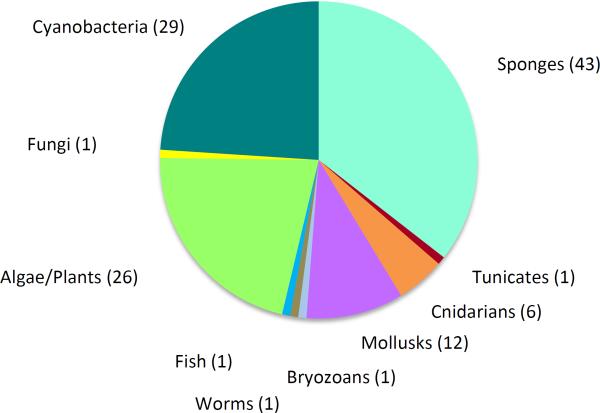
While the “holy grail” of drug discovery efforts is to bring an actual product to the clinic, marine natural products have a rich history of providing a wealth of toxins and other agents that are very useful in cell biology and pharmacological research. Early discoveries along these lines include kainic acid from the red alga Digenia simplex which has become the defining ligand of this receptor, saxitoxin from Alaskan butter clams (but deriving from their diet of microalgae) and tetrodotoxin from the Puffer fish (deriving from symbiotic bacteria); both of these latter agents were critical in defining sodium channel function. Subsequently, a number of commercial providers offer marine natural products for sale as research biochemicals. Figure 2 details the “collected” origin of these products (121 different products), and reveals that sponges, cyanobacteria and algae/plants (microalgae, macroalgae and aquatic plants) are the sources of most of these useful tool compounds.
Figure 2.
Pie chart illustrating the collected sources of marine natural products that are available commercially for their useful pharmacological properties in biomedical research (121 total).
Success stories from marine chemical biology
We have witnessed a transformation in marine natural product biosynthesis over the past decade that has moved the field of one of basic curiosity of how complex organic molecules are constructed to one of application in which basic discoveries are now addressing supply issues and delivering designer analogs to address structure-activity relationships. While there have been many success stories over the past years (Lane and Moore, 2011), we highlight here two examples, one involving the proteasome inhibitor salinosporamide A from a cultured marine bacterium and the second involving the anticancer agent ET-743 from an invertebrate-associated, uncultured marine bacterium.
One of the most promising proteasome inhibitor drug candidates currently in human clinical trials is salinosporamide A (Marizomib; Potts et al., 2011), a natural product first isolated from the marine bacterium S. tropica (Gulder and Moore, 2010). Salinosporamide A possesses a complex, densely functionalized γ-lactam-β-lactone pharmacophore that is chemically distinct from bortezomib (Velcade™) and other peptide-based proteasome inhibitors that as a group bind to the catalytic β-subunit of the 20S proteasome to inhibit its function and thereby the cell's primary route of regulated proteolysis (Borissenko and Groll, 2007). Significantly, salinosporamide A triggers apoptosis in bortezomib-resistant multiple myeloma cells and has prolonged inhibitory activity in tumors and packed whole blood versus other tissues (Potts et al., 2011). Early data from the on-going phase I trial of salinosporamide A (NPI-0052, Nereus Pharmaceuticals) report potential clinical benefit in patients with relapsed and relapsed/refractory multiple myeloma, thereby demonstrating the exciting clinical promise of this natural product drug candidate.
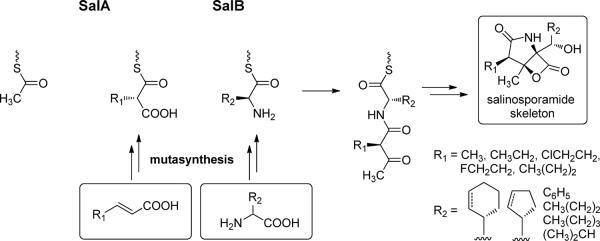
Salinosporamide A is active as a potent and selective irreversible inhibitor against all three proteasomal activities of the 20S catalytic core in which its cyclohexenyl moiety is important for its preferential binding in the S1 site of the proteasomal β-subunit, while its chloroethyl group is fundamental to its irreversible activity (Gulder and Moore, 2010). Isotope tracer studies followed by the interrogation of the biosynthetic gene cluster revealed the biogenesis of salinosporamide A in which the three building blocks acetyl-CoA, chloroethylmalonyl-CoA and cyclohexenylalanine jointly contribute to the core bicycle while uniquely providing the three side chains (Figure 5). This observation provided a convenient strategy for the mutasynthesis and engineered biosynthesis of a focused library of novel analogs to evaluate some aspects of structure-activity relationships in this molecular series. Among these, fluorosalinosporamide displays potent “slowly reversible” proteasome inhibitory activity (Eustaquio and Moore, 2008), while the cyclopentenyl derivative salinosporamide X7 exhibits superior in vivo activity (Nett et al., 2009). Knowledge of the biosynthetic machinery further enabled the selective increase in salinosporamide A production over the other natural non-chlorinated analogs by control of the pathway regulatory gene salR2, which activates transcription of two divergent operons containing genes specific to the chloroethylmalonyl-CoA pathway (Lechner et al., in press). While traditional manipulations to the strain and fermentation conditions dramatically improved the production yield of salinosporamide A over 100-fold to 450 mg/L (Potts and Lam, 2010), the addition of recombinant techniques may be needed if this drug candidate is ultimately approved for clinical use.
Figure 5.
Assembly line biosynthesis of salinosporamide and library development of structure analogs via mutasynthesis and other genetic engineering approaches.
Beyond enabling biosynthesis, the genetic machinery can often provide clues to how an organism counters autotoxicity. In the case of the actinobacterium S. tropica, which possesses a 20S proteasome, the question of self-resistance was addressed again by having access to the complete genome sequence. Peripheral to the sal biosynthesis genes resides a redundant proteasome β-subunit, salI, whose protein product has an altered substrate specificity profile with 30-fold resistance to salinosporamide A, and cross resistance to the FDA-approved proteasome inhibitor bortezomib (Kale et al., 2011). The molecular basis for evolved resistance in S. tropica was correlated to an A49V mutation in SalI that coincidently correlates to bortezomib acquired resistance in the human proteasome β5-subunit (Franke et al., 2011), suggesting that intrinsic resistance to natural proteasome inhibitors may predict clinical outcomes as well as predict the discovery of new proteasome inhibitors via genome mining.
As discussed earlier in this article, the discovery and clinical development of the tunicate-derived ET-743 (Yondelis®) is a success story in the marine natural product field for its clinical approval in Europe as an antitumor agent. Its striking structural resemblance to the saframycin family of microbial products was immediately recognized and hinted at a bacterial origin. Metagenomic sequencing of total DNA from the tunicate Ecteinascidia turbinata and its associated microbes recently provided a 35 kb contig containing the majority of the putative ET-743 biosynthetic gene cluster (Rath et al., 2011) that closely mirrored the previously characterized saframycin A locus from Streptomyces lavendulae (Koketsu et al., 2010). This work suggested that the individual genes were likely of microbial origin from the γ-proteobacterium Candidatus Endoecteinascidia frumentensis, a prevalent microbial member of the community (Perez-Matos et al., 2007), rather that the host tunicate due to codon usage characteristics. Functional support for the putative ET-743 gene locus was achieved through metaproteomic analysis of several biosynthetic proteins and enzymatic confirmation of a key nonribosomal peptide synthetase assembly line reaction (Rath et al, 2011). While the ET-743 biosynthetic gene cluster is incomplete and the functional work has only just begun, the preliminary discovery work clearly establishes the molecular basis for its assembly and lays the foundation for future bioengineering efforts to establish a renewable path to solve the long-term supply of ET-743 through microbial fermentation.
F. Problems encountered and lessons learned in marine drug discovery
The above success stories demonstrate that the marine world has much to offer human society in the way of pharmaceutical agents, agrochemicals, and research biochemicals. Moreover, a substantial number of marine-derived agents are progressing through the development process, primarily with indications in cancer, but also in pain and inflammation. Beneath this group, however, is an even larger cadre of quite promising leads. The experiences of the marine natural products community over the past 40 years has elevated the approaches and strategies that are currently employed, making compound discovery more focused and structure elucidation a much more efficient process. The recent trend in phenotypic and high content screening appears to be very effective in revealing the potentially useful biological properties of natural products, and has the increased benefit of identifying new targets by which to treat human disease while at the same time being appropriately disease focused. Indeed, the current success rate of discovery from the marine world, namely seven clinically useful and approved drugs from 22,000 (J. Blunt, personal communication) discovered molecular entities (e.g. 1 drug per 3,140 natural products described) is approximately 1.7- to 3.3-fold better than the industry average (1 in 5,000–10,000 tested compounds) (http://www.phrma.org/sites/default/files/159/rd_brochure_022307.pdf). As one considers further the very rich pipeline of clinical trial agents as well as preclinical candidates, several of which will almost certainly become approved drug agents, the marine world has indeed been an exceptional resource in which to search for new pharmaceutical agents.
High-throughput screening technologies have fundamentally changed the face of biology in the industrial setting. Hundreds of thousands of assays can be performed in a matter of days using 1534-well assay plates, and entire screening campaigns can be completed in a few short weeks. Gearing up for such an effort in terms of robot programming, reagents, assay protocols, and control conditions is time consuming, and not easily re-activated once a screening campaign is completed (Mayr and Bojanic, 2009). Thus, the screening of impure materials wherein the value is only obtained subsequent to a reiterative process of progressive chromatographic separation of components and re-testing in the assay does not integrate well into the timeline created by modern HTS (Koehn and Carter, 2005). Indeed, the concept of HTS screening is well suited to screening of pure compound libraries, and thus the pharmaceutical industry has placed emphasis on creation of large libraries of synthetic scaffolds. Unfortunately, this appetite of HTS for large numbers of chemical substances has placed emphasis on quantity rather than quality, and most synthetic libraries are populated by planar molecules of low chemodiversity, and more importantly, do not match well to drug chemical space (Feher and Schmidt, 2003; Rouhi, 2003)
Marine drug discovery efforts of the 1970–2000 period worked with relatively large biological sample sizes. This was necessary due to the practicalities of structure elucidation of new and oftentimes unprecedented structure types, thus requiring multi-milligram quantities of the compound. At the end of such an exercise, substantial quantities of isolated compound remained, and these could be funneled into biological screening programs. While miniaturization of assays has continued, the reduction in the quantity of needed material for structure elucidation has kept pace to HTS advances (Mayr and Bojanic, 2009). Using microcryoprobe or flow-cell NMR spectrometers in conjunction with ever more powerful MS methods and algorithms, such as that recently reported for de novo sequencing of cyclic peptides (Ng et al., 2009), structures are being solved on the nanomolar and even smaller scale (Molinski, 2010). These technological advances have allowed ever smaller amounts of biological material to be collected which in turn has allowed the exploration of marine species existing in very low biomass in nature, such as thinly encrusting invertebrates or microalgal slimes. With only microgram quantities being isolated, and some being sacrificed to destructive analytical procedures or synthetic modification, an insufficient amount remains to drive a meaningful biological evaluation of these new products. Thus, such studies become more academic enterprises except in cases where total chemical synthesis or biosynthesis can provide a resupply of the compound for meaningful biological assay. Ultimately, it is critical to connect biological properties to these unique natural product structures; otherwise, they come to represent chemical oddities lacking in scientific or biomedical relevance and significance. Indeed, some journals have articulated the publication requirement that new natural products should be evaluated for their biological properties (http://pubs.acs.org/paragonplus/submission/jnprdf/jnprdf_scope.pdf).
A long standing and related issue to the above nanomole scale isolation problem has been termed “crossing the valley of death”. While there are several `valley's' in the complex process of drug development, in this context the difficulty is how to move `early stage discoveries' into `late stage preclinical candidates'. A disappointingly large number of marine natural products (and other natural products), some with astounding structures and biological properties, are published in the scientific literature as “exciting leads” but never advance beyond this embryonic stage. The needs to advance an agent to the next level include determination of mechanism and site of action, development of structure activity relationships, formulation and in vivo evaluations in animal models for toxicity and efficacy, and characterization of pharmacokinetic parameters and pharmaceutical properties including improvements through medicinal chemistry. Critical to these pursuits is an abundant source of the compound or an improved analog, and this usually requires chemical synthesis or fermentation with the overarching goal of substantial compound production (e.g. gram scale)(Smith et al., 1999). Unfortunately, despite great advances in total chemical synthesis methodology, this goal is all too often not reached, and development of compounds languish due to lack of supply and momentum in the project. Compounding these problems and regardless of the approach undertaken to bridge this gap, there is a general lack of financial resources and mechanisms available for advancing early stage leads across this abyss.
The pharmaceutical industry has fundamentally changed over the last 20 years, with large pharma being ever more risk adverse in their research strategies. Correspondingly, there is some perception that the main site of innovation in drug discovery has shifted to small start-up companies and academia (Kaitin, 2010). One model large pharma has employed is to license compounds or purchase entire companies that have successfully navigated the “valley of death” and brought an innovative new therapy to clinical trials. Unfortunately, the financial resources of small companies are necessarily limited, and thus, ironically, success of a research program to advance a candidate is oftentimes intimately linked with the termination of these very same research departments with attendant layoffs of the scientific staff. These structural changes in industry have the net effect of introducing further impediments to forwarding early stage leads to become true clinical candidates.
The lack of development of early stage leads has been a major issue in natural products drug discovery research for some time, and was part of the rationale for formulating Drug Discovery Groups (DDG) through the NCIs very successful National Cooperative DDG program. In this program, groups of academic investigators teamed together with partner pharmaceutical companies and the NCI so as to improve the focus of the cancer drug discovery process as well as to have development capacity inherently built into the endeavor. During the 23 year period of the NCDDG program (1984–2007), there were 21 New Investigational Drugs that went into clinical trial (12 small molecules and 9 biologics) that derived from substantial input from this program. Indeed, this is widely perceived as the single most effective granting mechanism by which to bring fundamentally new cancer treatments into existence, and with its disappearance, we have taken a giant step backwards. Academic investigators are once again formulating these crucial connections with industry on an individual one-by-one basis. Combined with a greater sensitivity to druggable targets, broad knowledge of disease biology, and the capacity of meaningful development of leads across the valley of death and into the clinic, large Pharma is an invaluable partner to the highly innovative yet risky approaches of academic natural products laboratories. Indeed, the NCDDG structure is an inspired concept that needs to be renewed in some form (Crews et al., 2003).
While isolation and structure elucidation technology has advanced enormously in recent years, as noted above, this process when coupled to a biological assay can sometimes take weeks or months. This is simply too slow to compete with the screening of pure compounds with known structures. To surmount this, two options exist. The first is to purify unique natural compounds based on chemical diversity criteria, and then screen these as pure materials of defined composition (Abel et al, 2002). Alternatively, even more rapid techniques of structure elucidation need to be developed so that screening hits can be transformed into compound hits in a matter of minutes to hours. To this end, a growing number of approaches and algorithms using MS data, largely coming from the metabolomics field, are developing de novo structures based on high resolution data and fragmentation pathways. For example, using the complete MS2 and MS3 dataset, the Cyclic Peptide Sequencing web-based program can construct, in a completely automatic fashion and with high accuracy, the structures of cyclic peptide natural products (Mohimani et al., 2011; (http://rofl.ucsd.edu/nrp/index.html). Advances in Ion Mobility and other techniques suggest the possibility that even stereochemical features will be able to be deduced using mass spectrometry alone (Bai et al., 2011). A vision of the not too distant future, similar to the then unrealistic scene in the 1982 movie “Medicine Man”, is that we will define the structures of novel molecules on-line, in real time and by entirely automatic processes! Necessarily, the questions we will ask will shift away from the structure elucidation process to issues of chemical interaction of organisms and their biochemical pathways, systems biology, molecular pharmacology, and biotechnological developments.
Another exciting development from the Mass Spectrometry world is the ability to image biological tissues and cells for their occurrence of specific secondary metabolites. MALDI, DESI and IMS are three such techniques, each with their assets and drawbacks (Esquenazi et al., 2009). However, the instrumental advances in this area are staggering, and soon we will be looking at the metabolomes of single cells or even of sub-cellular compartments. This type of MS imaging technology has great utility in locating specific secondary metabolites to particular filament types in complex consortia of bacteria (e.g. cyanobacteria), secondary metabolites associated with morphological features of tissues in macroalgae, and chemical interactions of competing microbial species on agar surfaces (Esquenazi et al., 2009; Gonzalez, 2011). These innovative types of experiments and tools are giving fundamentally new insights into how organisms interact with one another through their suite of secondary metabolites.
As it should, the field of natural products is rapidly evolving to intersect powerfully with other fields of science, and one such area that should prove very exciting in the coming years is computational chemistry and structural biology. Natural products, for whatever reason, have not often been including in in silico screening programs, however, this is rapidly changing as the uniqueness of these natural scaffolds are better understood and appreciated by a wider range of chemists. These approaches can include evaluations for structural matches with the shapes of enzyme active sites as well as protein-protein association domains, allosteric sites, and other drug binding folds of proteins. When the binding site can be identified or reasonably inferred in a target, the more than 50,000 protein structures now available in the Protein Data Bank permits rapid and effective structural biology approaches to be applied. However, there is a continuing need to improve and make more reliable the force field parameters for calculating both protein and small molecule energies in various conformations. Medicinal chemistry approaches in concert with biophysical methods such as crystallography, NMR or isothermal calorimetry, along with computational binding interactions, using entire natural product structures or pharmacophoric fragments, are a proven method for enhancing the binding affinity of small molecules to their protein targets (Coyne et al., 2010).
Challenges and opportunities in marine chemical biology
As with all new emerging technologies, many challenges await the field of marine natural product biotechnology before it can reach its full potential of providing practical approaches to supplying complex marine organic molecules for clinical evaluation and development. Current limitations that are actively being addressed in academia include developing universal expression systems for high-yielding small molecule biosynthesis, constructing genetic tools to access the in vivo potential of cultured marine microorganisms such as the metabolically-rich marine filamentous cyanobacteria, and the regulatory awakening of orphan or “silent” biosynthetic pathways for small molecule discovery.
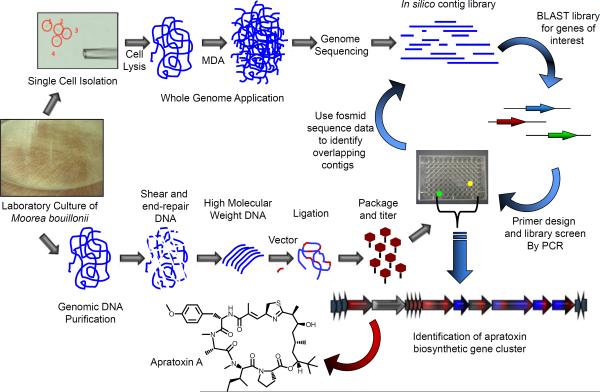
If the successes of the past decade are any indication, the future is indeed bright with new methods applied to natural products research becoming available at a dizzying pace. Two new approaches are worth mentioning in this light. First, single-cell genomics (Laskin, 2007) now allow for the genomic analysis at the cellular level in which mixtures or communities can be sorted and the genomes of individual members analyzed. In the field of marine natural products, two reports recently revealed the power of the technique. Multiple displacement amplification of the marine sponge-associated candidate phylum “Poribacteria” revealed PKS genes that may be involved in methyl-branched fatty acids (Siegl and Hentschel, 2010; Siegl et al., 2011) while separation of filamentous L. majuscula cells from its sheath covered with unicellular bacteria provided a clean draft genome sequence that revealed the molecular basis of apratoxin biosynthesis (Figure 6) (Grindberg et al., 2011). These examples highlight the ability to effectively work with small amounts of material from the environment to capture the invaluable genomic information encapsulated within that may hold new opportunities for small molecule discovery. A second, and in some way complementary, method recently reported involves the coordination of mass spectrometry and genomics as a new experiment-based genome mining technique called peptidogenomics that rapidly speeds up the discovery rate of new ribosomal and nonribosomal natural product peptides from sequenced organisms (Kersten et al., 2011). This orthogonal natural product discovery approach to traditional assay-based isolation schemes has the potential for automation that could dramatically alter the way in which peptides and possibly other natural product classes are discovered in nature.
Figure 6.
Parallel strategy employed by Grindberg et al. (2011) to rapidly access the biosynthetic gene cluster for apratoxin A, a promising anticancer lead compound from the marine cyanobacterium Moorea bouillonii. On the top arm, single cells are obtained by microdissection from non-axenic cultures of cyanobacteria, and DNA is extracted and amplified by Multiple Displacement Amplification (MDA) for partial genome sequencing. The sequences of recognizable gene motifs associated with natural product pathways are then used to construct PCR probes to screen a fosmid library which is produced in the normal fashion (lower arm). Fosmids probing positively by this process can be further characterized for desired gene motifs, and then sequenced. The melding of these approaches can accelerate the process of biosynthetic gene cluster discovery and description, such as is illustrated here for apratoxin A, especially in cases of non-axenic cultures or environmental samples.
Conclusions
Efforts of the past 30 years have been highly productive in defining major trends in the secondary metabolism of diverse classes of marine organisms. From these, seven agents have entered the clinic as approved drugs, mostly in the area of cancer, but also for viral, pain and hyperlipidemia indications. When combined with the rich pipeline of agents in clinical trial and preclinical evaluation, it can be surmised that the marine environment has performed extremely well in yielding new medicines as well as pharmacological tools. However, there has been growing recognition in the past decade that the most productive source of unique secondary metabolites actually derives from metabolic processes at work in microorganisms, including actinomycetes, cyanobacteria, microalgae such as dinoflagellates, and others. In a sense, this is quite fortunate, for the biosynthetic pathways that code for secondary metabolites in prokaryotes are better understood and more amenable for study than those in eukaryotes. Progress in understanding how these biosynthetic pathways operate at the genetic and biochemical levels is opening new doors for harnessing this potential, and the future looks very optimistic for realizing tangible benefits from such efforts, such as new designer molecules with improved biological and pharmaceutical properties.
Highlights
Seven marine-derived or inspired drugs have been approved for clinical use
A rich pipeline of clinical, preclinical and tool compounds come from marine life
Marine microorganisms are the dominant source of bioactive marine natural products
Marine chemical biology is making substantial contributions to drug discovery
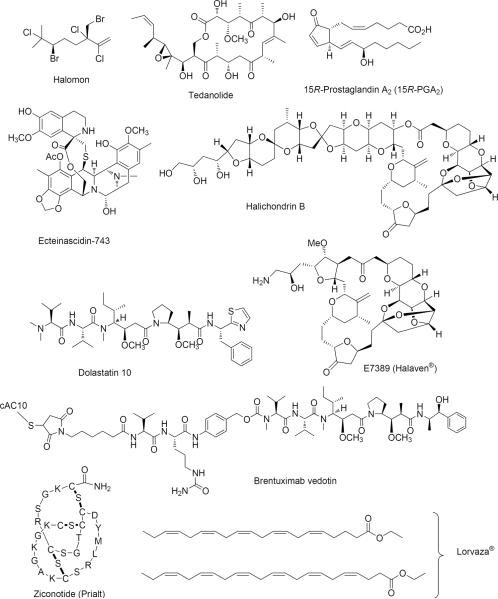
Figure 3.
Chemical structures of the approved drugs deriving from or inspired by a marine natural product and other marine metabolites discussed in the text (one letter amino acid codes are used for depicting the structure of ziconotide).
Acknowledgements
We gratefully acknowledge the assistance of T. Byrum and E. Monroe in the construction of this manuscript, and helpful discussion with M. Gilson (all of UCSD). Research in the authors laboratories have been supported with NIH grants TW006634, NS053398, CA108874 and CA100851 (W.H.G.) and AI47818, CA127622 and GM857770 (B.S.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abel U, Koch C, Speitling M, Hansske FG. Modern methods to produce natural-product libraries. Curr. Op. Chem. Biol. 2002;6:453–458. doi: 10.1016/s1367-5931(02)00338-1. [DOI] [PubMed] [Google Scholar]
- Bai L, Romanova EV, Sweedler JV. Distinguishing endogenous D-amino acid-containing neuropeptides in individual neurons using tandem mass spectrometry. Anal. Chem. 2011;83:2794–2800. doi: 10.1021/ac200142m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barter P, Ginsberg HN. Effectiveness of combined statin plus omega-3 fatty acid therapy for mixed dyslipidemia. Am. J. Cardiol. 2008;102:1040–1045. doi: 10.1016/j.amjcard.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann W, Watkins JC, Stempien MF., Jr. Contribution to the study of marine products. XLV. Sponge nucleic acids. J. Org. Chem. 1957;22:1308–13. [Google Scholar]
- Borissenko L, Groll M. 20S proteasome and its inhibitors: Crystallographic knowledge for drug development. Chem. Rev. 2007;107:687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- Brunton LL, Chabner BA, Knollmann BC. Goodman & Gilman's The Pharmacological Basis of Therapeutics. 12th ed. McGraw-Hill Companies; New York: 2011. [Google Scholar]
- Bugni TS, Harper MK, McCulloch MWB, Reppart J, Ireland CM. Fractionated marine invertebrate extract libraries for drug discovery. Molecules. 2008;13:1372–1383. doi: 10.3390/molecules13061372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, Flatt P, Gerwick WH, Nguyen VA, Willis CL, Sherman DH. The barbamide biosynthetic gene cluster: a novel marine cyanobacterial system of mixed polyketide synthase (PKS)-non-ribosomal peptide synthetase (NRPS) origin involving an unusual trichloroleucyl starter unit. Gene. 2002:235–247. doi: 10.1016/s0378-1119(02)00860-0. [DOI] [PubMed] [Google Scholar]
- Chang Z, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia J, Sherman DH, Gerwick WH. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2004;67:1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- Coyne AG, Scott DE, Abell C. Drugging challenging targets using fragment-based approaches. Curr. Opin. Chem. Biol. 2010;14:299–307. doi: 10.1016/j.cbpa.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Crews P, Gerwick W, Schmitz F, France D, Bair K, Wright A, Hallock Y. Molecular approaches to discover marine natural products anticancer leads - an update from a drug discovery group collaboration. Pharm. Biol. 2003;41:39–52. [Google Scholar]
- Cuevas C, Pérez M, Martín MJ, Chicharro JL, Fernández-Rivas C, Flores M, Francesch A, Gallego P, Zarzuelo M, de La Calle F, et al. Synthesis of ecteinascidin ET-743 and phthalascidin Pt-650 from cyanosafracin. B. Org. Lett. 2000;2:2545–8. doi: 10.1021/ol0062502. [DOI] [PubMed] [Google Scholar]
- Daly NL, Craik DJ. Conopeptides as novel options for pain management. Drug Future. 2011;36:25–32. [Google Scholar]
- Dayspring TD. Understanding hypertriglyceridemia in women: clinical impact and management with prescription omega-3-acid ethyl esters. Int. J. Womens Health. 2011;3:87–97. doi: 10.2147/IJWH.S16702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat. Chem. Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engene N, Rottacker EC, Kastovsky J, Byrum T, Choi H, Ellisman MH, Komárek J, Gerwick WH. Moorea producta gen. nov., sp. nov. and Moorea bouillonii comb. nov., tropical marine cyanobacteria rich in bioactive secondary metabolites. Int. J. Syst. Evol. Microbiol. 2011 doi: 10.1099/ijs.0.033761-0. doi: 10.1099/ijs.0.033761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquenazi E, Yang Y, Watrous J, Gerwick WH, Dorrestein PC. Imaging mass spectrometry of natural products. Nat. Prod. Rep. 2009;26:1521–1534. doi: 10.1039/b915674g. [DOI] [PubMed] [Google Scholar]
- Eustáquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, et al. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-CoA from S-adenosyl-L-methionine. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12295–12300. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustáquio AS, Moore BS. Mutasynthesis of fluorosalinosporamide, a potent and reversible inhibitor of the proteasome. Angew. Chem. Int. Ed. 2008;47:3936–3938. doi: 10.1002/anie.200800177. [DOI] [PubMed] [Google Scholar]
- Eustaquio AS, Nam S-J, Penn K, Lechner A, Wilson MC, Fenical W, Moore BS. The discovery of salinosporamide K from the marine bacterium “Salinispora pacifica” by genome mining gives insight into pathway evolution. ChemBioChem. 2011;12:61–64. doi: 10.1002/cbic.201000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner DJ. Marine natural products. Nat. Prod. Rep. 2002;19:1–48. doi: 10.1039/b009029h. [DOI] [PubMed] [Google Scholar]
- Feher MF, Schmidt JM. Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. J. Chem. Inf. Comput. Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- Fenical W, Jensen PR. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat. Chem. Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- Fisch KM, Gurgui C, Heycke N, van der Sar SA, Anderson SA, Webb VL, Taudien S, Platzer M, Rubio BK, Robinson SJ, et al. Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat. Chem. Biol. 2009;5:494–501. doi: 10.1038/nchembio.176. [DOI] [PubMed] [Google Scholar]
- Franke NE, Niewerth D, Assaraf YG, van Meerloo J, Vojtekova K, van Zantwijk CH, Zweegman S, Chan ET, Kirk CJ, Geerke DP, et al. Impaired bortezomib binding to mutant β5 subunit of the proteasome is the underlying basis for bortezomib resistance in leukemia cell. Leukemia. 2011 doi: 10.1038/leu.2011.256. doi: 10.1038/leu.2011.1256. [DOI] [PubMed] [Google Scholar]
- Gonzalez DJ, Haste NM, Hollands A, Fleming TC, Hamby M, Pogliano K, Nizet V, Dorrestein PC. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology. 2011;157:2485–2492. doi: 10.1099/mic.0.048736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindberg RV, Ishoey T, Brinza D, Esquenazi E, Coates RC, Liu WT, Gerwick L, Dorrestein PC, Pevzner P, Lasken R, et al. Single cell genome amplification accelerates identification of the apratoxin biosynthetic pathway from a complex microbial assemblage. PLoS One. 2011;6:e18565. doi: 10.1371/journal.pone.0018565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindberg RV, Shuman CF, Sorrels CM, Wingerd J, Gerwick WH. Neurotoxic Alkaloids from Cyanobacteria. In: Fattorusso E, Taglialatela-Scafati O, editors. Modern Alkaloids, Structure, Isolation, Synthesis and Biology. Wiley-VCH Verlang GmbH & Co.; Weinheim: 2008. pp. 139–170. [Google Scholar]
- Gulder TAM, Moore BS. Chasing the treasures of the sea-bacterial marine natural products. Curr. Opin. Microbiol. 2009;12:252–260. doi: 10.1016/j.mib.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulder TAM, Moore BS. The salinosporamide natural product family: Potent 20S proteasome inhibitors as promising cancer chemotherapeutics. Angew. Chem. Int. Ed. 2010;49:9346–9367. doi: 10.1002/anie.201000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haygood MG, Schmidt EW, Davidson SK, Faulkner DJ. Microbial symbionts of marine invertebrates: opportunities for microbial biotechnology. J. Mol. Microbiol. Biotechnol. 1999;1:33–43. [PubMed] [Google Scholar]
- Hirata Y, Uemura D. Halichondrins-antitumor polyether macrolides from a marine sponge. Pure Appl. Chem. 1986;58:701–10. [Google Scholar]
- Johnson TA, Morgan MVC, Aratow NA, Estee SA, Sashidhara KV, Loveridge ST, Segraves NL, Crews P. Assessing pressurized liquid extraction for the high-throughput extraction of marine-sponge-derived natural products. J. Nat. Prod. 2010;73:359–364. doi: 10.1021/np900565a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AC, Monroe EA, Podell S, Hess WR, Klages S, Esquenazi E, Niessen S, Hoover H, Rothmann M, Lasken RS, et al. Genomic insights into the physiology and ecology of the marine filamentous cyanobacterium Lyngbya majuscula. Proc. Natl. Acad. Sci. U.S.A. 2011;107:10430–11435. doi: 10.1073/pnas.1101137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MA, Kamath K, Manna T, Okouneva T, Miller HP, Davis C, Littlefield BA, Wilson L. The primary antimitotic mechanism of action of the synthetic halichondrin E7389 is suppression of microtubule growth. Mol. Cancer Ther. 2005;4:1086–1095. doi: 10.1158/1535-7163.MCT-04-0345. [DOI] [PubMed] [Google Scholar]
- Kaitin KI. Deconstructing the drug development process: the new face of innovation. Clin. Pharmacol. Ther. 2010;87:356–361. doi: 10.1038/clpt.2009.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale AJ, McGlinchey RP, Lechner A, Moore BS. Bacterial self-resistance to the natural proteasome inhibitor salinosporamide A. ACS Chem. Biol. 2011;6:1257–1264. doi: 10.1021/cb2002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J, Janik JE, Younes A. Brentuximab Vedotin (SGN-35) Clin. Cancer Res. 2011;17:6428–6436. doi: 10.1158/1078-0432.CCR-11-0488. [DOI] [PubMed] [Google Scholar]
- Kersten RD, Yang Y-L, Xu Y, Cimermancic P, Nam S-J, Fenical W, Fischbach MA, Moore BS, Dorrestein PC. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- Koketsu K, Watanabe K, Suda H, Oguri H, Oikawa H. Reconstruction of the saframycin core scaffold defines dual Pictet-Spengler mechanisms. Nat. Chem. Biol. 2010;6:408–410. doi: 10.1038/nchembio.365. [DOI] [PubMed] [Google Scholar]
- Lane AL, Moore BS. A sea of biosynthesis: Marine natural products meet the molecular age. Nat. Prod. Rep. 2011;28:411–428. doi: 10.1039/c0np90032j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner A, Eustáquio AS, Gulder TAM, Hafner M, Moore BS. Selective overproduction of the proteasome inhibitor salinosporamide A via precursor pathway regulation. Chem. Biol. doi: 10.1016/j.chembiol.2011.10.014. doi:10.1016/j.chembiol.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Sher D, Kelly L, Shi Y, Huang K, Knerr PJ, Joewono I, Rusch D, Chisholm SW, van der Donk WA. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesch H, Moore RE, Paul VJ, Mooberry SL, Corbett TH. Isolation of dolastatin 10 from the marine cyanobacterium symploca species VP642 and total stereochemistry and biological evaluation of its analogue symplostatin 1. J. Nat. Prod. 2001;64:907–910. doi: 10.1021/np010049y. [DOI] [PubMed] [Google Scholar]
- Mayer AMS, Glaser KB, Cuevas C, Jacobs RS, Kem W, Little RD, McIntosh JM, Newman DJ, Potts BC, Shuster DE. The odyssey of marine pharmaceuticals: a current pipeline perspective. Trends Pharmacol. Sci. 2010;31:255–265. doi: 10.1016/j.tips.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Mayr LM, Bojanic D. Novel trends in high-throughput screening. Curr. Opin. Pharmacol. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- McGlinchey RP, Nett M, Moore BS. Unraveling the biosynthesis of the sporolide cyclohexenone building block. J. Am. Chem. Soc. 2008;130:2406–2407. doi: 10.1021/ja710488m. [DOI] [PubMed] [Google Scholar]
- Menis J, Twelves C. Eribulin (Halaven): a new, effective treatment for women with heavily pretreated metastatic breast cancer. Breast Cancer: Targets Ther. 2011;3:101–111. doi: 10.2147/BCTT.S21741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanaga A, Janso JE, McDonald L, He M, Liu H, Barbieri L, Eustáquio AS, Fielding EN, Carter GT, Jensen PR, et al. Discovery and assembly-line biosynthesis of the lymphostin pyrroloquinoline alkaloid family of mTOR inhibitors in Salinispora bacteria. J. Am. Chem. Soc. 2011;133:13311–13313. doi: 10.1021/ja205655w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohimani H, Liu W-T, Mylne JS, Poth AG, Colgrave ML, Tran D, Selsted ME, Dorrestein PC, Pevzner PA. Cycloquest: Identification of cyclopeptides via database search of their mass spectra against genome databases. J Proteome Res. 2011;10:4505–4512. doi: 10.1021/pr200323a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinski TF. NMR of natural products at the nanomole-scale. Nat. Prod. Rep. 2010;27:321–329. doi: 10.1039/b920545b. [DOI] [PubMed] [Google Scholar]
- Molinski TF, Dalisay DS, Lievens SL, Saludes JP. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009;8:69–85. doi: 10.1038/nrd2487. [DOI] [PubMed] [Google Scholar]
- Moore BS. Biosynthesis of marine natural products: microorganisms (part A) Nat. Prod. Rep. 2005;22:580–593. doi: 10.1039/b404737k. [DOI] [PubMed] [Google Scholar]
- Moore BS. Biosynthesis of marine natural products: macroorganisms (part B) Nat. Prod. Rep. 2006;23:615–629. doi: 10.1039/b508781n. [DOI] [PubMed] [Google Scholar]
- Nett M, Gulder TAM, Kale AJ, Hughes CC, Moore BS. Function-oriented biosynthesis of β-lactone proteasome inhibitors in Salinispora tropica. J. Med. Chem. 2009;52:6163–6167. doi: 10.1021/jm901098m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products, derivatives and mimics as antitumor agents. In: Wrigley S, Thomas R, Nicholson N, Bedford C, editors. Functional Molecules from Natural Sources. RSC Publishing; Cambridge, England: 2010. pp. 3–36. [Google Scholar]
- Ng J, Bandeira N, Liu W, Ghassemian M, Simmons TL, Gerwick WH, Linington R, Dorrestein PC, Pevzner PA. Dereplication and de novo sequencing of nonribosomal peptides. Nat. Methods. 2009;6:596–599. doi: 10.1038/nmeth.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou KC, Snyder SA. Chasing molecules that were never there: Misassigned natural products and the role of chemical synthesis in modern structure elucidation. Angew. Chem. Int. Edit. 2005;44:1012–1044. doi: 10.1002/anie.200460864. [DOI] [PubMed] [Google Scholar]
- Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen EE, et al. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 2009;3:1193–1203. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit GR, Kamano Y, Herald CL, Tuinman AA, Boettner FE, Kizu H, Schmidt JM, Baczynskyj L, Tomer KB, Bontems RJ. The isolation and structure of a remarkable marine animal antineoplastic constituent: dolastatin 10. J. Am. Chem. Soc. 1987;109:6883–6885. [Google Scholar]
- Piel J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2004;21:519–538. doi: 10.1039/b310175b. [DOI] [PubMed] [Google Scholar]
- Piel J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- Piel J. Approaches to capturing and designing biologically active small molecules produced by uncultured microbes. Annu. Rev. Microbiol. 2011;65:431–453. doi: 10.1146/annurev-micro-090110-102805. [DOI] [PubMed] [Google Scholar]
- Piel J, Hertweck C, Shipley P, Hunt DS, Newman MS, Moore BS. Cloning, sequencing and analysis of the enterocin biosynthesis gene cluster from the marine isolate “Streptomyces maritimus”: Evidence for the derailment of an aromatic polyketide synthase. Chem. Biol. 2000;7:943–955. doi: 10.1016/s1074-5521(00)00044-2. [DOI] [PubMed] [Google Scholar]
- Piel J, Hui DQ, Wen GP, Butzke D, Platzer M, Fusetani N, Matsunaga S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Matos AE, Rosado W, Govind NS. Bacterial diversity associated with the Caribbean tunicate Ecteinascidia turbinata. Antonie Van Leeuwenhoek. 2007;92:155–164. doi: 10.1007/s10482-007-9143-9. [DOI] [PubMed] [Google Scholar]
- Potts BC, Albitar MX, Anderson KC, Baritaki S, Berkers C, Bonavida B, Chandra J, Chauhan D, Cusack JCJ, Fenical W, et al. Marizomib, a proteasome inhibitor for all seasons: Preclinical profile and a framework for clinical trials. Curr. Cancer Drug Targets. 2011;11:254–284. doi: 10.2174/156800911794519716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts BC, Lam KS. Generating a generation of proteasome inhibitors: From microbial fermentation to total synthesis of salinosporamide A (Marizomib) and other salinosporamides. Mar. Drugs. 2010;8:835–880. doi: 10.3390/md8040835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath CM, Janto B, Earl J, Ahmed A, Hu FZ, Hiller L, Dahlgren M, Kreft R, Yu F, Wolff JJ, et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem. Biol. 2011;6:1244–1256. doi: 10.1021/cb200244t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinehart K, Holt TG, Fregeau NL, Stroh JG, Keifer PA, Sun F, Li LH, Martin DG. Ecteinascidins 729, 743, 745, 759A, 759B, and 770: potent antitumor agents from the Caribbean tunicate Ecteinascidia turbinata. J. Org. Chem. 1990;55:4512–15. [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, Johnson, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Rouhi AM. Rediscovering natural products. Chem. Eng. News. 2003;81:77–78. 82–83, 86, 88–91. [Google Scholar]
- Sagar S, Kaur M, Minneman KP. Antiviral lead compounds from marine sponges. Mar. Drugs. 2010;8:2619–38. doi: 10.3390/md8102619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashidhara KV, White KN, Crews P. A selective account of effective paradigms and significant outcomes in the discovery of inspirational marine natural products. J. Nat. Prod. 2009;72:588–603. doi: 10.1021/np800817y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeberle TF, Goralski E, Neu E, Erol O, Hoelzl G, Doermann P, Bierbaum G, Koenig GM. Marine myxobacteria as a source of antibiotics - comparison of physiology, polyketide-type genes and antibiotic production of three new isolates of Enhygromyxa salina. Mar. Drugs. 2010;8:2466–2479. doi: 10.3390/md8092466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AB, III, Kaufman MD, Beauchamp TJ, LaMarche MJ, Arimoto H. Gram-scale synthesis of (+)-discodermolide. Org. Lett. 1999;1:1823–1826. doi: 10.1021/ol9910870. [DOI] [PubMed] [Google Scholar]
- Soares DS, Machado MS, Rocca CJ, Poindessous V, Ouaret D, Sarasin A, Galmarini CM, Henriques JAP, Escargueil AE, Larsen AK. Trabectedin and its C subunit modified analogue PM01183 attenuate nucleotide excision repair and show activity toward platinum-resistant cells. Mol. Cancer Ther. 2010;10:1481–1489. doi: 10.1158/1535-7163.MCT-11-0252. [DOI] [PubMed] [Google Scholar]
- Sudek S, Haygood MG, Youssef DT, Schmidt EW. Structure of trichamide, a cyclic peptide from the bloom-forming cyanobacterium Trichodesmium erythraeum, predicted from the genome sequence. Appl. Environ. Microbiol. 2006;72:4382–4387. doi: 10.1128/AEM.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, Liu HB, Patel A, Sherman DH, Haygood MG. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J. Nat. Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Vairappan CS. Halogenated secondary metabolites from Japanese species of the red algal genus Laurencia (Rhodomelaceae, Ceramiales) Curr. Top. Phytochem. 2005;7:1–34. [Google Scholar]
- Swinney DC, Anthony J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- Taylor RE. Tedanolide and the evolution of polyketide inhibitors of eukaryotic protein synthesis. Nat. Prod. Rep. 2008;25:854–861. doi: 10.1039/b805700c. [DOI] [PubMed] [Google Scholar]
- Teichart RW, Olivera BM. Natural products and ion channel pharmacology. Future Med. Chem. 2010;2:731–744. doi: 10.4155/fmc.10.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa FA, Gerwick L. Marine natural product drug discovery: leads for treatment of inflammation, cancer, infections, and neurological disorders. Immunopharm. Immunot. 2010;32:228–237. doi: 10.3109/08923970903296136. [DOI] [PubMed] [Google Scholar]
- Weinheimer AJ. Discovery of 15-epi PGA2 in Plexaura homomalla. Stud. Trop. Oceanogr. 1974;12:17–21. [Google Scholar]
- Williams PG. Panning for chemical gold: marine bacteria as a source of new therapeutics. Trends Biotechnol. 2008;27:45–52. doi: 10.1016/j.tibtech.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Wright AE, Forleo DA, Gunawardana GP, Gunasekera SP, Koehn FE, McConnell OJ. Antitumor tetrahydroisoquinoline alkaloids from the colonial ascidian Ecteinascidia turbinata. J. Org. Chem. 1990;55:4508–4512. [Google Scholar]
- Yeung BKS. Natural product drug discovery: the successful optimization of ISP-1 and halichondrin B. Curr. Opin. Chem. Biol. 2011;15:523–528. doi: 10.1016/j.cbpa.2011.05.019. [DOI] [PubMed] [Google Scholar]