Abstract
Bacterial muropeptides are soluble peptidoglycan structures central to recycling of the bacterial cell wall, and messengers in diverse cell-signaling events. Bacteria sense muropeptides as signals that antibiotics targeting cell-wall biosynthesis are present, and eukaryotes detect muropeptides during the innate immune response to bacterial infection. This review summarizes the roles of bacterial muropeptides as messengers, with a special emphasis on bacterial muropeptide structures and the relationship of structure to the biochemical events that the muropeptides elicit. Muropeptide sensing and recycling in both Gram-positive and Gram-negative bacteria is discussed, followed by muropeptide sensing by eukaryotes as a crucial event to the innate immune response of insects (via peptidoglycan-recognition proteins) and mammals (through Nod-like receptors) to bacterial invasion.
A bacterium must engage its environment and yet preserve its structural integrity during the processes of growth and cell division. A key structural component of this integrity is the bacterial envelope. The envelope comprises the bacterial cell surface as a tightly organized series of layers including the cell wall, membrane(s), and the intervening space(s) between these layers.1,2 The organization of these layers demarcates the prokaryotes. For a monoderm (single membrane) bacterium such as Staphylococcus aureus (a Gram-positive-staining bacterium), the outer surface of its envelope is the cell wall, and the cell wall surrounds a single membrane. For diderm bacteria such as Mycobacterium tuberculosis (also Gram-positive staining), the outer surface is comprised of mycolic acid lipids attached to the cell wall, while for the Gram-negative-staining diderm Escherichia coli the layer organization is an outer-membrane surface (with a lipopolysaccharide external leaflet), followed by an intervening space (the periplasm, within which is the cell wall), and lastly an inner membrane. Because many of the components of the bacterial envelope are structurally unique to the bacteria, the biochemical pathways relating to the bacterial envelope (and especially to the cell wall) are molecular targets of antibiotics.3,4 In order to preserve the structural integrity of this incredibly complex envelope, bacteria have developed mechanisms to detect and respond to circumstances that jeopardize its integrity. Likewise, when the food source for a bacterium is an unwilling eukaryote, the eukaryote responds, and does so in part by detecting the presence of structures uniquely bacterial. One particular structural class, used in both detection events, is the bacterial cell wall-derived muropeptide.
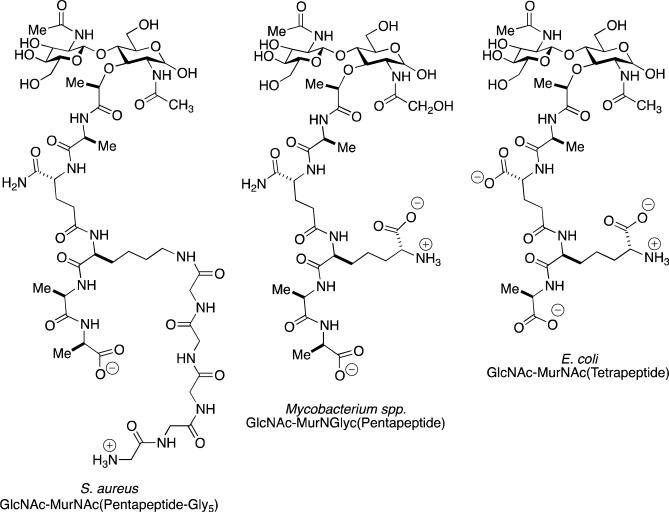
The bacterial cell wall is a peptidoglycan polymer—a single molecule, called the sacculus—formed by the cross-linking of oligopeptide stems presented by neighboring glycan strands. A synonym for the peptidoglycan polymer is “murein”, derived from murus, Latin for “wall”. The structure of the polysaccharide is a repeating β-(1→4)-(N-acetylglucosamine-β-(1→4)-N-acetylmuramic) disaccharide possessing interconnected oligopeptide stems attached to the lactyl group of the muramic saccharide. During cell-wall biosynthesis, the oligopeptide stem of one glycan strand is cross-linked by acyl transfer to an amine on the stem of an adjacent strand. Notwithstanding the seeming implication of a repetitive pattern for the peptidoglycan structure, the sacculus shows substantial “internal” variability in terms of glycan strand length, oligopeptide stem structure, and degree of cross-linking under different growth conditions. The term “muropeptide” refers to the ensemble of structures obtained from enzymatic digestion of the peptidoglycan polymer. This digestion uses both glycan-cleaving and stem-cleaving (amidase) enzymes, giving the representative (N-acetylglucosamine-β-(1→4)-N-acetylmuramic) (GlcNAc-MurNAc) muropeptide structures of Scheme 1. The ensemble of structures that results from these enzymatic events are identified typically by mass spectrometric interpretation of the peaks obtained from HPLC analysis.
Scheme 1.
Left: A representative muropeptide of the monoderm Gram-positive bacterium Staphylococcus aureus. The GlcNAc-MurNAc disaccharide is shown as the “free” MurNAc glycoside. This reducing saccharide is customarily transformed by chemical reduction prior to chromatographic analysis. The sequence of the peptide stem attached to the lactyl carboxylate of the MurNAc is l-Ala-d-iGln-l-Lys-d-Ala-d-Ala. A (Gly)5 “bridge” is biosynthetically added to the ε-amine of the l-Lys. In the stem cross-linking event catalyzed by the bifunctional penicillin-binding protein enzymes, the ultimate d-Ala residue is lost as a leaving group as an acyl-enzyme is formed. This d-alanyl acyl-enzyme is transferred to the amine of the terminal glycine of the (Gly)5 bridge of an adjacent peptidoglycan strand to complete the cross-link. Center: A representative muropeptide of a Gram-positive Mycobacterium spp., with an l-Ala-d-iGln-meso-DAP-d-Ala-d-Ala stem peptide sequence. Crosslinking occurs from the d-alanyl acyl-enzyme derived from one strand to the amine terminus of the meso-diaminopimelate of an adjacent strand. Right: A representative muropeptide of the diderm Gram-negative bacterium Escherichia coli.
Muropeptide structures are also obligatory intermediates during cell-wall biosynthesis and remodeling by bacteria, and during infection as a result of the immune response. The biological relevance of muropeptide structures to both events has been known for some time.5 However, as a result of the confluence of technological advances, including an improved understanding of the molecular bases for immune recognition, the routine availability of robust methods to determine muropeptide structure, and the development of methods for muropeptide synthesis, the specific roles of particular muropeptide structures as messengers are at last coming into focus. Muropeptides serve as messenger molecules to the bacterium that cell-wall-targeting antibiotics are present, to spore formers and persisters that time for outgrowth is optimal as communicated by neighbouring cells, and to the eukaryote that a bacterial infection is in progress. In this perspective we evaluate these roles, with particular emphasis on the relationship of muropeptide structure to the key biochemical events within each.
MUROPEPTIDE SENSING BY GRAM-POSITIVE BACTERIA
The term antibiotic has a strong anthropocentric connotation: a molecule isolated from Nature that has the ability to engage a bacterial target so as to prevent bacterial growth, or to act as a bactericide, and thus having value for the chemotherapeutic control of infection. Efforts to understand the roles of the natural products in Nature, and of the “antibiotics” in particular, from disinterested perspectives embrace their potential additional roles in intraspecies and interspecies communication.6-12 Nonetheless, antibiotics are indisputably functional molecules, and a bacterium that encounters an antibiotic must respond whether for the purposes of evasion or mobilization of a defense. A consequence of the targeting by many antibiotics of the enzymes of cell-wall biosynthesis and remodeling is perturbation of the steady-state concentrations of biosynthetic intermediates, and now in two examples—the β-lactam antibiotic-dependent induction of vancomycin resistance in the Gram-positive bacterium methicillin-resistant S. aureus and induction of β-lactam resistance in Gram-negative bacteria—muropeptides act as messenger molecules.
The Muropeptide Messenger in β-Lactam-Dependent Induction of Vancomycin Resistance in Methicillin-resistant S. aureus
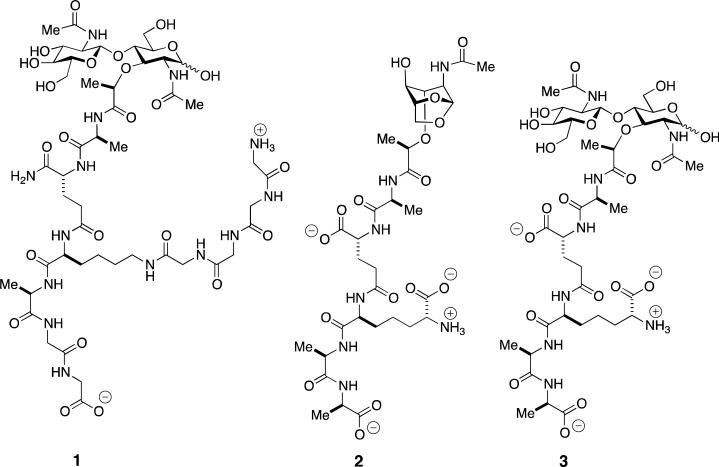
Vancomycin is a glycopeptide antibiotic that is most active against Gram-positive bacteria.13,14 Its mechanism is disruption of cell-wall biosynthesis by occlusion of the key structures in cell-wall biosynthesis, especially the biosynthetic intermediate Lipid II. This occlusion occurs by formation of a stable, non-covalent complex between vancomycin and cell-wall structures exhibiting a ...d-Ala-d-Ala stem terminus (see Scheme 1) as a key recognition motif.15 Two resistance mechanisms to vancomycin are known. In the Enterococci an alternative biosynthetic pathway operates wherein the ...d-Ala-d-Ala stem structure is replaced with a depsipeptide ...d-Ala-d-Lac structure, for which vancomycin has poor affinity.16,17 Vancomycin-resistant Enterococci result. Intermediate-level vancomycin-resistance occurs in another Gram-positive bacterium, S. aureus, as a result of increased peptidoglycan biosynthesis giving a thicker and more extensively cross-linked peptidoglycan. While vancomycin·d-Ala-d-Ala complexes are still formed, they are unproductive with respect to inhibition of cell-wall biosynthesis.18 A new variation on this second mechanism to attain vancomycin resistance is now described for a strain of methicillin-resistant S. aureus (MRSA).19 MRSA has high-level resistance to the β-lactam class of antibiotics (this class includes the extensively clinically used penicillins, cephalosporins and carbapenems). β-Lactam antibiotics act as ...d-Ala-d-Ala mimetics to achieve irreversible inactivation of the transpeptidase domain of the bifunctional penicillin-binding protein (PBP) enzymes of peptidoglycan biosynthesis. β-Lactam resistance by MRSA results from the replacement of a β-lactam-susceptible PBP by a PBP enzyme that is not susceptible. Because MRSA-infected patients are often also infected with Gram-negative bacteria, combined vancomycin and β-lactam therapy is common. This combination therapy has given rise to β-lactam antibiotic-induced vancomycin-resistant MRSA (BIVR-MRSA). In the presence of β-lactam antibiotics, BIVR-MRSA cells survive by a structural transformation that enables the rapid depletion of vancomycin from their environment, eventually allowing the resumption of cell growth when the vancomycin is sufficiently depleted.19 A hypothesis to explain this phenomenon, based on the vancomycin-resistant Enterococci, is increased Lipid II biosynthesis induced by the β-lactam antibiotics giving more abundant d-Ala-d-Ala-containing muropeptides. Ikeda et al. show that in the presence of both vancomycin and the β-lactam antibiotic ceftizoxime, BIVR cells released 10-fold more muropeptides into the medium compared to either MRSA or methicillin-susceptible S. aureus.20 Induction of BIVR resistance correlated with the presence of a particular muropeptide structure, GlcNAc-MurNAc-l-Ala-d-iGln-l-Lys-(ε-Gly4)-d-Ala-Gly2 (1, Scheme 2), identified by in vitro enzymatic degradation of the cell wall and exhaustive muropeptide purification. When this purified muropeptide is added to a vancomycin-containing culture of BIVR-MRSA in the absence of a β-lactam, the bacteria grow more rapidly than bacteria treated with vancomycin alone. In this same assay, the muropeptide derivative of 1 lacking the tetraglycine substitution to the ε-amine of the lysine was equipotent compared to 1. The ability of these muropeptides at 50–100 μg mL–1 to promote BIVR-MRSA cell growth exceeded that of 1 μg mL–1 ceftizoxime.
Scheme 2.
Muropeptide messengers implicated in the induction of antibiotic resistance: (1): BIVR-MRSA S. aureus responds to the presence of muropeptide 1 in the medium with the creation of a cell wall better able to deplete vancomycin from the medium. (2): Following β-lactam challenge of Gram-negative bacteria having a complete ampR-ampC system, the structural homeostasis of the free muropeptide pool entering the cytoplasm for recycling is altered. Among the enzymes controlling the structural identities of the entering muropeptides are a low-molecular-mass PBP (found in the cytoplasm, and presumed to be inhibited by the β-lactam), the NagZ β-glucosaminidase of the cytoplasm, and the AmpD amidase of the cytoplasm (see Scheme 5). Anhydromuropeptide 2 is the probable structure that engages the LysR-type transcription regulator AmpR so as to derepress the ampC gene and allow AmpC β-lactamase expression. (3): Following β-lactam challenge of the Gram-negative bacterium Aeromonas hydrophila, the presence of muropeptide 3 is increased (whether as part of the cell wall, or as the free muropeptide). This increase is sensed by a BlrB kinase system, and results in the induction of β-lactamase expression.
The mechanism by which 1 induces this BIVR phenomenon is not fully understood. Ikeda et al.20 exclude the simplest explanation—complex formation between 1 and vancomycin is not observed—and hence 1 itself does not act as a vancomycin trap. Previous studies on the muropeptide composition of the peptidoglycan polymer of MRSA show alterations in its biosynthesis, likely as a result of the use of a different ensemble of biosynthetic muropeptides.21 Since the thickened cell wall observed in vancomycin-intermediate S. aureus correlates to a vancomycin-trapping mechanism,22 a mechanism whereby released muropeptides are internalized so as to “activate” cell-wall biosynthesis (for example, by altering muropeptide structure through biosynthetic gene repression and expression, so as to favor peptidoglycans with improved vancomycin binding) is possible.23 A muropeptide-sensing mechanism is used to induce β-lactam resistance in Gram-negative bacteria (vide infra). In contrast, the signaling messenger in the VanS histidine kinase-dependent expression of the vancomycin-resistance pathway of Streptomyces coelicolor is vancomycin itself.24 Both observations attest to the ability of these Gram-positives to internalize complex structures from their environment, and to act on this internalization with a resistance response. In contrast, the molecular mechanism for the expression of vancomycin resistance in Enterococcal pathogens is not proven. Vancomycin resistance has a substantial fitness cost that correlates to a requirement for its inducible regulation through a vanR/vanS system.25 However, there are substantial genetic variations in this operon—for example,26-28 encompassing differential responses by vancomycin, televancin, and teicoplanin—leaving open the possibilities of regulation by the antibiotic, or by changes in the cell wall, or by changes in the levels of cell wall intermediates.23
Muropeptide Recycling in Gram-positive bacteria
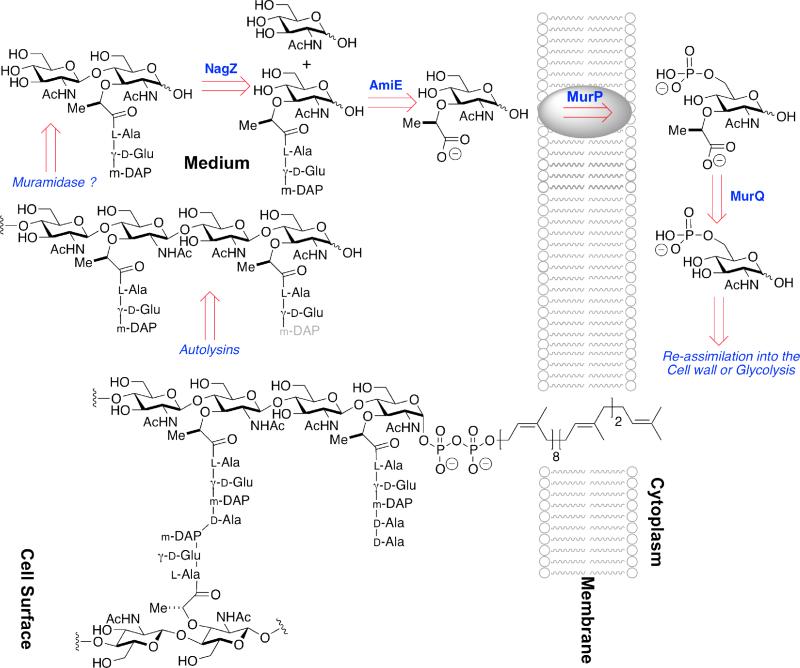
The ability of Gram-negative bacteria to efficiently recycle their muropeptides is long recognized, and interpreted in terms of the evolutionary advantage with respect to these nutrients, and in the minimization of the immune response of a host to bacterial infection (vide infra).29,30 Recognition that muropeptide recycling occurs in Gram-positive bacteria is, however, new.29,31 While the peptidoglycan of Gram-negative bacteria is assembled between two membranes, and thus the muropeptides they used for both cell-wall biosynthesis and cell-wall remodeling are confined to their periplasmic space, the cell wall of the monoderm Gram-positive bacterium is exposed to the medium. The apparent absence of a Gram-positive periplasmic space and the observation that as much as 50% of the cell wall is released during the growth of the model Gram-positive bacterium B. subtilis suggested the absence of efficient muropeptide recycling.32 As a complement to a study of N-acetylmuramic acid catabolism in E. coli,33 Mayer et al. identified a cluster of six genes in B. subtilis encoding muropeptide-recycling enzymes. The identity of the gene products of five is known: MurQ, a MurNAc-6-phosphate etherase; MurR, the transcriptional regulator; MurP, a MurNAc-specific phosphotransferase; AmiE, an amidase hydrolytically cleaving the MurNAc-l-Ala amide bond; and NagZ, a cell-wall associated β-N-acetylglucosaminidase.34 The suggested operation of four of the enzymes in this gene cluster,31 in concert with other cell-wall autolysins released into the medium, is summarized in Scheme 3. The four B. subtilis enzymes are orthologs of enzymes found in the muropeptide-recycling pathway of E. coli. Additional orthologs of other enzymes, and of a phosphotransferase used by E. coli for GlcNAc recycling, were also identified in B. subtilis.31,35 Although our present understanding suggests that nutrient recovery is a primary role, the evidence implicating a role for muropeptides in β-lactam-dependent induction of vancomycin resistance in BIVR-MRSA and Gram-positive spore resuscitation (discussed below) suggests the possibility that Gram-positive muropeptide recovery may be used for transcriptional regulation of resistance responses.
Scheme 3.
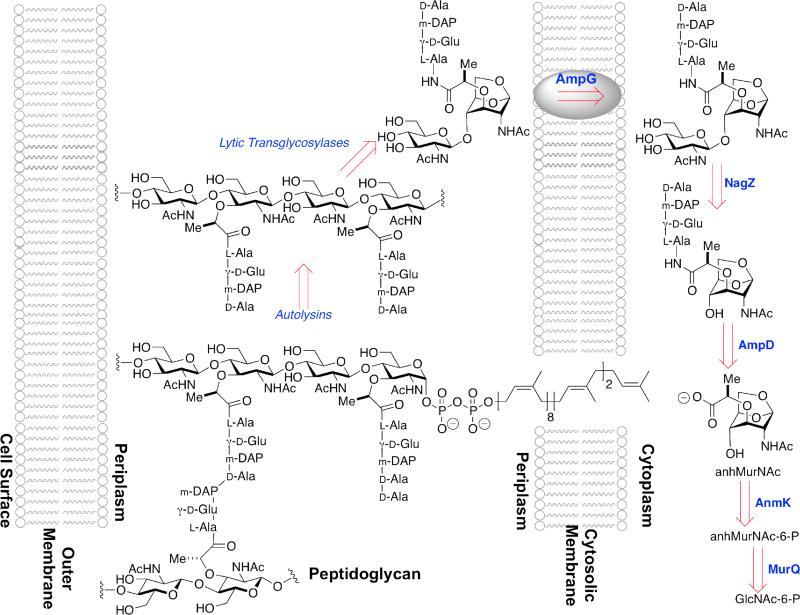
A schematic overview of MurNAc recycling in the Gram-positive monoderm B. subtilis. The single membrane of the bacterium is depicted as a cartoon running vertically to the right of center. The cytoplasm of the bacterium is found to the right of the membrane, while the cell surface is found to the left. In Gram-positive bacteria the cell surface consists of thick (relative to Gram-negative bacteria) peptidoglycan (in the example of B. subtilis, approximately 35 nm),36 to which proteins are both covalently and non-covalently attached. The membrane-bound peptidoglycan-lipid (lower structure) represents a biosynthetic intermediate, here shown with one uncross-linked peptide stem and with one cross-linked (to an adjacent glycan strand) peptide stem. Autolysin (including a not-yet-characterized muramidase) releases the GlcNAc-MurNAc disaccharide (upper left). NagZ-dependent glycosylase activity splits the disaccharide, following which AmiE (possibly in concert with other amidases) releases the peptide. MurNAc is internalized by the MurP phosphotransferase, after which the lactyl ether is cleaved by the MurQ etherase. This schematic for B. subtilis should not be taken as general: Reith and Mayer have obtained strong evidence for a wholly cytoplasmic recycling pathway in the Gram-positive bacterium Clostridium acetobutylicum.37,38
Muropeptide Sensing in Gram-positive Resuscitation
Bacteria have sophisticated mechanisms to cope with environmental stress, including the formation of persister39 and dormant states (notably seen for mycobacteria),40 and spore formation by other Gram-positive bacteria (notably the Gram-positive Clostridiae and Bacilli spp).41-43 Entry into dormant and spore states, and resuscitation from these states, is of particular interest due to the relationship of these states to human infection by (for example) Mycobacterium tuberculosis and Bacillus anthracis.44,45 Peptidoglycan structure is a key aspect of these dormant states, and especially with respect to resuscitation. The structures of the peptidoglycans of the dormant M. tuberculosis and the B. anthracis spore are altered for the purposes of structural integrity, stability toward muramidase degradation, and possible differentiation of the structure of the dormant peptidoglycan from the peptidoglycan made during bacterial growth. A key feature of the dormant mycobacterial peptidoglycan is altered cross-linking46,47 of an N-glycolylmuramic acid-containing peptidoglycan,48,49 and for the Bacilli the extensive appearance in the spore peptidoglycan of the so-called muramic δ-lactam.50
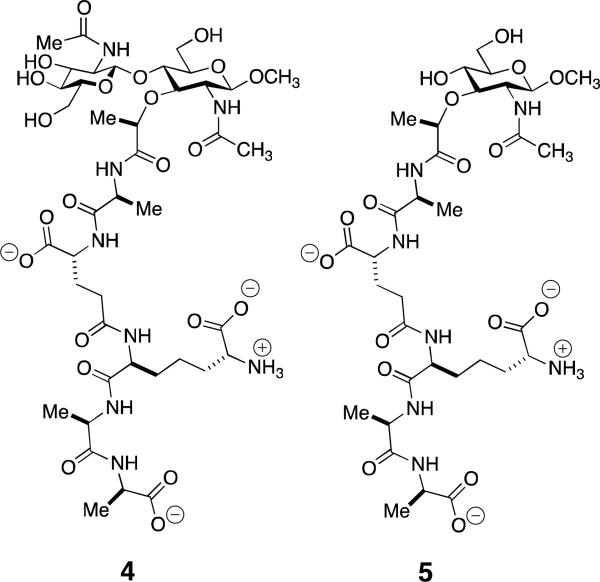
A central issue to the understanding of both dormant bacteria and bacterial spores is the mechanism(s) by which they sense the presence of an advantageous environment for the resumption of growth. While there is no doubt that the sensing process is multi-step and involves concurrence of multiple signaling pathways, substantial evidence now implicates muropeptides as key signaling molecules during the resuscitation pathways of both dormant mycobacteria and Bacilli spores.51,52 The targets of these muropeptides are specific bacterial membrane-bound eukaryotic-like serine/threonine protein kinases (STK).53-55 Dworkin et al. have correlated environmental muropeptides, released as a result of peptidoglycan digestion by a secreted peptidoglycan hydrolase,56 acting as germinants toward B. subtilis spores through activation of the PrkC STK kinase.57,58 Evaluation of a set of synthetic muropeptides59 as stimulants of the germinant activity of B. subtilis spores identified the GlcNAc-MurNAc muropeptide 4 (Scheme 4) as substantially more effective (EC50 10 nM) than either MurNAc monosaccharide 5 or GlcNAc-anhMurNAc disaccharide 2 (Scheme 2). A tetrasaccharide (GlcNAc-MurNAc)2 muropeptide was equipotent to 4. The molecular target of muropeptide 4 as the PrkC STK was proven elegantly by substitution of the B. subtilis PrkC STK gene with that of the homologous gene for S. aureus PrkC. B. subtilis synthesizes a meso-diaminopimelate-containing peptidoglycan, and its PrkC STK does not recognize lysine-containing muropeptides. In contrast, S. aureus synthesizes a lysine-containing peptidoglycan and its PrkC recognizes both meso-diaminopimelate- and lysine-containing muropeptides. Thus spores obtained from B. subtilis with the S. aureus STK would be expected (as long as the S. aureus STK remains competent for signaling in B. subtilis) to respond to a lysine-containing muropeptide. Indeed, the S. aureus STK is competent in B. subtilis, and the spores with the S. aureus STK resuscitate in response to the S. aureus muropeptide.59 Among the pathways presumed activated in spore germination are four (in B. anthracis) lytic enzymes required for the remodeling of the cortex peptidoglycan of the spore.60-62
Scheme 4.
Structure of the active GlcNAc-MurNAc muropeptide 4 for muropeptide-dependent resuscitation of B. subtilis spores, via activation of the B. subtilis STK serine/threonine kinase, and compared to the much less active MurNAc muropeptide 5. The two synthetic muropeptides59,63 show β-methyl glycosidic termini at their MurNAc saccharide.
A complete understanding of the relationship between muropeptide binding to the STK and activation of the STK is not fully in place. The structural organization of these two PrkC proteins is identical: an intracellular serine/threonine kinase domain, a transmembrane domain, and an extracellular domain consisting of a three-fold repeat of a penicillin binding-associated and serine/threonine kinase associated (PASTA) domain.53,64 PASTA domains were first encountered in the structure of the highly β-lactam antibiotic-resistant penicillin-binding protein 2x,65,66 an enzyme used by methicillin-resistant Streptococcus pneumoniae to complete the transpeptidation step of cell-wall biosynthesis in the presence of otherwise inhibitory concentrations of β-lactam antibiotics. The presumption that the muropeptide (and polymeric peptidoglycan)67 bind to the PASTA domain(s) is supported by numerous studies.58,59 In contrast to the compacted PASTA domains seen in the penicillin-binding protein 2x crystal structures, recent STK structural studies on the M. tuberculosis PknB kinase,68 the S. aureus Stk1 kinase,69 and the S. aureus PrkC kinase70 implicate an extended orientation of the PASTA domains. Notwithstanding the sub-micromolar muropeptide activity of synthetic muropeptides in B. subtilis spore resuscitation, the in vitro affinity of these STKs for muropeptide structure is not high. A GlcNAc-MurNAc-pentapeptide muropeptide showed millimolar affinity for an engineered tryptophan reporter group-containing S. pneumoniae StkP kinase,67 millimolar affinity for the B. subtilis PrkC,58 and millimolar binding of more than one MurNAc-tripeptide to the three PASTA-domains of the S. aureus PrkC kinase.69 Given the limited diversity of available muropeptide structures, and our ignorance as to the exact structures their respective kinases have evolved to respond, these initial determinations of relatively weak in vitro affinity cannot be interpreted. This challenge is exemplified by the observation that peptidoglycan from growing bacteria, as opposed to the peptidoglycan from stationary phase bacteria or the peptidoglycan from the spore cortex, is optimal as a germinant.52 The expected mechanism for STK activation is muropeptide-induced dimerization,71-73 and proposals have been made for the role of the PASTA domains in this dimerization.68-70 Muropeptide binding is suggested to occur at the hinge region between individual PASTA domains.70
A conceptually similar pathway may operate in the transition from dormancy to active growth in the non-sporulating actinobacteria, exemplified by Micrococcus luteus and M. tuberculosis. As noted above M. tuberculosis has a PASTA domain-containing STK, PknD. A possible relationship of muropeptide signaling to its activation is the resuscitation-promoting (Rpf) class of enzymes, of which M. luteus has one and M. tuberculosis has five. In contrast to spore resuscitation, however, both the Rpf and STK enzymes function within the broad scope of dormancy rescue, growth, cell division and pathogenicity.45,53 A molecular understanding of the presumed involvement of muropeptide-dependent signaling is less developed. The seminal observation was the identification of the M. luteus Rpf as a secreted protein74 with muralytic activity.51,75,76 Neither its requirements with respect to its peptidoglycan as substrate, nor the products of its catalytic activity (whether lysozyme-like or lytic transglycosylase-like), is established. A similar uncertainty as to molecular mechanisms carries forward to five Rpf proteins of M. tuberculosis, notwithstanding structure studies on the one of the five (RpfB) that is absolutely essential to resuscitation.77,78 Adding to the challenge of understanding the roles for RpfB is the discovery of its complexation with RipA, an enzyme essential for cell division and believed to have amidase activity toward the d-iGln–m-DAP amide bond of the peptidoglycan stem,79 and which also interacts with the high-molecular-mass biosynthetic penicillin-binding protein of this bacterium so as to antagonize the muralytic activity of RpfB.80 Given the importance of the M. tuberculosis Rpf proteins to both the resuscitation and virulence (but not growth) of this bacterium,81 there is a compelling opportunity to clarify the molecular basis of their peptidoglycan-dependent signaling. This effort might coincide either toward inhibiting their activity82 or alternatively to stimulating the Rpf-dependent pathways, following the observation that nutrient resuscitation of persister bacteria can restore antibiotic susceptibility.83
MUROPEPTIDE SENSING BY GRAM-NEGATIVE BACTERIA
A characteristic of the growth of many Gram-negative bacteria is robust turnover of the their cell wall via muropeptide formation.30 A single generation of growth liberates approximately 40–45% of the E. coli peptidoglycan as anhydromuropeptides, which subsequently are efficiently recovered and reused. The dynamics of cell-wall growth and turnover are relevant to the understanding of cell-wall biosynthesis during growth and division, and to the mechanism used by many Gram-negative bacteria to mount a resistance response to β-lactam antibiotics (antibiotics which interfere with peptidoglycan synthesis by inactivation of the transpeptidase catalytic domain of the penicillin-binding proteins). Many Gram-negative bacteria sense a disruption in the recycling pathway as a consequence of the β-lactams as antibiotics, and as a result of this sensing initiate expression of resistance enzyme(s). In particular, the high-level β-lactamase expression that results from this disruption achieves clinical levels of β-lactam resistance.84 The seemingly paradoxical observation that the integrity of the peptidoglycan-recycling pathway is neither essential to cell growth under laboratory conditions, nor correlates to an observable in vitro phenotype, suggests that a preeminent purpose for muropeptide recycling is the regulation of resistance and virulence responses.85
The high-level expression of these inducible AmpC β-lactamases confers high-level β-lactam resistance to many Gram-negative pathogens. AmpC expression is induced by the presence of β-lactam antibiotics, regulated by the ampR-ampC system. AmpR is the 32-kDa LysR-type transcriptional regulator of this system. The ampR gene is absent from the chromosome of some Gram-negatives, notably E. coli. As a result expression of the E. coli AmpC β-lactamase (the gene for this enzyme is present) is constitutive but attains only very low levels of AmpC. The complete ampR system is present in Citrobacter freundii, where it is best studied. The structure of the effector binding domain (lacking the N-terminal DNA binding domain) of C. freundii AmpR has been solved in a conformational state that although lacking a bound effector, is suggested by circumstantial evidence to correspond to the state wherein AmpC expression is repressed.86 This same study confirmed the sensitivity of ampR to point mutation, which can result either in the loss of AmpC expression or in its constitutive expression.
The central question is the regulatory relationship between the ampR-ampC system and peptidoglycan biosynthesis and recycling. Peptidoglycan biosynthesis uses a biosynthetic precursor (Lipid II) that is fully assembled in the cytoplasm and exported to the periplasm to serve as the substrate of the PBP enzymes for the sequential glycan-strand elongation and peptide stem cross-linking events of cell-wall biosynthesis. New peptidoglycan is incorporated uniformly into existing peptidoglycan during sidewall elongation of the rod-shaped E. coli bacterium,87,88 and this incorporation coordinates with substantial liberation of portions of the existing peptidoglycan as soluble anhydromuropeptides.30,89 While the identity of the full ensemble of enzyme catalysts used for cell-wall biosynthesis is uncertain, anhydromuropeptide release involves both non-hydrolytic glycan cleavage catalyzed by the lytic transglycosylase enzymes,90,91 and hydrolytic amidase-catalyzed cleavage of the peptide stem cross-links.92,93 The resulting anhydromuropeptides (Scheme 5)—the anhydro prefix refers to the bridging ether ring that results from the action of the lytic transglycosylases toward the MurNAc–GlcNAc glycosidic bond—and oligopeptides are released into the periplasm. A portion of the amidase-released oligopeptides is released from the periplasm to the medium.89 These oligopeptides are recovered for reuse by complexation to the MppA binding protein for subsequent import through the Opp oligopeptide permease system.94-96 The anhydromuropeptides are themselves recovered for reuse by entry into the cytoplasm through the AmpG permease protein. Indeed, the central roles for both the lytic transglycosylases97,98 and the AmpG permease99-101 in muropeptide recycling were first identified through correlation to β-lactam-dependent β-lactamase expression.102,103
Scheme 5.
A schematic overview of peptidoglycan recycling in Gram-negative bacteria. The cytoplasmic membrane of the bacterium is depicted as a cartoon at the right, and the outer membrane at the left. The periplasmic space between these two membranes contains the peptidoglycan. After initial processing by autolysins, lytic transglycosylases cleave the glycosidic bond in a non-hydrolytic fashion giving a GlcNAc-anhydroMurNAc disaccharide. Transport into the cytoplasm by the AmpG permease is followed by NagZ-dependent glycosylase activity, which splits the disaccharide. AmpD then cleaves the peptide stem as shown, AnmK hydrolyzes the hemiacetal and phosphorylates the 6-position of MurNAc, and MurQ removes the lactyl moiety.
As it is now understood, the muropeptide-recycling pathway coordinates to de novo peptidoglycan biosynthesis by a concise series of events. Muropeptides liberated during peptidoglycan biosynthesis are “sized” by the lytic transglycosylases localized to the inner leaflet of the outer membrane to give the GlcNAc-anhydroMurNAc structures shown in Scheme 5.104 Translocation of this muropeptide to the cytoplasm occurs through the AmpG permease. Within the cytoplasm, the glycosidic bond of this muropeptide is hydrolytically cleaved by the NagZ β-N-acetylglucosaminidase.105,106 The action of NagZ to provide anhydroMurNAc products—in all of their tri-, tetra, and pentapeptide forms—constitutes the entry point to the pool of effector structures believed to govern the structure and/or affinity of the DNA-ampR complex. The precise composition of this effector pool is further controlled by at least three other enzymes.107 NagZ is a cytosolic β-glucosaminidase that acts on the anhydromuropeptide as it enters the cytoplasm (Scheme 5). AmpD is a cytosolic zinc-dependent amidohydrolase that cleaves the lactyl amide of the muropeptides, releasing as products the peptide stem and an anhydroMurNAc muropeptide. Both products enter the de novo Lipid II biosynthesis pathway. The third enzyme is a low-molecular-mass PBP, which in vitro (and possibly in vivo) acts as a d,d-carboxypeptidase with respect to the d-Ala-d-Ala residues of the peptide stems found on some of the muropeptides of the peptidoglycan polymer, and on some of the anhydromuropeptides released by the lytic transglycosylases. As this PBP activity is not central to the peptidoglycan-recycling pathway, its role with respect to composition of the effector pool is considered following a closer consideration of NagZ and AmpD.107
The hypothesis that the relative levels of the catabolic and anabolic muropeptides, as directly influenced by AmpG and these enzymes, govern AmpC transcription is supported by complementary experiments from several laboratories. Muropeptides originate through sequential lytic transglycosylase and amidase excision in the periplasm, and pass into the cytoplasm through the AmpG permease. Genetic deletion of the lytic transglycosylases of E. coli,98 or of the permease in β-lactam-resistant P. aeruginosa,108,109 restores significant β-lactam susceptibility as a result of the inability of these mutants to induce AmpC expression through loss of muropeptides in the cytoplasm. The structural basis of AmpC expression is suggested, but not proven, by additional observations. Muropeptides which enter the cytoplasm through the AmpG permease are acted upon by the NagZ and AmpD enzymes. Mutational loss-of-function of AmpD is a long-known clinical event that results in high-level constitutive AmpC expression.110,111 Our current mechanistic understanding assigns AmpD the role of dissipating structures within the anhydromuropeptide pool that would otherwise bind to AmpR and repress AmpC expression. A detailed examination of the substrate specificity of the C. freundii AmpD enzyme shows a strong preference for an anhydromuramic acid (monosaccharide) substrate, and insensitivity (comparing the tri- to pentapeptide) with respect to peptide stem length.112 A crystallographic study of this same AmpD shows an open conformation capable of substrate accommodation, and verifies its close structural similarity to the peptidoglycan-recognition proteins (PGRPs) of eukaryotic innate immunity.113 A corollary to the observation that AmpD null mutations confer a resistance phenotype is that inhibition of NagZ should potentiate β-lactam efficacy. This expectation is confirmed.114-118 The optimization of NagZ inhibitors with respect to potency, selectivity (relative to human enzymes), and accessibility (with respect to access to the cytoplasmic NagZ enzyme) so as to attain clinical relevancy is a promising strategy against high-level Gram-negative β-lactam resistance.107 Moreover, these same inhibitors will enable clarification of the possible relationship between muropeptide recycling and resistance in Gram-positive bacteria.
The simplest explanation for control of AmpR regulation of AmpC expression through the ebb and flow of the metabolic/catabolic muropeptide pool in the cytoplasm cannot be, however, fully correct. The direct participation of a low-molecular-mass PBP suggests the likelihood of specific, regulatory muropeptide structures. It has been long known to medicinal chemists that optimization of β-lactam antibiotic structure required awareness of their spectrum of PBP inhibition, so as to secure preferential inactivation of high-molecular-mass PBPs (antibiotic activity) while avoiding inactivation of low-molecular-mass PBPs (induction of β-lactamase expression).119 Oliver et al. have directly connected PBP4 of P. aeruginosa to the muropeptide recycling pathway.109,120,121 The presumption is that the enzymatic activity of this PBP alters the -d-Ala-d-Ala pentapeptide stem structures so as to enable an anhydromuropeptide-AmpR complex that coincides with repressed AmpC expression. The precise anhydromuropeptide structure used in AmpR complexation is uncertain.107,122 When a β-lactam inactivates PBP4, the enzymatic activity is abolished, and the composition of the entering muropeptides alters so as to result in an AmpR complex that derepresses the ampC gene. PBP4 (and presumably a low-molecular-mass PBP of other Gram-negatives with the ampR-ampC system) thus signals the presence of β-lactam antibiotics through control of the anhydromuropeptide structures entering the cytoplasm. A candidate for the effector structure that binds to AmpR, so as to derepress the ampC gene encoding the AmpC β-lactamase, is shown as structure 2 of Scheme 2.
Although assigning to this PBP a role as a sentinel enzyme is attractive—an assignment that harmonizes fully with the consequences of AmpG deletion, AmpD mutational inactivation, and NagZ inhibition—other observations suggest a more complicated β-lactamase induction pathway in these Gram-negatives. As one unexplained observation, most Gram-negatives have several low-molecular-mass PBPs (E. coli has seven, P. aeruginosa at least three), all of which are ascribed d,d-carboxypeptidase and/or -endopeptidase activities. The P. aeruginosa PBP4 is not the most abundant. While there are temporal patterns with respect to the appearance of these enigmatic low-molecular-mass PBPs during culture, it seems unlikely that the abrupt loss of the activity of one minor PBP would substantively alter the composition of the robust stream of muropeptides entering the cytoplasm for recycling. Uncertainty as to the true enzymatic activity (and the preferred substrates) of these enigmatic low-molecular-mass PBPs123 complicates efforts to assign to one of these PBPs full responsibility for a specific structural transformation in the muropeptide recycling pathway. One emerging possibility is clearer recognition that bacterial peptidoglycans may incorporate d-amino acids other than d-alanine, and that the -d-Ala-d-Ala terminus of the peptidoglycan stem may not be as unique as was once thought.124-126 Modulation of the ampR gene may occur through interaction of AmpR with structurally unprecedented anhydromuropeptides of the recycling pool.
Additional observations attest to the complexity of Gram-negative muropeptide recycling. P. aeruginosa has muropeptide permeases apart from AmpG.108,127 Although Neisseria gonorrhoeae has a fully functional AmpG (but not apparently ampR), it has an unknown pathway that it uses to divert its muropeptides into the medium as an apparent virulence mechanism.110 As discussed below, muropeptide-induced toxicity is characteristic of yet other Gram-negative pathogens such as Shigella and Bordetella.128 An interrelationship, by an as-yet-unproven mechanism, between the ampR-ampC and BlrAB (CreBC) resistance systems also occurs in P. aeruginosa120 and Aeromonas hydrophila.129 In P. aeruginosa, the BlrAB two-component regulatory system and the AmpR system are simultaneously activated as a result of PBP4 inactivation.120A. hydrophila PBP inactivation by β-lactam antibiotics alters the homeostasis of the muropeptide pool. This alteration is sensed by a periplasmic BlrB two-component kinase, and is expressed through auto-phosphorylation of the BlrB cytoplasmic domain. Phosphate transfer to the cytoplasmic BlrA protein enables phospho-BlrA binding upstream to the blr regulon gene promoters so as to recruit an RNA polymerase to initiate β-lactamase expression.129 Analysis of the concurrent change in the muropeptide composition of the A. hydrophila peptidoglycan polymer showed a four-fold increased abundance of a single muropeptide sub-structure 3 (Scheme 2). Verification of a direct interaction between this muropeptide (whether as the free muropeptide, or as a sub-structure within the murein polymer) and BlrB is awaited. Last, Stenotrophomonas maltophilia has two chromosomal β-lactamases whose expression is controlled by an ampR-ampC-type system. While the expression of these two β-lactamases is muropeptide-dependent (expression requires the AmpG permease, and with high-level expression upon AmpD deletion), evaluation of different β-lactam structures and deletion of a high-molecular-mass PBP implicate a NagZ-independent pathway for β-lactamase induction in addition to induction through its intact ampR-ampC system.130
MUROPEPTIDES IN EUKARYOTIC IMMUNE RECOGNITION OF BACTERIA
The germline-encoded innate-immune system detects and defends against microbial pathogens, and as the first line of defense in vertebrates precedes activation of the acquired immune system. The innate-immune system uses immune sensors, known as pattern-recognition receptors (PRRs),131 that are activated by microbial-derived structures known as pathogen-associated molecular patterns (PAMPs).132 Mammalian PRRs include the extracellular Toll-like receptors,133-137 the intracellular nucleotide-binding domain/leucine-rich repeat receptors (NLRs),138-140 and the retinoic acid-inducible gene-I-like receptors (RLRs).141,142 The immune responses that follow activation of the PRRs reflect extensive receptor cross-talk, presumably in order to validate the focus and extent of an immune response.143,144 Due to the relevance of PRR recognition with respect to (for example) infection, autoimmune disease, and microbiome equilibrium with differential commensal organism versus pathogen recognition,145 an understanding of the PRRs is an emerging area of immunology. These aspects are beyond the scope of this review. Our focus is the structural aspects of PRR recognition of muropeptides, and how this recognition is relevant to the ensuing (or absence of an) immune response.146
Bacterial PAMPs include peptidoglycan-derived muropeptides from both Gram-positive and Gram-negative bacteria,147 lipoprotein-derived lipopeptides,148 lipopolysaccharide segments derived from the outer membrane of Gram-negative bacteria (LPS),149 and lipoteichoic acid segments derived from the peptidoglycan of Gram-positive bacteria.150 While the immunostimulatory activities of muropeptides have been known for decades,5 it is only recently that the molecular basis for muropeptide immune recognition has emerged, in parallel with methodologies enabling the synthesis of muropeptides (as well as the other PAMPs) having defined structure. A critical limitation of prior efforts using materials from biological sources was insufficient reagent purity. The extraordinary progress with respect to both advances in muropeptide synthesis, as well as improved methods for the isolation and purification of endogenous muropeptides, is exemplified by the correlation of specific chemical structure to the activation of the NOD1 and NOD2 NLR receptors.151-154 NOD1 and NOD2 recognize the muropeptides resulting from degradation of bacterial peptidoglycan. These receptors have three domains: a C-terminal leucine-rich repeat (LRR) responsible for ligand recognition, a central NOD domain that facilitates oligomerization and exhibits ATPase activity, and an N-terminal caspase-recruitment domain (CARD). Interest in the NOD receptors is driven by numerous studies correlating their expression with human diseases, such as intestinal homeostasis and other allergic diseases,155 and more recently to their critical role in the autophagic response to bacterial infection.156,157
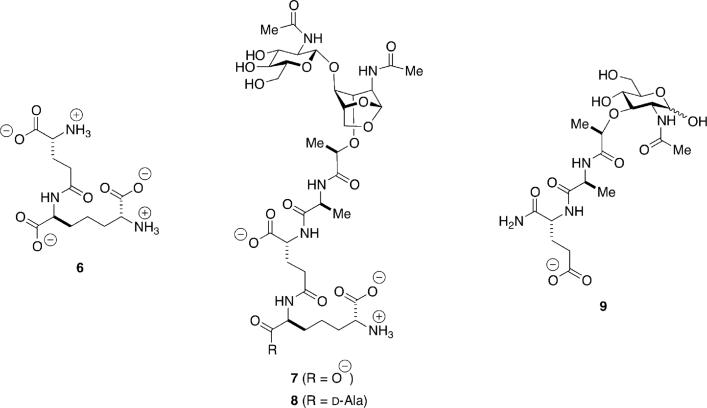
The minimal structure recognized by NOD1 is γ-d-glutamyl-meso-diaminopimelic acid (6, Scheme 6).158,159 This structure is characteristic of the peptidoglycan of Gram-negative bacteria, as further exemplified by muropeptides 7 and 8. NOD2 recognizes muramyl dipeptide (MDP, 9).160,161 1,6-Anhydromuramyl moieties such as 7 and 8 are generated by the action of lytic transglycosylases, discussed previously with respect to their central role in peptidoglycan recycling. Fukase and co-workers showed that a synthetic sample of 7 exhibits human NOD1 stimulatory activity that is higher than synthetic 6.132,162 They also showed that the compound obtained from the most active fraction from RP-HPLC purification of the supernatant of E. coli K-12 cells is identical to 7.163 Muropeptide 8 was originally isolated from the Gram-negative organism Bordetella pertussis,164 the causative agent of pertussis or whooping cough, and is known as tracheal cytotoxin (TCT) for its causative role in the destruction of the cilia of tracheal cells during B. pertussis infection. Synthetic 8 displayed very weak human NOD1 stimulatory activity,132,162 which was consistent with results using a sample of 8 from natural sources.165 Interestingly, 8 is able to stimulate the innate immune systems of other species. It activates murine NOD1165 and peptidoglycan-recognition protein-LC (PGRP-LC) from Drosophila.166,167 Muramyl dipeptide (9) is a product of enzymatic degradation of the peptidoglycan, and represents a minimal structural motif for both Gram-positive and Gram-negative bacteria. It has been used as an adjuvant since the 1970s when it was shown to be the minimal active structure of peptidoglycan enhancing the immune response to Freund's incomplete adjuvant. Stimulation of NOD2 by MDP activates the proinflammatory NF-κB signaling pathway.
Scheme 6.
Structures recognized by the intracellular receptors NOD1 (6–8) and NOD2 (9).
Although the molecular nature of the interaction between MDP and NOD2 is not known, several conclusions can be made in terms of the structural requirements for stimulation of the receptor. NOD2 can detect an MDP variant in which d-iGln has been substituted with d-Glu. However, substitution of d-iGln for l-iGln or l-Ala for d-Ala abrogates activity.158,160,161 This result strongly suggests that recognition of MDP by NOD2 is stereoselective. The dipeptide chain is also crucial for recognition, as synthetic GlcNAc-MurNAc dimers or tetramers missing the peptide portion do not stimulate NOD2-dependent NF-κB activation.160 Interestingly, NOD2 is also able to detect muramyl tripeptides containing either lysine or ornithine at the third position, but is unable to detect a muramyl tripeptide or tetrapeptide in which the third amino acid is meso-diaminopimelic acid (meso-DAP).158,161 Finally, NOD2 sensing requires an intact MurNAc ring, as reduction of this moiety by NaBH4 abolishes NOD2 recognition of the peptidoglycan fragment. In contrast, NOD1 can detect both native and reduced peptidoglycan fragments.161
Recent results suggest that NOD2 may detect other peptidoglycan fragments. It was shown by Volz et al. that a “peptidoglycan monomer” from S. aureus may be a ligand for NOD2, as NOD2-deficient dendritic cells fail to exhibit an increase in interleukins-12p70 and -23 upon activation with this monomer in concert with lipoteichoic acid or lipopolysaccharide.168 Mass spectral analysis suggests the structure of this monomer to be that of muropeptide 1 (and not the structure as suggested), thus identical to the active component in the BIVR phenomenon in MRSA. The N-glycolyl MDP derivative (as is generated by mycobacteria), is more potent than the N-acetyl parent in activation of NOD2-mediated signaling in vitro.169
Because MDP corresponds to the minimal structural motif found in peptidoglycan, NOD2 can be regarded as a general sensor of peptidoglycan from Gram-positive and Gram-negative bacteria. In contrast, NOD1 detects γ-d-Glu-meso-DAP (6), a structure mostly found in peptidoglycan from Gram-negative bacteria and suggesting a specific role for NOD1 in Gram-negative innate immune response. Two hypotheses are emerging regarding the mechanisms leading to PRR activation: one proposing a direct interaction between the PAMP and its associated PRR, leading to NF-κB activation, and the second arguing that the innate immune response to PAMPs casts a wider shadow, requiring perturbations of host metabolic and homeostatic functions and converging on the mitochondria as a signaling hub.170 In support of the former proposal, surface plasmon resonance and atomic force microscopy show a direct interaction between l-Ala-d-iGln-meso-DAP and NOD1, with a Kd value of 35 μM.171 This interaction is abolished when a truncated NOD1 lacking its LRR is used. Furthermore, binding between NOD1 and receptor-interacting serine/threonine protein kinase 2 (RICK) was increased when the tripeptide was pre-incubated with NOD1, as well as RICK-phosphorylation activity. RICK is thought to be recruited by NOD1 upon binding by the tripeptide, and to mediate NF-κB activation.172
The emergence of mitochondria as a checkpoint for innate immune signaling is in part due to the identification of several innate-immune receptors associated with the mitochondria. In particular, NLRX is a member of the NLR family that localizes to the mitochondrial matrix,173 as the first and only PRR in this environment. The function of NLRX is not yet understood, but it has been argued that because of its confined localization it may not be involved in the direct detection of PAMPs.170 Another class of PRRs, known as the peptidoglycan-recognition proteins (PGRPs), also plays a role in the innate-immune response to invading bacteria.174,175 PGRPs are conserved from invertebrates to vertebrates. Unlike NOD receptors, which bind to peptidoglycan monomers, mammalian PGRPs bind to polymeric peptidoglycan.176-180 Several studies elucidate specific interactions between human PGRPs and peptidoglycan using synthetic peptidoglycan monomers.178,181-183
In insects, PGRPs act through signaling pathways mediated by Toll receptors (in response to Gram-positive bacteria) and the immune-deficiency pathway (as a result of Gram-negative bacterial infection) to induce expression of antimicrobial peptides.174,175,184 A thorough discussion of insect PGRPs is beyond the scope of this review. However, there are certain instances in which muropeptides are involved in this signaling, namely the extracellular binding of the Gram-negative cell-wall recycling product TCT (8) to a heterodimer formed by the Drosophila melanogaster PGRPs PGRP-LCx and PGRP-LCa,166,185-188 as well as its intracellular binding to full-length PGRP-LE (PGRP-LEfl).189-191 Both these binding events lead to activation of the immune deficiency pathway.
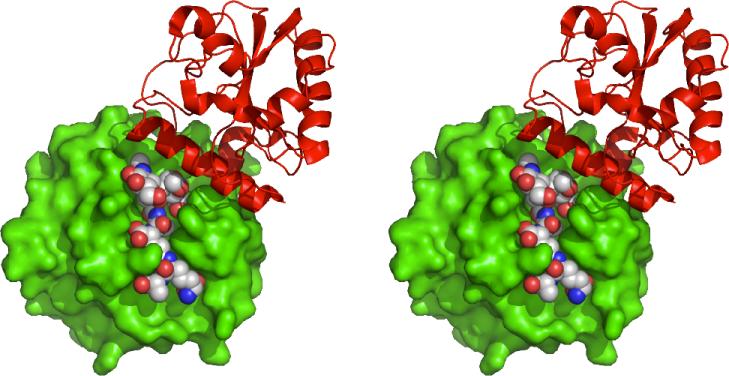
TCT induces the heterodimerization of PGRP-LCx and PGRP-LCa upon binding to their extracellular domains,186,192 and the structure of the TCT-heterodimer complex has been reported.188 The structure reveals that TCT binds in the LCx docking groove. Its exposed GlcNAc-1,6-anhydroMurNAc moiety as well as several residues of LCx engage the surface of LCa (Figure 1). The crystal structure reveals why this signaling pathway is specific for DAP-type peptidoglycan: the meso-DAP of TCT forms an electrostatic interaction with the side-chain of Arg413 of LCx and a hydrogen bond to the main-chain amide of its Asp395.188 Along with Trp394, the residues form a binding pocket to accommodate the meso-DAP moiety.
Figure 1.
Stereoview of the complex of TCT with the extracellular domains of PGRP-LCx (green, space-filling) and PGRP-LCa (red, ribbon). Adapted from the PDB file 2F2L188
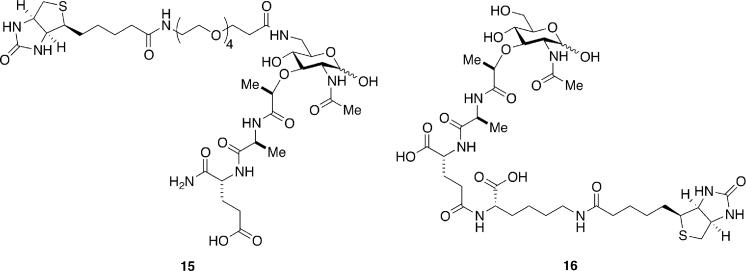
The crystal structure of TCT with PGRP-LE (Figure 2) reveals similar interactions as in its complex with the PGRP-LCx and PGRP-LCa heterodimer.190 TCT induces oligomerization of PGRP-LE, and this is achieved through the interactions of the GlcNAc-1,6-anhydroMurNAc group of bound TCT to a second molecule of PGRP-LE. Furthermore, the meso-DAP moiety of TCT forms a salt bridge with Arg254 of PGRP-LE, underscoring its preference for DAP-type peptidoglycan. The results of these structural studies are consistent with previous reports that both the GlcNAc-1,6-anhydroMurNAc and DAP moieties are required for activation of the immune deficiency pathway.166,167 As with other areas covered in this review, studies of the immune recognition of bacteria have been hampered by the lack of well defined, homogeneous muropeptide samples, as isolates from natural sources may contain traces of other immunostimulatory compounds (e.g. LPS). With the advent of new synthetic methodologies, access to these ligands in pure form is now possible.149,193 Although a clearer understanding of how muropeptide structure affects its interactions with PGRPs is slowly emerging, less is known in the case of NOD receptors. What is the nature of the interactions of these ligands with their putative receptors? How do the subtle changes in ligand structure modulate their activity and specificity? The answers to these questions will be obtained in part with the synthesis of immunostimulatory peptidoglycan-based structures. For example, a recent report detailed the structure-activity relationship of γ-d-Glu-meso-DAP (6) through the synthesis of several analogues and evaluation of their activity as NOD1 agonists.194 With the goal of determining whether or not MDP interacts directly with NOD2, syntheses of several active biotinylated versions of this muropeptide, such as 15195 and 16196 (Scheme 7) have been reported. Similar MDP derivatives have already demonstrated value to the study of the cell biology of NOD signaling.197
Figure 2.
Stereoview of the complex of TCT with PGRP-LE. Adapted from the PDB file 2CB3190
Scheme 7.
Biotinylated muramyl dipeptides for studies of NOD2 binding.
CONCLUSIONS
The relevance of cell wall-derived muropeptides as messengers in processes such as bacterial detection of the presence of antibiotics and eukaryotic detection of bacterial infections is now well established. Although the structures of many of the muropeptides responsible for these phenomena have been elucidated, there remains much to be discovered in terms of the precise mechanisms of action of these molecules, exemplified by their interactions with putative receptors. For example, the understanding of muropeptide binding to the PASTA domain of STKs in Gram-positive sporulation resuscitation requires additional structural information. A similar scenario exists in the case of eukaryotic immune recognition of bacterial muropeptides, where the putative muropeptide receptors, NOD1 and NOD2, have been identified, but a clear picture of the specific interactions of the cell-wall fragments with these proteins is not yet in focus. Finally, the mechanism of the muropeptide-induced BIVR phenomenon in MRSA may not be a direct result of muropeptides themselves, but may occur due to sequestration of vancomycin, for example by the extra lipid II that is biosynthesized in the presence of these cell-wall fragments. As a first step, the syntheses of muropeptides and their analogues have allowed correlations between structure and activity to be made. Further syntheses of muropeptides containing affinity tags or fluorescent reporters will provide new tools with which to study the interactions of these small molecules with their putative receptors, providing a more complete picture of their roles as second messengers.
Acknowledgments
Funding
This work was supported by National Institutes of Health Grant GM61629 (S.M.).
ABBREVIATIONS
- BIVR-MRSA
β-lactam antibiotic-induced vancomycin-resistant MRSA
- meso-DAP
meso-diaminopimelic acid
- LPS
lipopolysaccharide
- MDP
muramyl dipeptide
- MRSA
methicillin-resistant S. aureus
- NLR
intracellular nucleotide-binding domain/leucine-rich repeat receptor
- NLRX
mitochondrial NLR
- NOD
nucleotide oligomerization domain
- PAMP
pathogen-associated molecular pattern
- PASTA
penicillin binding-associated and serine/threonine kinase associated
- PBP
penicillin-binding protein
- PGRP
peptidoglycan recognition protein
- PRR
pattern recognition receptor
- RICK
receptor-interacting serine/threonine protein kinase 2, also known as RIP-2
- STK
serine-threonine kinase
- TCT
tracheal cytotoxin
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutcliffe IC. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 2010;18:464–470. doi: 10.1016/j.tim.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Schneider T, Sahl H-G. An oldie but a goodie—cell wall biosynthesis as antibiotic target pathway. Int. J. Med. Microbiol. 2010;300:161–169. doi: 10.1016/j.ijmm.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Bugg TD, Braddick D, Dowson CG, Roper DI. Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol. 2011;29:167–173. doi: 10.1016/j.tibtech.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Adam A, Lederer E. Muramyl peptides: immunomodulators, sleep factors, and vitamins. Med. Res. Rev. 1984;4:111–152. doi: 10.1002/med.2610040202. [DOI] [PubMed] [Google Scholar]
- 6.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falconer SB, Czarny TL, Brown ED. Antibiotics as probes of biological complexity. Nat. Chem. Biol. 2011;7:415–423. doi: 10.1038/nchembio.590. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien J, Wright GD. An ecological perspective of microbial secondary metabolism. Curr. Opin. Biotechnol. 2011;22:552–558. doi: 10.1016/j.copbio.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Ratcliff WC, Denison RF. Microbiology. Alternative actions for antibiotics. Science. 2011;332:547–548. doi: 10.1126/science.1205970. [DOI] [PubMed] [Google Scholar]
- 10.Romero D, Traxler MF, Lopez D, Kolter R. Antibiotics as Signal Molecules. Chem. Rev. 2011;111:5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies J. How to discover new antibiotics: harvesting the parvome. Curr. Opin. Chem. Biol. 2011;15:5–10. doi: 10.1016/j.cbpa.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Davies J, Ryan KS. Introducing the parvome: bioactive compounds in the microbial world. ACS Chem. Biol. 2012;7:252–259. doi: 10.1021/cb200337h. [DOI] [PubMed] [Google Scholar]
- 13.Zhanel GG, Calic D, Schweizer F, Zelenitsky S, Adam H, Lagace-Wiens PR, Rubinstein E, Gin AS, Hoban DJ, Karlowsky JA. New lipoglycopeptides: a comparative review of dalbavancin, oritavancin and telavancin. Drugs. 2010;70:859–886. doi: 10.2165/11534440-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 14.Allen NE. From vancomycin to oritavancin: The discovery and development of a novel lipoglycopeptide antibiotic. Anti-Infect. Agents Med. Chem. 2011;9:23–47. [Google Scholar]
- 15.Fisher JF, Mobashery S. Host-guest chemistry of the peptidoglycan. J. Med. Chem. 2010;53:4813–4829. doi: 10.1021/jm100086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crowley BM, Boger DL. Total synthesis and evaluation of [Ψ[CH2NH]Tpg4]vancomycin aglycon: Reengineering vancomycin for dual d-Ala-d -Ala and d-Ala- d-Lac binding. J. Am. Chem. Soc. 2006;128:2885–2892. doi: 10.1021/ja0572912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie J, Pierce JG, James RC, Okano A, Boger DL. A redesigned vancomycin engineered for dual d-Ala-d-Ala and d-Ala-d-Lac binding exhibits potent antimicrobial activity against vancomycin-resistant bacteria. J. Am. Chem. Soc. 2011;133:13946–13949. doi: 10.1021/ja207142h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira PM, Filipe SR, Tomasz A, Pinho MG. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2007;51:3627–3633. doi: 10.1128/AAC.00431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanagisawa C, Hanaki H, Matsui H, Ikeda S, Nakae T, Sunakawa K. Rapid depletion of free vancomycin in medium in the presence of β-lactam antibiotics and growth restoration in Staphylococcus aureus strains with β-lactam-induced vancomycin resistance. Antimicrob. Agents Chemother. 2009;53:63–68. doi: 10.1128/AAC.00762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda S, Hanaki H, Yanagisawa C, Ikeda-Dantsuji Y, Matsui H, Iwatsuki M, Shiomi K, Nakae T, Sunakawa K, Omura S. Identification of the active component that induces vancomycin resistance in MRSA. J. Antibiot. (Tokyo) 2010;63:533–538. doi: 10.1038/ja.2010.75. [DOI] [PubMed] [Google Scholar]
- 21.de Jonge BLM, Chang YS, Gage D, Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 1992;267:11248–11254. [PubMed] [Google Scholar]
- 22.Sieradzki K, Tomasz A. Inhibition of the autolytic system by vancomycin causes mimicry of vancomycin-intermediate Staphylococcus aureus-type resistance, cell concentration dependence of the MIC, and antibiotic tolerance in vancomycin-susceptible S. aureus. Antimicrob. Agents Chemother. 2006;50:527–533. doi: 10.1128/AAC.50.2.527-533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bisicchia P, Bui NK, Aldridge C, Vollmer W, Devine KM. Acquisition of VanB-type vancomycin resistance by Bacillus subtilis: the impact on gene expression, cell wall composition and morphology. Mol. Microbiol. 2011;81:157–178. doi: 10.1111/j.1365-2958.2011.07684.x. [DOI] [PubMed] [Google Scholar]
- 24.Koteva K, Hong HJ, Wang XD, Nazi I, Hughes D, Naldrett MJ, Buttner MJ, Wright GD. A vancomycin photoprobe identifies the histidine kinase VanSsc as a vancomycin receptor. Nat. Chem. Biol. 2010;6:327–329. doi: 10.1038/nchembio.350. [DOI] [PubMed] [Google Scholar]
- 25.Foucault ML, Depardieu F, Courvalin P, Grillot-Courvalin C. Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16964–16969. doi: 10.1073/pnas.1006855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong HJ. Studying gene induction of glycopeptide resistance using gene swapping. Methods Mol. Biol. 2010;642:45–62. doi: 10.1007/978-1-60327-279-7_4. [DOI] [PubMed] [Google Scholar]
- 27.Hill CM, Krause KM, Lewis SR, Blais J, Benton BM, Mammen M, Humphrey PP, Kinana A, Janc JW. Specificity of induction of the vanA and vanB operons in vancomycin-resistant enterococci by telavancin. Antimicrob. Agents Chemother. 2010;54:2814–2818. doi: 10.1128/AAC.01737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gagnon S, Levesque S, Lefebvre B, Bourgault AM, Labbe AC, Roger M. vanA-containing Enterococcus faecium susceptible to vancomycin and teicoplanin because of major nucleotide deletions in Tn1546. J. Antimicrob. Chemother. 2011;66:2758–2762. doi: 10.1093/jac/dkr379. [DOI] [PubMed] [Google Scholar]
- 29.Reith J, Mayer C. Peptidoglycan turnover and recycling in Gram-positive bacteria. Appl. Microbiol. Biotechnol. 2011;92:1–11. doi: 10.1007/s00253-011-3486-x. [DOI] [PubMed] [Google Scholar]
- 30.Park JT, Uehara T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 2008;72:211–227. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litzinger S, Duckworth A, Nitzsche K, Risinger C, Wittmann V, Mayer C. Muropeptide rescue in Bacillus subtilis involves sequential hydrolysis by β-N-acetylglucosaminidase and N-acetylmuramyl-L-alanine amidase. J. Bacteriol. 2010;192:3132–3143. doi: 10.1128/JB.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mauck J, Chan L, Glaser L. Turnover of the cell wall of Gram-positive bacteria. J. Biol. Chem. 1971;246:1820–1827. [PubMed] [Google Scholar]
- 33.Jaeger T, Mayer C. The transcriptional factors MurR and catabolite activator protein regulate N-acetylmuramic acid catabolism in Escherichia coli. J. Bacteriol. 2008;190:6598–6608. doi: 10.1128/JB.00642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Litzinger S, Fischer S, Polzer P, Diederichs K, Welte W, Mayer C. Structural and kinetic analysis of Bacillus subtilis N-acetylglucosaminidase reveals a unique Asp-His dyad mechanism. J. Biol. Chem. 2010;285:35675–35684. doi: 10.1074/jbc.M110.131037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plumbridge J. An alternative route for recycling of N-acetylglucosamine from peptidoglycan involves the N-acetylglucosamine phosphotransferase system in Escherichia coli. J. Bacteriol. 2009;191:5641–5647. doi: 10.1128/JB.00448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matias VRF, Beveridge TJ. Lipoteichoic acid is a major component of the Bacillus subtilis periplasm. J. Bacteriol. 2008;190:7414–7418. doi: 10.1128/JB.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reith J, Berking A, Mayer C. Characterization of an N-acetylmuramic acid/N-acetylglucosamine kinase of Clostridium acetobutylicum. J. Bacteriol. 2011;193:5386–5392. doi: 10.1128/JB.05514-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reith J, Mayer C. Characterization of a glucosamine/glucosaminide N-acetyltransferase of Clostridium acetobutylicum. J. Bacteriol. 2011;193:5393–5399. doi: 10.1128/JB.05519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis K. Persister cells. Annu. Rev. Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 40.Ghosh J, Larsson P, Singh B, Pettersson BM, Islam NM, Sarkar SN, Dasgupta S, Kirsebom LA. Sporulation in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10781–10786. doi: 10.1073/pnas.0904104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Errington J. From spores to antibiotics via the cell cycle. Microbiology. 2010;156:1–13. doi: 10.1099/mic.0.035634-0. [DOI] [PubMed] [Google Scholar]
- 42.Paredes-Sabja D, Setlow P, Sarker MR. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 2011;19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 43.Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 2012;36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Driks A. The Bacillus anthracis spore. Mol. Aspects Med. 2009;30:368–373. doi: 10.1016/j.mam.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 45.Kana BD, Mizrahi V. Resuscitation-promoting factors as lytic enzymes for bacterial growth and signaling. FEMS Immunol. Med. Microbiol. 2010;58:39–50. doi: 10.1111/j.1574-695X.2009.00606.x. [DOI] [PubMed] [Google Scholar]
- 46.Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l,d-transpeptidation. J. Bacteriol. 2008;190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat. Med. 2010;16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahapatra S, Scherman H, Brennan PJ, Crick DC. N-Glycolylation of the nucleotide precursors of peptidoglycan biosynthesis of Mycobacterium spp. is altered by drug treatment. J. Bacteriol. 2005;187:2341–2347. doi: 10.1128/JB.187.7.2341-2347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahapatra S, Yagi T, Belisle JT, Espinosa BJ, Hill PJ, McNeil MR, Brennan PJ, Crick DC. Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. J. Bacteriol. 2005;187:2747–2757. doi: 10.1128/JB.187.8.2747-2757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Popham DL. Specialized peptidoglycan of the bacterial endospore: the inner wall of the lockbox. Cell. Mol. Life Sci. 2002;59:426–433. doi: 10.1007/s00018-002-8435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keep NH, Ward JM, Cohen-Gonsaud M, Henderson B. Wake up! Peptidoglycan lysis and bacterial non-growth states. Trends Microbiol. 2006;14:271–276. doi: 10.1016/j.tim.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 52.Dworkin J, Shah IM. Exit from dormancy in microbial organisms. Nat. Rev. Microbiol. 2010;8:890–896. doi: 10.1038/nrmicro2453. [DOI] [PubMed] [Google Scholar]
- 53.Jones G, Dyson P. Evolution of transmembrane protein kinases implicated in coordinating remodeling of Gram-positive peptidoglycan: inside versus outside. J. Bacteriol. 2006;188:7470–7476. doi: 10.1128/JB.00800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Setlow P. Dormant spores receive an unexpected wake-up call. Cell. 2008;135:410–412. doi: 10.1016/j.cell.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 55.Pereira SF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol. Mol. Biol. Rev. 2011;75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shah IM, Dworkin J. Induction and regulation of a secreted peptidoglycan hydrolase by a membrane Ser/Thr kinase that detects muropeptides. Mol. Microbiol. 2010;75:1232–1243. doi: 10.1111/j.1365-2958.2010.07046.x. [DOI] [PubMed] [Google Scholar]
- 57.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Squeglia F, Marchetti R, Ruggiero A, Lanzetta R, Marasco D, Dworkin JE, Petoukhov M, Molinaro A, Berisio R, Silipo A. Chemical basis of peptidoglycan discrimination by PrkC, a key kinase Involved in bacterial resuscitation from dormancy. J. Am. Chem. Soc. 2011;133:20676–20679. doi: 10.1021/ja208080r. [DOI] [PubMed] [Google Scholar]
- 59.Lee M, Hesek D, Shah IM, Oliver AG, Dworkin J, Mobashery S. Synthetic peptidoglycan motifs for germination of bacterial spores. ChemBioChem. 2010;11:2525–2529. doi: 10.1002/cbic.201000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giebel JD, Carr KA, Anderson EC, Hanna PC. The germination-specific lytic enzymes SleB, CwlJ1, and CwlJ2 each contribute to Bacillus anthracis spore germination and virulence. J. Bacteriol. 2009;191:5569–5576. doi: 10.1128/JB.00408-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heffron JD, Lambert EA, Sherry N, Popham DL. Contributions of four cortex lytic enzymes to germination of Bacillus anthracis spores. J. Bacteriol. 2010;192:763–770. doi: 10.1128/JB.01380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heffron JD, Sherry N, Popham DL. In vitro studies of peptidoglycan binding and hydrolysis by the Bacillus anthracis germination-specific lytic enzyme SleB. J. Bacteriol. 2011;193:125–131. doi: 10.1128/JB.00869-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hesek D, Lee M, Morio K.-i., Mobashery S. Synthesis of a fragment of bacterial cell wall. J. Org. Chem. 2004;69:2137–2146. doi: 10.1021/jo035583k. [DOI] [PubMed] [Google Scholar]
- 64.Yeats C, Finn RD, Bateman A. The PASTA domain: a β-lactam-binding domain. Trends Biochem. Sci. 2002;27:438. doi: 10.1016/s0968-0004(02)02164-3. [DOI] [PubMed] [Google Scholar]
- 65.Gordon E, Mouz N, Duee E, Dideberg O. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 2000;299:477–485. doi: 10.1006/jmbi.2000.3740. [DOI] [PubMed] [Google Scholar]
- 66.Dessen A, Mouz N, Gordon E, Hopkins J, Dideberg O. Crystal structure of PBP2x from a highly penicillin-resistant Streptococcus pneumoniae clinical isolate: a mosaic framework containing 83 mutations. J. Biol. Chem. 2001;276:45106–45112. doi: 10.1074/jbc.M107608200. [DOI] [PubMed] [Google Scholar]
- 67.Maestro B, Novakova L, Hesek D, Lee M, Leyva E, Mobashery S, Sanz JM, Branny P. Recognition of peptidoglycan and β-lactam antibiotics by the extracellular domain of the Ser/Thr protein kinase StkP from Streptococcus pneumoniae. FEBS Lett. 2011;585:357–363. doi: 10.1016/j.febslet.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barthe P, Mukamolova GV, Roumestand C, Cohen-Gonsaud M. The structure of PknB extracellular PASTA domain from Mycobacterium tuberculosis suggests a ligand-dependent kinase activation. Structure. 2010;18:606–615. doi: 10.1016/j.str.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 69.Paracuellos P, Ballandras A, Robert X, Kahn R, Hervé M, Mengin-Lecreulx D, Cozzone AJ, Duclos B, Gouet P. The extended conformation of the 2.9 Å crystal structure of the three-PASTA domain of a Ser/Thr kinase from the human pathogen Staphylococcus aureus. J. Mol. Biol. 2010;404:847–858. doi: 10.1016/j.jmb.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 70.Ruggiero A, Squeglia F, Marasco D, Marchetti R, Molinaro A, Berisio R. X-ray structural studies of the entire extra-cellular region of the Ser/Thr kinase PrkC from Staphylococcus aureus. Biochem. J. 2011;435:33–41. doi: 10.1042/BJ20101643. [DOI] [PubMed] [Google Scholar]
- 71.Madec E, Laszkiewicz A, Iwanicki A, Obuchowski M, Seror S. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 2002;46:571–586. doi: 10.1046/j.1365-2958.2002.03178.x. [DOI] [PubMed] [Google Scholar]
- 72.Pallova P, Hercik K, Saskova L, Novakova L, Branny P. A eukaryotic-type serine/threonine protein kinase StkP of Streptococcus pneumoniae acts as a dimer in vivo. Biochem. Biophys. Res. Commun. 2007;355:526–530. doi: 10.1016/j.bbrc.2007.01.184. [DOI] [PubMed] [Google Scholar]
- 73.Greenstein AE, Echols N, Lombana TN, King DS, Alber T. Allosteric activation by dimerization of the PknD receptor Ser/Thr protein kinase from Mycobacterium tuberculosis. J. Biol. Chem. 2007;282:11427–11435. doi: 10.1074/jbc.M610193200. [DOI] [PubMed] [Google Scholar]
- 74.Mukamolova GV, Turapov OA, Kazarian K, Telkov M, Kaprelyants AS, Kell DB, Young M. The rpf gene of Micrococcus luteus encodes an essential secreted growth factor. Mol. Microbiol. 2002;46:611–621. doi: 10.1046/j.1365-2958.2002.03183.x. [DOI] [PubMed] [Google Scholar]
- 75.Cohen-Gonsaud M, Keep NH, Davies AP, Ward J, Henderson B, Labesse G. Resuscitation-promoting factors possess a lysozyme-like domain. Trends Biochem. Sci. 2004;29:7–10. doi: 10.1016/j.tibs.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 76.Mukamolova GV, Murzin AG, Salina EG, Demina GR, Kell DB, Kaprelyants AS, Young M. Muralytic activity of Micrococcus luteus Rpf and its relationship to physiological activity in promoting bacterial growth and resuscitation. Mol. Microbiol. 2006;59:84–98. doi: 10.1111/j.1365-2958.2005.04930.x. [DOI] [PubMed] [Google Scholar]
- 77.Cohen-Gonsaud M, Barthe P, Bagneris C, Henderson B, Ward J, Roumestand C, Keep NH. The structure of a resuscitation-promoting factor domain from Mycobacterium tuberculosis shows homology to lysozymes. Nat. Struct. Mol. Biol. 2005;12:270–273. doi: 10.1038/nsmb905. [DOI] [PubMed] [Google Scholar]
- 78.Ruggiero A, Tizzano B, Pedone E, Pedone C, Wilmanns M, Berisio R. Crystal structure of the resuscitation-promoting factor (ΔDUF)RpfB from M. tuberculosis. J. Mol. Biol. 2009;385:153–162. doi: 10.1016/j.jmb.2008.10.042. [DOI] [PubMed] [Google Scholar]
- 79.Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, Rubin EJ. A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Mol. Microbiol. 2007;66:658–668. doi: 10.1111/j.1365-2958.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- 80.Hett EC, Chao MC, Rubin EJ. Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLoS Pathog. 2010;6:e1001020. doi: 10.1371/journal.ppat.1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kana BD, Gordhan BG, Downing KJ, Sung N, Vostroktunova G, Machowski EE, Tsenova L, Young M, Kaprelyants A, Kaplan G, Mizrahi V. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol. Microbiol. 2008;67:672–684. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Demina GR, Makarov VA, Nikitushkin VD, Ryabova OB, Vostroknutova GN, Salina EG, Shleeva MO, Goncharenko AV, Kaprelyants AS. Finding of the low molecular weight inhibitors of resuscitation promoting factor enzymatic and resuscitation activity. PLoS One. 2009;4:e8174. doi: 10.1371/journal.pone.0008174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tuomanen E, Lindquist S, Sande S, Galleni M, Light K, Gage D, Normark S. Coordinate regulation of β-lactamase induction and peptidoglycan composition by the amp operon. Science. 1991;251:201–204. doi: 10.1126/science.1987637. [DOI] [PubMed] [Google Scholar]
- 85.Kong KF, Jayawardena SR, Indulkar SD, Del Puerto A, Koh CL, Hoiby N, Mathee K. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB β-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob. Agents Chemother. 2005;49:4567–4575. doi: 10.1128/AAC.49.11.4567-4575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Balcewich MD, Reeve TM, Orlikow EA, Donald LJ, Vocadlo DJ, Mark BL. Crystal structure of the AmpR effector binding domain provides insight into the molecular regulation of inducible AmpC β-lactamase. J. Mol. Biol. 2010;400:998–1010. doi: 10.1016/j.jmb.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 87.Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 88.Olrichs NK, Aarsman ME, Verheul J, Arnusch CJ, Martin NI, Herve M, Vollmer W, de Kruijff B, Breukink E, den Blaauwen T. A Novel in vivo cell-wall labeling approach sheds new light on peptidoglycan synthesis in Escherichia coli. ChemBioChem. 2011;12:1124–1133. doi: 10.1002/cbic.201000552. [DOI] [PubMed] [Google Scholar]
- 89.Goodell EW, Schwarz U. Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J. Bacteriol. 1985;162:391–397. doi: 10.1128/jb.162.1.391-397.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scheurwater E, Reid CW, Clarke AJ. Lytic transglycosylases: Bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 2008;40:586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 91.Scheurwater EM, Clarke AJ. The C-terminal domain of Escherichia coli YfhD functions as a lytic transglycosylase. J. Biol. Chem. 2008;283:8363–8373. doi: 10.1074/jbc.M710135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 93.van Heijenoort J. Peptidoglycan Hydrolases of Escherichia coli. Microbiol. Mol. Biol. Rev. 2011;75:636–663. doi: 10.1128/MMBR.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park JT, Raychaudhuri D, Li H, Normark S, Mengin-Lecreulx D. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide l-alanyl-γ-d-glutamyl-meso-diaminopimelate. J. Bacteriol. 1998;180:1215–1223. doi: 10.1128/jb.180.5.1215-1223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maqbool A, Levdikov VM, Blagova EV, Herve M, Horler RS, Wilkinson AJ, Thomas GH. Compensating stereochemical changes allow murein tripeptide to be accommodated in a conventional peptide-binding protein. J. Biol. Chem. 2011;286:31512–31521. doi: 10.1074/jbc.M111.267179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Das D, Herve M, Feuerhelm J, Farr CL, Chiu HJ, Elsliger MA, Knuth MW, Klock HE, Miller MD, Godzik A, Lesley SA, Deacon AM, Mengin-Lecreulx D, Wilson IA. Structure and function of the first full-length murein peptide ligase (Mpl) cell wall recycling protein. PLoS One. 2011;6:e17624. doi: 10.1371/journal.pone.0017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kraft AR, Prabhu J, Ursinus A, Holtje JV. Interference with murein turnover has no effect on growth but reduces β-lactamase induction in Escherichia coli. J. Bacteriol. 1999;181:7192–7198. doi: 10.1128/jb.181.23.7192-7198.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Korsak D, Liebscher S, Vollmer W. Susceptibility to antibiotics and β-lactamase induction in murein hydrolase mutants of Escherichia coli. Antimicrob. Agents Chemother. 2005;49:1404–1409. doi: 10.1128/AAC.49.4.1404-1409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schmidt H, Korfmann G, Barth H, Martin HH. The signal transducer encoded by ampG is essential for induction of chromosomal AmpC β-lactamase in Escherichia coli by β-lactam antibiotics and ‘unspecific’ inducers. Microbiology. 1995;141:1085–1092. doi: 10.1099/13500872-141-5-1085. [DOI] [PubMed] [Google Scholar]
- 100.Lindquist S, Weston-Hafer K, Schmidt H, Pul C, Korfmann G, Erickson J, Sanders C, Martin HH, Normark S. AmpG, a signal transducer in chromosomal β-lactamase induction. Mol. Microbiol. 1993;9:703–715. doi: 10.1111/j.1365-2958.1993.tb01731.x. [DOI] [PubMed] [Google Scholar]
- 101.Chahboune A, Decaffmeyer M, Brasseur R, Joris B. Membrane topology of the Escherichia coli AmpG permease required for recycling of cell wall anhydromuropeptides and AmpC β-lactamase induction. Antimicrob. Agents Chemother. 2005;49:1145–1149. doi: 10.1128/AAC.49.3.1145-1149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jacobs C, Huang LJ, Bartowsky E, Normark S, Park JT. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 1994;13:4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jacobs C, Frere JM, Normark S. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in Gram-negative bacteria. Cell. 1997;88:823–832. doi: 10.1016/s0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 104.Suvorov M, Lee M, Hesek D, Boggess B, Mobashery S. Lytic transglycosylase MltB of Escherichia coli and its role in recycling of peptidoglycan strands of bacterial cell wall. J. Am. Chem. Soc. 2008;130:11878–11879. doi: 10.1021/ja805482b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheng Q, Li H, Merdek K, Park JT. Molecular characterization of the β-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J. Bacteriol. 2000;182:4836–4840. doi: 10.1128/jb.182.17.4836-4840.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Votsch W, Templin MF. Characterization of a β-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and β-lactamase induction. J. Biol. Chem. 2000;275:39032–39038. doi: 10.1074/jbc.M004797200. [DOI] [PubMed] [Google Scholar]
- 107.Mark BL, Vocadlo DJ, Oliver A. Providing β-lactams a helping hand: targeting the AmpC β-lactamase induction pathway. Future Microbiol. 2011;6:1415–1427. doi: 10.2217/fmb.11.128. [DOI] [PubMed] [Google Scholar]
- 108.Zhang Y, Bao Q, Gagnon LA, Huletsky A, Oliver A, Jin S, Langaee T. ampG gene of Pseudomonas aeruginosa and its role in β-lactamase expression. Antimicrob. Agents Chemother. 2010;54:4772–4779. doi: 10.1128/AAC.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zamorano L, Reeve TM, Juan C, Moya B, Cabot G, Vocadlo DJ, Mark BL, Oliver A. AmpG inactivation restores susceptibility of pan-β-lactam-resistant Pseudomonas aeruginosa clinical strains. Antimicrob. Agents Chemother. 2011;55:1990–1996. doi: 10.1128/AAC.01688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Garcia DL, Dillard JP. Mutations in ampG or ampD affect peptidoglycan fragment release from Neisseria gonorrhoeae. J. Bacteriol. 2008;190:3799–3807. doi: 10.1128/JB.01194-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schmidtke AJ, Hanson ND. Role of ampD homologs in overproduction of AmpC in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008;52:3922–3927. doi: 10.1128/AAC.00341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee M, Zhang W, Hesek D, Noll BC, Boggess B, Mobashery S. Bacterial AmpD at the crossroads of peptidoglycan recycling and manifestation of antibiotic resistance. J. Am. Chem. Soc. 2009;131:8742–8743. doi: 10.1021/ja9025566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Carrasco-López C, Rojas-Altuve A, Zhang W, Hesek D, Lee M, Barbe S, André I, Ferrer P, Silva-Martin N, Castro GR, Martínez-Ripoll M, Mobashery S, Hermoso JA. Crystal structures of bacterial peptidoglycan amidase AmpD and an unprecedented activation mechanism. J. Biol. Chem. 2011;286:31714–31722. doi: 10.1074/jbc.M111.264366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stubbs KA, Balcewich M, Mark BL, Vocadlo DJ. Small molecule inhibitors of a glycoside hydrolase attenuate inducible AmpC-mediated β-lactam resistance. J. Biol. Chem. 2007;282:21382–21391. doi: 10.1074/jbc.M700084200. [DOI] [PubMed] [Google Scholar]
- 115.Stubbs KA, Scaffidi A, Debowski AW, Mark BL, Stick RV, Vocadlo DJ. Synthesis and use of mechanism-based protein-profiling probes for retaining β-D-glucosaminidases facilitate identification of Pseudomonas aeruginosa NagZ. J. Am. Chem. Soc. 2008;130:327–335. doi: 10.1021/ja0763605. [DOI] [PubMed] [Google Scholar]
- 116.Balcewich MD, Stubbs KA, He Y, James TW, Davies GJ, Vocadlo DJ, Mark BL. Insight into a strategy for attenuating AmpC-mediated β-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ. Protein Sci. 2009;18:1541–1551. doi: 10.1002/pro.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Asgarali A, Stubbs KA, Oliver A, Vocadlo DJ, Mark BL. Inactivation of the glycoside hydrolase NagZ attenuates antipseudomonal β-lactam resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009;53:2274–2282. doi: 10.1128/AAC.01617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zamorano L, Reeve TM, Deng L, Juan C, Moya B, Cabot G, Vocadlo DJ, Mark BL, Oliver A. NagZ inactivation prevents and reverts β-lactam resistance, driven by AmpD and PBP 4 mutations, in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2010;54:3557–3563. doi: 10.1128/AAC.00385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schurek KN, Wiebe R, Karlowsky JA, Rubinstein E, Hoban DJ, Zhanel GG. Faropenem: review of a new oral penem. Expert Rev. Anti-Infect. Ther. 2007;5:185–198. doi: 10.1586/14787210.5.2.185. [DOI] [PubMed] [Google Scholar]
- 120.Moya B, Dotsch A, Juan C, Blazquez J, Zamorano L, Haussler S, Oliver A. β-Lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 2009;5:e1000353. doi: 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zamorano L, Moya B, Juan C, Oliver A. Differential β-lactam resistance response driven by ampD or dacB (PBP4) inactivation in genetically diverse Pseudomonas aeruginosa strains. J. Antimicrob. Chemother. 2010;65:1540–1542. doi: 10.1093/jac/dkq142. [DOI] [PubMed] [Google Scholar]
- 122.Dietz H, Pfeifle D, Wiedemann B. The signal molecule for β-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob. Agents Chemother. 1997;41:2113–2120. doi: 10.1128/aac.41.10.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ghosh AS, Chowdhury C, Nelson DE. Physiological functions of d-alanine carboxypeptidases in Escherichia coli. Trends Microbiol. 2008;16:309–317. doi: 10.1016/j.tim.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 124.Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. Distinct pathways for modification of the bacterial cell wall by non-canonical d-amino acids. EMBO J. 2011;30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cava F, Lam H, de Pedro MA, Waldor MK. Emerging knowledge of regulatory roles of d-amino acids in bacteria. Cell. Mol. Life Sci. 2011;68:817–831. doi: 10.1007/s00018-010-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kong KF, Aguila A, Schneper L, Mathee K. Pseudomonas aeruginosa β-lactamase induction requires two permeases, AmpG and AmpP. BMC Microbiol. 2010;10:328. doi: 10.1186/1471-2180-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nigro G, Fazio LL, Martino MC, Rossi G, Tattoli I, Liparoti V, De Castro C, Molinaro A, Philpott DJ, Bernardini ML. Muramylpeptide shedding modulates cell sensing of Shigella flexneri. Cell. Microbiol. 2008;10:682–695. doi: 10.1111/j.1462-5822.2007.01075.x. [DOI] [PubMed] [Google Scholar]
- 129.Tayler AE, Ayala JA, Niumsup P, Westphal K, Baker JA, Zhang L, Walsh TR, Wiedemann B, Bennett PM, Avison MB. Induction of β-lactamase production in Aeromonas hydrophila is responsive to β-lactam-mediated changes in peptidoglycan composition. Microbiology. 2010;156:2327–2335. doi: 10.1099/mic.0.035220-0. [DOI] [PubMed] [Google Scholar]
- 130.Huang YW, Hu RM, Lin CW, Chung TC, Yang TC. NagZ-dependent and NagZ-independent Mechanisms for the β-lactamases expression in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2012;56 doi: 10.1128/AAC.05645-11. DOI: 10.1128/AAC.05645-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 132.Fujimoto Y, Fukase K. Structures, synthesis, and human Nod1 stimulation of immunostimulatory bacterial peptidoglycan fragments in the environment. J. Nat. Prod. 2011;74:518–525. doi: 10.1021/np100795d. [DOI] [PubMed] [Google Scholar]
- 133.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 134.Ting JP-Y, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, Flavell RA, Girardin SE, Godzik A, Harton JA, Hoffman HM, Hugot JP, Inohara N, Mackenzie A, Maltais LJ, Nunez G, Ogura Y, Otten LA, Philpott D, Reed JC, Reith W, Schreiber S, Steimle V, Ward PA. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Beutler BA. TLRs and innate immunity. Blood. 2009;113:1399–1407. doi: 10.1182/blood-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 137.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 138.Inohara N, Chamaillard M, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 139.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 140.Franchi L, Warner N, Viani K, Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yoneyama M, Onomoto K, Fujita T. Cytoplasmic recognition of RNA. Adv. Drug Deliv. Rev. 2008;60:841–846. doi: 10.1016/j.addr.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 142.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 143.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 145.Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 2010;8:171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 146.Sorbara MT, Philpott DJ. Peptidoglycan: a critical activator of the mammalian immune system during infection and homeostasis. Immunol. Rev. 2011;243:40–60. doi: 10.1111/j.1600-065X.2011.01047.x. [DOI] [PubMed] [Google Scholar]
- 147.Fujimoto Y, Tanaka K, Shimoyama A, Fukase K. Self and non-self recognition with bacterial and animal glycans, surveys by synthetic chemistry. Methods Enzymol. 2010;478:323–342. doi: 10.1016/S0076-6879(10)78016-2. [DOI] [PubMed] [Google Scholar]
- 148.Ingale S, Wolfert MA, Buskas T, Boons GJ. Increasing the antigenicity of synthetic tumor-associated carbohydrate antigens by targeting Toll-like receptors. ChemBioChem. 2009;10:455–463. doi: 10.1002/cbic.200800596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kusumoto S, Fukase K, Shiba T. Key structures of bacterial peptidoglycan and lipopolysaccharide triggering the innate immune system of higher animals: chemical synthesis and functional studies. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010;86:322–337. doi: 10.2183/pjab.86.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schmidt RR, Pedersen CM, Qiao Y, Zahringer U. Chemical synthesis of bacterial lipoteichoic acids: an insight on its biological significance. Org. Biomol. Chem. 2011;9:2040–2052. doi: 10.1039/c0ob00794c. [DOI] [PubMed] [Google Scholar]
- 151.Kufer TA, Sansonetti PJ. Sensing of bacteria: NOD a lonely job. Curr. Opin. Microbiol. 2007;10:62–69. doi: 10.1016/j.mib.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 152.Chen G, Shaw MH, Kim YG, Nunez G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu. Rev. Pathol. 2009;4:365–398. doi: 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- 153.Lecat A, Piette J, Legrand-Poels S. The protein Nod2: An innate receptor more complex than previously assumed. Biochem. Pharmacol. 2010;80:2021–2031. doi: 10.1016/j.bcp.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 154.Magalhaes JG, Sorbara MT, Girardin SE, Philpott DJ. What is new with Nods? Curr. Opin. Immunol. 2011;23:29–34. doi: 10.1016/j.coi.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 155.Werts C, Rubino S, Ling A, Girardin SE, Philpott DJ. Nod-like receptors in intestinal homeostasis, inflammation, and cancer. J. Leukoc. Biol. 2011;90:474–482. doi: 10.1189/jlb.0411183. [DOI] [PubMed] [Google Scholar]
- 156.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 157.Shaw MH, Kamada N, Warner N, Kim YG, Nunez G. The ever-expanding function of NOD2: autophagy, viral recognition, and T cell activation. Trends Immunol. 2011;32:73–79. doi: 10.1016/j.it.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, Tedin K, Taha MK, Labigne A, Zahringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ, Philpott DJ. Nod1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 159.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nunez G, Inohara N. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 160.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nunez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 161.Girardin SE, Travassos LH, Hervé M, Blanot D, Boneca IG, Philpott DJ, Sansonetti PJ, Mengin-Lecreulx D. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 2003;278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- 162.Kawasaki A, Karasudani Y, Otsuka Y, Hasegawa M, Inohara N, Fujimoto Y, Fukase K. Synthesis of diaminopimelic acid containing peptidoglycan fragments and tracheal cytotoxin (TCT) and investigation of their biological functions. Chem. Eur. J. 2008;14:10318–10330. doi: 10.1002/chem.200801121. [DOI] [PubMed] [Google Scholar]
- 163.Pradipta AR, Fujimoto Y, Hasegawa M, Inohara N, Fukase K. Characterization of natural human nucleotide-binding oligomerization domain protein 1 (Nod1) ligands from bacterial culture supernatant for elucidation of immune modulators in the environment. J. Biol. Chem. 2010;285:23607–23613. doi: 10.1074/jbc.M110.137893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Goldman WE, Klapper DG, Baseman JB. Detection, isolation, and analysis of a released Bordetella pertussis product toxic to cultured tracheal cells. Infect. Immun. 1982;36:782–794. doi: 10.1128/iai.36.2.782-794.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Magalhaes JG, Philpott DJ, Nahori MA, Jehanno M, Fritz J, Le Bourhis L, Viala J, Hugot JP, Giovannini M, Bertin J, Lepoivre M, Mengin-Lecreulx D, Sansonetti PJ, Girardin SE. Murine Nod1 but not its human orthologue mediates innate immune detection of tracheal cytotoxin. EMBO Rep. 2005;6:1201–1207. doi: 10.1038/sj.embor.7400552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, Silverman N. Monomeric and polymeric Gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 167.Stenbak CR, Ryu JH, Leulier F, Pili-Floury S, Parquet C, Herve M, Chaput C, Boneca IG, Lee WJ, Lemaitre B, Mengin-Lecreulx D. Peptidoglycan molecular requirements allowing detection by the Drosophila immune deficiency pathway. J. Immunol. 2004;173:7339–7348. doi: 10.4049/jimmunol.173.12.7339. [DOI] [PubMed] [Google Scholar]
- 168.Volz T, Nega M, Buschmann J, Kaesler S, Guenova E, Peschel A, Rocken M, Gotz F, Biedermann T. Natural Staphylococcus aureus-derived peptidoglycan fragments activate NOD2 and act as potent costimulators of the innate immune system exclusively in the presence of TLR signals. FASEB J. 2010;24:4089–4102. doi: 10.1096/fj.09-151001. [DOI] [PubMed] [Google Scholar]
- 169.Coulombe F, Divangahi M, Veyrier F, de Leseleuc L, Gleason JL, Yang Y, Kelliher MA, Pandey AK, Sassetti CM, Reed MB, Behr MA. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J. Exp. Med. 2009;206:1709–1716. doi: 10.1084/jem.20081779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Arnoult D, Soares F, Tattoli I, Girardin SE. Mitochondria in innate immunity. EMBO Rep. 2011;12:901–910. doi: 10.1038/embor.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Laroui H, Yan Y, Narui Y, Ingersoll SA, Ayyadurai S, Charania MA, Zhou F, Wang B, Salaita K, Sitaraman SV, Merlin D. L-Ala-γ-D-Glu-mesodiaminopimelic Acid (DAP) Interacts Directly with Leucine-rich Region Domain of Nucleotide-binding Oligomerization Domain 1, Increasing Phosphorylation Activity of Receptor-interacting Serine/Threonine-protein Kinase 2 and Its Interaction with NOD1. J. Biol. Chem. 2011;286:31003–31013. doi: 10.1074/jbc.M111.257501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Watanabe T, Asano N, Fichtner-Feigl S, Gorelick PL, Tsuji Y, Matsumoto Y, Chiba T, Fuss IJ, Kitani A, Strober W. NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. J. Clin. Invest. 2010;120:1645–1662. doi: 10.1172/JCI39481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Arnoult D, Soares F, Tattoli I, Castanier C, Philpott DJ, Girardin SE. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J. Cell. Sci. 2009;122:3161–3168. doi: 10.1242/jcs.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat. Rev. Microbiol. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- 175.Royet J, Gupta D, Dziarski R. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat. Rev. Immunol. 2011;11:837–851. doi: 10.1038/nri3089. [DOI] [PubMed] [Google Scholar]
- 176.Lu X, Wang M, Qi J, Wang H, Li X, Gupta D, Dziarski R. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J. Biol. Chem. 2006;281:5895–5907. doi: 10.1074/jbc.M511631200. [DOI] [PubMed] [Google Scholar]
- 177.Dziarski R, Gupta D. The peptidoglycan recognition proteins (PGRPs). Genome Biol. 2006;7:232. doi: 10.1186/gb-2006-7-8-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cho S, Wang Q, Swaminathan CP, Hesek D, Lee M, Boons GJ, Mobashery S, Mariuzza RA. Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8761–8766. doi: 10.1073/pnas.0701453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kietzman C, Tuomanen E. PGRPs kill with an ancient weapon. Nat. Med. 2011;17:665–666. doi: 10.1038/nm0611-665. [DOI] [PubMed] [Google Scholar]
- 180.Kashyap DR, Wang M, Liu LH, Boons GJ, Gupta D, Dziarski R. Peptidoglycan recognition proteins kill bacteria by activating protein-sensing two-component systems. Nat. Med. 2011;17:676–683. doi: 10.1038/nm.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Guan R, Malchiodi EL, Wang Q, Schuck P, Mariuzza RA. Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Iα. J. Biol. Chem. 2004;279:31873–31882. doi: 10.1074/jbc.M404920200. [DOI] [PubMed] [Google Scholar]
- 182.Guan R, Brown PH, Swaminathan CP, Roychowdhury A, Boons GJ, Mariuzza RA. Crystal structure of human peptidoglycan recognition protein I α bound to a muramyl pentapeptide from Gram-positive bacteria. Protein Sci. 2006;15:1199–1206. doi: 10.1110/ps.062077606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sharma P, Dube D, Sinha M, Mishra B, Dey S, Mal G, Pathak KM, Kaur P, Sharma S, Singh TP. Multi-ligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein. J. Biol. Chem. 2011;286:31723–31730. doi: 10.1074/jbc.M111.264374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Guan R, Mariuzza RA. Peptidoglycan recognition proteins of the innate immune system. Trends Microbiol. 2007;15:127–134. doi: 10.1016/j.tim.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 185.Mellroth P, Karlsson J, Hakansson J, Schultz N, Goldman WE, Steiner H. Ligand-induced dimerization of Drosophila peptidoglycan recognition proteins in vitro. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6455–6460. doi: 10.1073/pnas.0407559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chang CI, Ihara K, Chelliah Y, Mengin-Lecreulx D, Wakatsuki S, Deisenhofer J. Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10279–10284. doi: 10.1073/pnas.0504547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Werner T, Borge-Renberg K, Mellroth P, Steiner H, Hultmark D. Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J. Biol. Chem. 2003;278:26319–26322. doi: 10.1074/jbc.C300184200. [DOI] [PubMed] [Google Scholar]
- 188.Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science. 2006;311:1761–1764. doi: 10.1126/science.1123056. [DOI] [PubMed] [Google Scholar]
- 189.Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh BH, Kurata S, Silverman N. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- 190.Lim JH, Kim MS, Kim HE, Yano T, Oshima Y, Aggarwal K, Goldman WE, Silverman N, Kurata S, Oh BH. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 2006;281:8286–8295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- 191.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, Yoshimori T, Kurata S. Autophagic control of Listeria through intracellular innate immune recognition in Drosophila. Nat. Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mellroth P, Steiner H. PGRP-SB1: an N-acetylmuramoyl L-alanine amidase with antibacterial activity. Biochem. Biophys. Res. Commun. 2006;350:994–999. doi: 10.1016/j.bbrc.2006.09.139. [DOI] [PubMed] [Google Scholar]
- 193.Dzierzbicka K, Wardowska A, Trzonkowski P. Recent developments in the synthesis and biological activity of muramylpeptides. Curr. Med. Chem. 2011;18:2438–2451. doi: 10.2174/092986711795843173. [DOI] [PubMed] [Google Scholar]
- 194.Agnihotri G, Ukani R, Malladi SS, Warshakoon HJ, Balakrishna R, Wang X, David SA. Structure-activity relationships in nucleotide oligomerization domain 1 (Nod1) agonistic γ-glutamyldiaminopimelic acid derivatives. J. Med. Chem. 2011;54:1490–1510. doi: 10.1021/jm101535e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Grimes CL, Podolsky DK, O'Shea EK. Synthesis of biologically active biotinylated muramyl dipeptides. Bioorg. Med. Chem. Lett. 2010;20:6061–6063. doi: 10.1016/j.bmcl.2010.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gisch N, Buske B, Heine H, Lindner B, Zahringer U. Synthesis of biotinylated muramyl tripeptides with NOD2-stimulating activity. Bioorg. Med. Chem. Lett. 2011;21:3362–3366. doi: 10.1016/j.bmcl.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 197.Blanot D, Lee J, Girardin SE. Synthesis and biological evaluation of biotinyl hydrazone derivatives of muramyl peptides. Chem. Biol. Drug Des. 2012;79:2–8. doi: 10.1111/j.1747-0285.2011.01204.x. [DOI] [PubMed] [Google Scholar]