Abstract
We have previously introduced a general kinetic approach for comparative study of processivity, thermostability, and resistance to inhibitors of DNA polymerases (Pavlov et. al., (2002) Proc. Natl. Acad. Sci. USA 99, 13510–13515). The proposed method was successfully applied to characterize hybrid DNA polymerases created by fusing catalytic DNA polymerase domains with various non-specific DNA binding domains. Here we use the developed kinetic analysis to assess basic parameters of DNA elongation by DNA polymerases and to further study the interdomain interactions in both previously constructed and new chimeric DNA polymerases. We show that connecting Helix-hairpin-Helix (HhH) domains to catalytic polymerase domains can increase thermostability, not only of DNA polymerases from extremely thermophilic species, but also of the enzyme from a faculatative thermophilic bacterium Bacillus stearothermophilus. We also demonstrate that addition of TopoV HhH domains extends efficient DNA synthesis by chimerical polymerases up to 105°C by maintaining processivity of DNA synthesis at high temperatures. We also found that reversible high-temperature structural transitions in DNA polymerases decrease the rates of binding of these enzymes to the templates. Furthermore, activation energies and pre-exponential factors of the Arrhenius equation suggest that the mechanism of electrostatic enhancement of diffusion-controlled association plays a minor role in binding templates to DNA polymerases.
Thermostable DNA polymerases play a crucial role in current methods of DNA amplification and sequencing (1). Major improvements in these methods have been made in the past twenty years since the introduction of Taq DNA polymerase, largely as a result of a significant number of new DNA polymerases available to scientists, and a marked increase in the understanding of how DNA polymerases function in DNA sequencing and polymerase chain reactions. Many of the currently available thermostable polymerases were discovered through screening thermophilic organisms and phages for thermostable enzymes. The current trend in making new and improved DNA polymerases, however, is to apply rational redesign strategies based on introducing single mutations and/or domain swapping or tagging to construct new variants of the existing thermostable DNA polymerases (1–3).
Our research has for several years been directed towards creating new DNA polymerases by 'domain swapping' and 'domain tagging' techniques, which provide large modifications of protein structure unachievable by point mutations or by small sequence insertions/deletions (4–6). New DNA polymerases were obtained by the flexible attachment of Helix-hairpin-Helix (HhH) DNA binding domains of topoisomerase V (Topo V) of Methanopyrus kandleri (7–9) to catalytic domains of DNA polymerases (5). The potential of this domain fusion approach was demonstrated using the Taq and Pfu DNA polymerases, and recently using the phi29 DNA polymerase (10), which belong to different structural families and work in different temperature ranges. This method produced polymerases that retain high processivity at high concentrations of salts and other inhibitors of DNA synthesis, such as phenol, blood, and DNA intercalating dyes (5). In addition, it was demonstrated (6) that attaching additional domains has the potential to greatly increase the thermostability of chimeric DNA polymerases.
We have developed a novel kinetic assay, which is based on time-dependent elongation of fluorescent Primer Template Junction (PTJ) substrates by DNA polymerases (6). Quantitation of the products of polymerization on a DNA sequencer together with a corresponding procedure of computating initial rates of DNA polymerization allowed for a standard kinetic analysis of the elongation.
For comparative analysis of hybrid polymerases and their natural counterparts, we introduced a quantitative approach for assessment of the general effects of salt-containing buffers on the processivity of DNA polymerases. This approach is a further development of a model for processive DNA synthesis in replication described earlier by von Hippel and co-workers (11, 12). The processivity equivalence parameter, Pe, which was introduced in our previous work (6), can be calculated independently for all studied DNA polymerases, and thus can serve a quantitative measure of processivity for these enzymes.
In this paper we present a study and analysis of the effects of interdomain interactions in previously constructed hybrid enzymes and in new chimeric DNA polymerases, including the polymerase constructed by fusion of the Bacillus stearothermophilus (Bst) DNA polymerase and Topo V HhH domains.
MATERIALS AND METHODS
Taq DNA polymerase was purchased from Roche Applied Science (Indianapolis, IN), the Stoffel fragment of Taq DNA polymerase was obtained from Applied BioSystems (Foster City, CA), and Pfu DNA polymerase was from Stratagene Cloning Systems (La Jolla, CA). The recombinant large fragment of Bst DNA polymerase (IsoTherm™ DNA polymerase) was purchased from Epicentre Technologies (Madison, WI). Fluorescent M13 single stranded DNA and ALF M13 Universal Primers were purchased from Amersham Biosciences (Piscataway, NJ). All other primers were synthesized at Fidelity Systems (Gaithersburg, MD).
The gene of the DNA polymerase family B from Methanopyrus kandleri AV19 (Mka polB) was obtained through the PCR amplification of genomic DNA (13). The PCR product was digested with NcoI and EcoRI and cloned into expression vector pET21d. The integrity of the constructed vectors was verified by DNA sequencing.
Chimeric DNA polymerases
Construction, expression and isolation of chimeric DNA polymerases TopoTaq, TaqTopoC1, TaqTopoC2, TaqTopoC3, and PfuC2 are described in our previous paper (6). DNA polymerase TaqTopoC1Δ that differs from TaqTopoC1 by lacking the HhH repeats H-L, and TaqTopoC2Δ obtained from TaqTopoC2 by cutting off 16 C-terminal positively charged amino acid residues, were prepared as follows: DNA fragments containing truncated HhH C1 and C2 tails were produced by PCR amplification using expression vectors TaqTopoC1-pET21d and TaqTopoC2-pET21d (6), respectively, as templates, with the T7 promotor primer and primers that insert a stop codon with a HindIII site into defined regions of DNA. The PCR products were then cut with HindIII, and the DNA sequences coding the truncated tails were inserted into expression vector TaqTopoC2-pET21d, which was cleaved using HindIII. The orientation and sequence of the tails were verified by DNA sequencing.
Genomic DNA of Bacillus stearothermophilus Donc (ATCC12980D-5) was obtained from ATCC (Manassas, VA). Since sequences of DNA polymerase for various strains of B. stearothermophilus have considerable differences, we designed several inward directed PCR primers to the ends of the polymerase gene, using data from GenBank. The longest PCR product was selected and sequenced. In order to get the flanking sequences for the Bst DNA polymerase gene, two outward Fimers complimentary to the ends of the PCR product were synthesized and used for direct genomic sequencing (14). Then, the region of DNA encoding the Bst polymerase domain was amplified by PCR with the following two primers:
CGAAAAGCCGCTCGCCGCCATGGATTTTGC (Primer 1) and
GGCAGGAAGCTTAATTTGGCGTCGTACCACG (Primer 2).
Primer 1 creates an NcoI restriction site followed by initiation codon for Large Fragment (LF) of the Bst DNA polymerase, and primer 2 incorporates a HindIII site downstream the termination codon of the Bst DNA polymerase gene. The PCR product was varified by DNA sequencing. As the amplified region had internal NcoI and HindIII sites, the following cloning strategy was applied:
Expression vector TaqTopoC2-pET21d was digested with NcoI and HindIII and two DNA fragments were purified: a HindIII-HindIII fragment encoding the C2 tail of TopoTaqC2 and an NcoI-HindIII fragment of vector pET21d.
The PCR-amplified DNA encoding LF Bst DNA polymerase was cut with NcoI and two large pieces were separated. The fragment encoding the C-terminal fragment of the LF Bst polymerase gene was then treated with HindIII and inserted between sites NcoI and HindIII of the NcoI-HindIII fragment of pET21d.
The resulting construct was then cut with HindIII, the C2 domain-coding region was inserted between two HindIII sites, and its orientation was verified by sequencing.
The plasmid was then cut with NcoI, and the NcoI – NcoI fragment of the PCR product encoding the N-terminal part of the LF Bst polymerase gene was inserted between two NcoI sites. The final construct, expression vector BstC2-pET21d, was verified by sequencing.
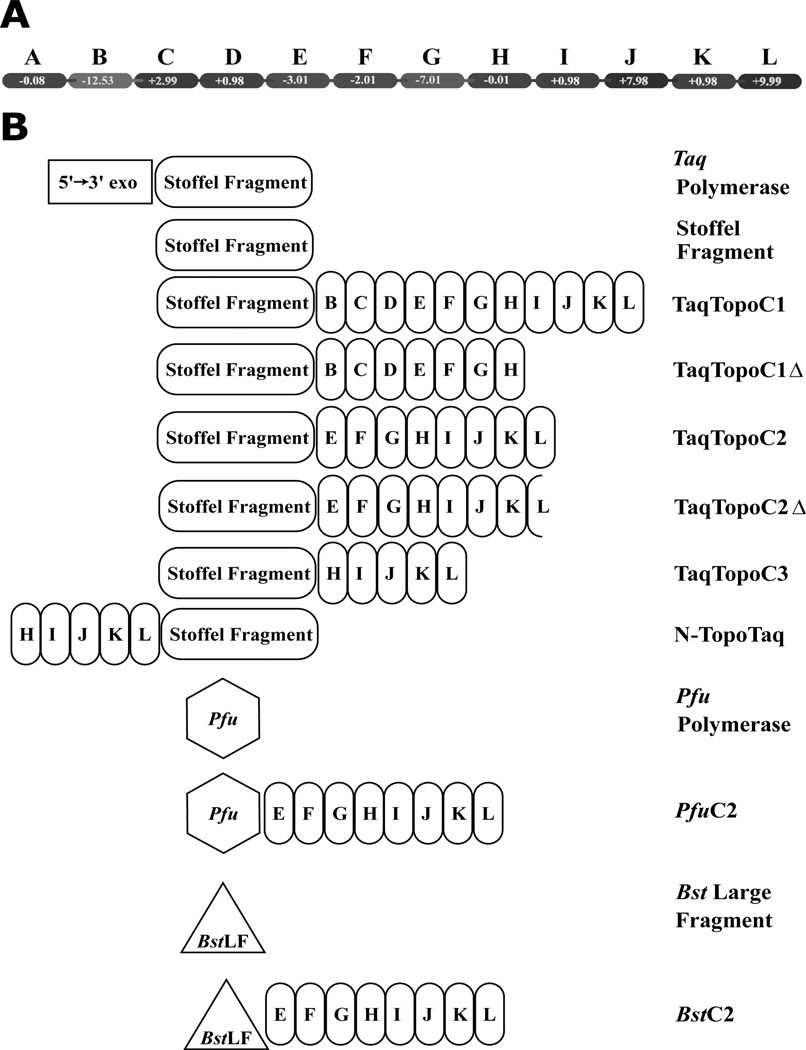
TaqTopoC1Δ, TaqTopoC2Δ, Mka PolB, and BstC2 were expressed in E. coli strain BL21 pLysS and purified to apparent homogeneity (> 95% purity, according to SDS polyacrylamid gel electrophoresis analysis (14). The cell lysates, prepared as described earlier (6), were heated at 75°C for 20 min (for TaqTopoC1Δ, TaqTopoC2Δ, and MkaPolB) or 60°C for 20 min (for BstC2), cleared by ultracentrifugation, and then applied onto a heparin High Trap column (Amersham Pharmacia Biotech) equilibrated with 0.25 M NaCl in 50 mM Tris-HCl buffer, pH 7.5 at 25°C. The proteins were eluted by a linear gradient of 0.25–1 M NaCl. Fractions containing the BstC2 protein were combined, diluted to 0.15 M NaCl, and applied onto Q-Sepharose Hi Trap 1 ml column equilibrated with a buffer containing 50 mM Tris-HCl, pH 8.5, 0.15 M NaCl, and 2 mM β-mercaptoethanol. The polymerases were eluted using a linear gradient of 0.15–1 M NaCl and concentrated with Centricon YM-30 (Millipore). Figure 1 presents a sketch of the catalytic and DNA binding domains for chimeric and natural polymerases used in this study.
Figure 1.
Schematic representation of chimeric DNA polymerases. Panel A displays HhH domains of TopoV with calculated partial charges at pH 7.0. Panel B illustrates chimeric DNA polymerases, their catalytical domains, and original enzymes used in the study. HhH repeats are designated as in (6, 7).
Protein concentrations
The concentrations of the expressed and purified DNA polymerases were determined by the Modified Lowry Protein Assay (Pierce, USA) and compared to the quantitation of the protein bands resolved by Disc SDS PAGE (15), using different concentrations of Bovine Serum Albumin (Sigma, USA) as a reference in both methods. The protein concentration of commercial DNA polymerases was determined by the PAGE followed by quantitation in the gels.
Melting Temperatures of Duplexes
The melting temperatures of duplexes were measured on a spectrophotometer Varian 300 with the heated block in primer extension reaction mixtures, using temperature gradients 0.2°C/min and continuous record of optical density at 254 nm, and calculated as mediums of the inflection points on the heating and cooling curves. Each melting experiment was carried out in triplicate.
Primer Extension Assays
DNA polymerase activity, the initial rates of extension (v1) and the processivity parameters for the reactions (Pe) were measured and calculated as described earlier (6). Briefly, DNA polymerase reaction mixtures (15–20 µl) contained dATP, dTTP, dCTP, and dGTP (1 mM each), 4.5 mM MgCl2, detergents Tween 20 and Nonidet P-40 (0.2% each), fixed concentrations of PTJ – duplexes, other additions, as indicated, and appropriate amounts of DNA polymerases in 30 mM Tris-HCl buffer pH 8.0 (25°C). The background reaction mixtures contained all components except the polymerases. Primer extensions were carried out for a preset time in PTC-150 Minicycler (MJ Research). For each reaction, 5 µl samples were removed after 2, 4, and 8 min. of incubation and chilled to 4°C, followed by immediate addition of 20 µl of 20 mM EDTA. The samples were desalted by centrifugation through Sephadex G-50 spun columns, diluted, and analyzed on an ABI Prism 377 DNA sequencer (Applied BioSystems). The products of elongation were quantitated after integration of fluorescent profiles of the electrophoresis, and the initial rates of extension were calculated from the progressive accumulation of the products, as described by E.A. Boeker (16). For calculation of the processivity parameters, after determination of the amount of extended products, the initial rates for appearance of each elongated product were calculated.
The substrate for the standard assay was prepared by annealing a 5’-end, labeled with a fluorescein 20-nt long primer, with a 30-nt long template:
The standard assay was carried out at 70°C, and the reactions were started by the addition of DNA polymerases.
Studies of Thermostability of DNA Polymerases
Proteins, concentrated at 0.1 mg/ml, in 25 µl of 20 mM Tris-HCl buffer (pH 8.0 at 25°C), containing 1 M potassium glutamate (K-Glu) and 1 M betaine, were incubated in a PTC-150 Minicycler (MJ Research) at fixed temperatures. Samples were removed at defined incubation times and assayed for primer extension activity with the standard substrate PTJ1.
Dependencies of the Rates and Processivity of Primer Extension Reactions on the Temperature
The duplex for high-temperature primer extension assays was prepared by annealing a 5’-end, labeled with a fluorescein 40-nt long primer, with a 50-nt long template:
The melting temperatures of the duplex was found to be higher than 80°C, even when no salts were added.
DNA polymerase reactions mixtures contained 0.32 µM PTJ2, betaine and NaCl or K-Glu, as indicated, and appropriate amounts of DNA polymerases (final concentrations of 14–200 nM were chosen depending on the activity of the polymerases to obtain similar initial rates of primer extension at 70°C). The control (background) reaction mixtures contained all components except the polymerases. The reactions were started by addition of PTJ2 to the reaction mixture preheated for 10 s at the specified temperatures. Primer extensions were carried out in the range of temperatures between 50–105°C. For comparison, the reactions at 37–75°C were carried out with the standard substrate PTJ1.
RESULTS
Construction of Chimeric Polymerases
DNA polymerase constructs used in the present study are shown in Figure 1. We found earlier that catalytic polymerase domains of thermostable DNA polymerases of both polA and polB structural families are able to produce functional hybrids with HhH domains of topoisomerase V from the hyperthermophile M. kandleri (6). We suggested that structural domains with high internal stability could fold independently in chimeras, maintaining the high catalytic activity of such constructs. All previously analyzed hybrids included the entire C3 domain of TopoV, which contained HhH repeats H-L (10 HhH motifs). The C3 domain was earlier produced as an individual protein, called Topo34, and its properties have been studied (8). In active chimeras, the C3 domain is attached either to the N-terminal or C-terminal ends of the Taq polymerase catalytic domain; that is, it folds before or after the polymerase domain gets formed. However, it is unknown if the presence, or the integrity, of the C3 domain plays any role in the stabilization of the polymerase catalytic domains of the hybrid enzymes. Because of a strong positive charge the C3 domain carries on its C-termini (Fig. 1A) it is particularly interesting to know if those positively charged amino acid residues could mediate domain interactions in chimeras. It is also important to see if the extreme thermal stability of the catalytic polymerase domains used in our previous experiments (6) could be a major factor for successful formation of the active highly thermostable hybrid polymerases. In order to answer these and other questions, we have created three additional chimeras:
TaqTopoC1Δ, which encompasses the Taq polymerase domain and a domain formed by HhH repeats B-H of Topo V(8).
TaqTopoC2Δ, which was obtained from TaqTopoC2 (6) by removing the last 16 amino acid residues that contribute to the very strong positive charge of the original chimera.
BstC2, which has the C2 domain attached to the C-terminus of polymerase domain of moderately thermolabile Bst DNA polymerase.
The successful expression of active and stable chimeric DNA polymerases in E. coli, taken together with previously published data (6), provides strong support to our hypothesis of the independent folding of the catalytic and DNA binding HhH domains in natural DNA processing proteins.
As expected, Taq hybrids with the truncated tails have lower affinities to heparin Sepharose in comparison to to the chimeras with the intact C3 domain. Surprisingly, the BstC2 polymerase has a much lower affinity than Bst LF, indicating that a strong interdomain interaction hinders either part of the molecule to efficiently intermingle with heparin groups.
Increased Thermostability of DNA Polymerases with Attached TopoV HhH Domains
Remarkable thermostability of chimeric DNA polymerases was demonstrated for DNA polymerases of structural families A and B (6). To further understand the mechanism of the stabilization of the polymerase catalytic domains at high temperatures facilitated by the introduced HhH domains, we designed new chimeras TaqTopoC1Δ and TaqTopoC2Δ, which are truncated versions of TaqTopoC1 and TaqTopoC2, respectively (see Materials and Methods and Fig. 1B), and measured the time-course of thermal inactivation of the chimeras and other DNA polymerases.
Analysis of the inactivation kinetics suggested single exponential decrease of the polymerase activities. The calculated rate constants of thermal inactivation for the studied DNA polymerases are presented in Table 1. As shown in the table, fusion of C1, C2, and C3 domains with the Taq polymerase catalytic domain, as well as fusion of the C2 domain with the Pfu DNA polymerase, maintains full enzymatic activity after incubation for 1h at 100°C, making the studied DNA polymerases as stable as the Methanopyrus PolB.
Table 1.
Apparent rate constants for thermal inactivation of DNA polymerases in the presence of 1 M potassium glutamate and 1 M betaine.
| DNA polymerase | kin ± S.E. (× 104 s−1) |
|---|---|
| Incubation at 100°C | |
| N-TopoTaq | <0.5 |
| TaqTopoC1 | <0.5 |
| TaqTopoC1Δ | 7.2 ± 0.8 |
| TaqTopoC2 | <0.5 |
| TaqTopoC2Δ | 1.7 ± 0.1 |
| TaqTopoC3 | <0.5 |
| Taq pol | 17.4 ± 2.1 |
| Stoffel Fragment | 12.7 ± 1.2 |
| Pfu pol | 1.0 ± 0.2 |
| PfuC2 | <0.5 |
| Mka PolB | <0.5 |
| Incubation at 95°C | |
| BstLF pol | 699.9 ± 57.1 |
| BstC2 | 441.7 ± 15.2 |
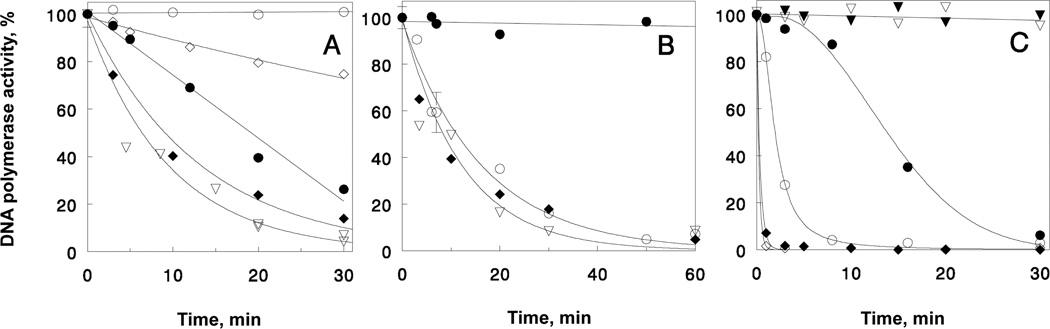
Figure 2A displays thermal inactivation of TaqTopoC1Δ and TaqTopoC2Δ at 100°C in the presence of 1 M potassium glutamate and 1 M betaine; also, data for thermal inactivation of TaqTopoC2, Taq polymerase, and the Stoffel fragment are shown for comparison. Clearly, trimming the C-terminal sequences reduces the thermostability of the chimeric polymerases. It is interesting that while the entire C3 domain alone, being attached to either termini of the Stoffel fragment (in chimeras TaqTopoC3 and N-TopoTaq) stabilizes the Taq polymerase catalytic domain at least for 1 h at 100°C (6); the removal of as many as 10 C3 HhH repeats in TaqTopoC1Δ does not completely abolish the aquired stability. The residual thermostability is provided by the remaining HhH repeats of domains B-H. As we suggested earlier (6), this effect of thermostabilization can be specifically attributed to the HhH repeats. In contrast, the natural exonuclease domains in Taq polymerase do not provide any stabilization (Fig. 2A, Table 1). It is especially important that the effect of domains B-H cannot be associated with the strong positive charge residing in the C3 domain, as domains B-H should have overall negative charge at neutral and alkaline pH (Fig. 1A). Furthermore, this observation is additionally supported by data on thermal stability of TaqTopoC2Δ, as the removal of the terminal 16 amino acids should make neutral the net charge of the HhH tail at low alkaline pH. Nonetheless, the remaining Topo V domains still exert a very strong effect on thermostability.
Figure 2.
Thermostability of chimeric DNA polymerases (see Materials and Methods). Panel A shows the stability of Taq polymerase chimeras with truncated C-terminal TopoV tails at 100°C in the presence of 1 M potassium glutamate and 1 M betaine: (○) –TaqTopoC2; (●) -TaqTopoC1Δ; (◇) –TaqTopoC2Δ; (▽) – Taq polymerase; (◆) – Stoffel Fragment. Panel B demonstrates the effect of NaCl on the thermostability at 100°C: (○) – N-TopoTaq in 0.5 M NaCl; (●) – N-TopoTaq in 0.5 M NaCl and 1 M betaine; (▽) – Taq polymerase; (◆) – Stoffel Fragment. Panel C compares inactivation of large fragment of Bst DNA polymerase and BstC2 chimera at different temperatures. 75°C: (▽) – BstLF; (▼) – BstC2, 85°C: (○) – BstLF; (●) – BstC2, 95°C: (◇) – BstLF; (◆) – BstC2.
Thus, it appears that the thermal stabilization of chimeric polymerases occurs due to charge-independant interactions of the polymerase domain and HhH domains. It is interesting that at 100°C the thermostabilities of the Taq polymerase and the Stoffel fragment are almost the same in 0.5 M NaCl, as well as in 1 M potassium glutamate with 1 M betaine. In contrast, the stabilizing effect of the C3 domain in N-TopoTaq completely disappears if K-Glu is replaced by 0.5 M NaCl, and the stability of N-TopoTaq does not differ from that of the catalytic domain alone (Fig. 2A, B). However, the addition of 1 M betaine restores the stabilization provided by the C3 domain (Fig. 2B). A similar decrease in the thermostability was observed in NaCl-containing buffers for TaqTopoC1-C3 even at 95°C (data not shown).
Figure 2C demonstrates the thermostability of the LF Bst polymerase with the attached TopoV’s HhH C2 domain. The half-life of the chimeric protein increases almost 8-fold, as compared to the Bst LF polymerase alone. The effect can be seen up to 95°C (Fig. 2C; Table 1), with the estimated half-lives 10 and 15.7 s for Bst LF and BstC2, respectively. It is worth noting, however, that the seemingly universal stabilization of different DNA polymerases domains by the fused TopoV HhH repeats is limited by stability of polymerase catalytic domains.
DNA Synthesis by Chimeric Polymerases in the Broad Range of Temperatures
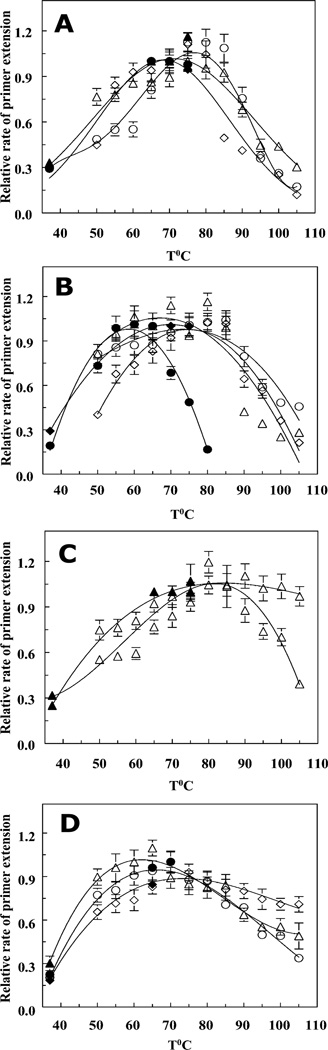
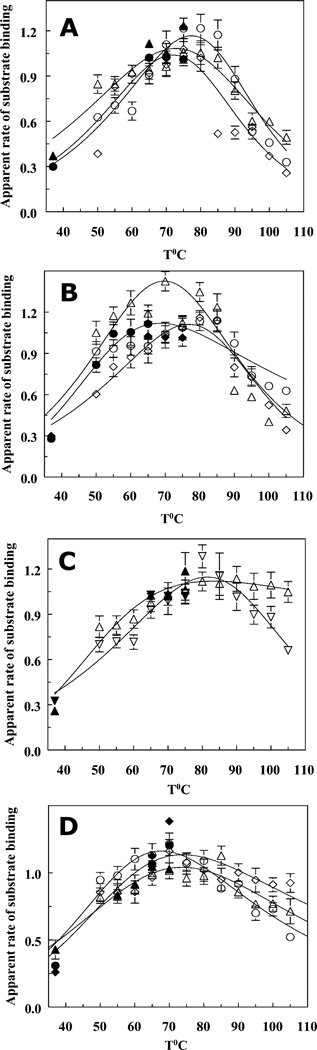
Using a specially designed duplex substrate with a high melting temperature (PTJ2), we studied primer extensions by Taq and Pfu DNA polymerases, fragments of Taq polymerase, and chimeric polymerases at temperatures from 37°C to 105°C (Fig. 3). Figures 3–5 show relative initial rates of DNA synthesis, apparent rates of substrate binding, and processivities on temperature for different DNA polymerases measured in the kinetic assays at 37–105°C. The catalytic domains of both Taq and Pfu DNA polymerases carry out DNA synthesis at least at 105°C with nearly the same efficiency as at lower temperatures (Fig. 3C,4C and 3D, 4D). At low temperatures (<70°C), the initial rates measured with PTJ1 display virtually the same dependencies of relative activity on temperature, although absolute values of the rates are different (e.g., at 70°C, the initial rate of extension of 0.32 µM PTJ2 is about two times higher than that of PTJ1 at the same concentration).
Figure 3.
Dependencies of DNA polymerase activity on temperature. The hollow symbols refer to the primer extension experiments carried out with PTJ1 substrate; the results obtained with PTJ2 are shown with filled symbols (see Materials and Methods). Panel A, (○, ●) – Taq polymerase; (△, ▲) – Stoffel Fragment; (◇, ◆) – Klentaq. Panel B, (○,●) – N-TopoTaq in potassium glutamate; (△) – N-TopoTaq in NaCl; (◇, ◆) – TaqTopoC3. Panel C, Taq chimeras with C1 (△, ▲) and C2 (▽, ▼) tails. Panel D, M.K. polB av19 (○, ●); Pfu DNA polymerase (△, ▲), and PfuC2 (◇, ◆). All kinetic measurements were carried out in triplicate, and the error bars represent standard deviations for each experimental value.
Figure 5.
Dependencies of apparent rate of substrate binding by DNA polymerases on temperature. Panel A, (○- PTJ1, ●- PTJ2) – Taq polymerase; (△- PTJ1, ▲- PTJ2) – Stoffel Fragment; (◇- PTJ1, ◆- PTJ2) – Klentaq. Panel B, (○- PTJ1,●- PTJ2) – N-TopoTaq in potassium glutamate; (△- PTJ1) – N-TopoTaq in NaCl; (◇- PTJ1, ◆- PTJ2) – TaqTopoC3. Panel C, Taq chimeras with C1 (△- PTJ1, ▲- PTJ2) and C2 (▽- PTJ1, ▼- PTJ2) tails. Panel D, Mka polB av19 (○- PTJ1, ●- PTJ2); Pfu DNA polymerase (△- PTJ1, ▲- PTJ2), and PfuC2 (◇- PTJ1, ◆- PTJ2). The solid lines are theoretical curves calculated from equation 7.
Figure 4.
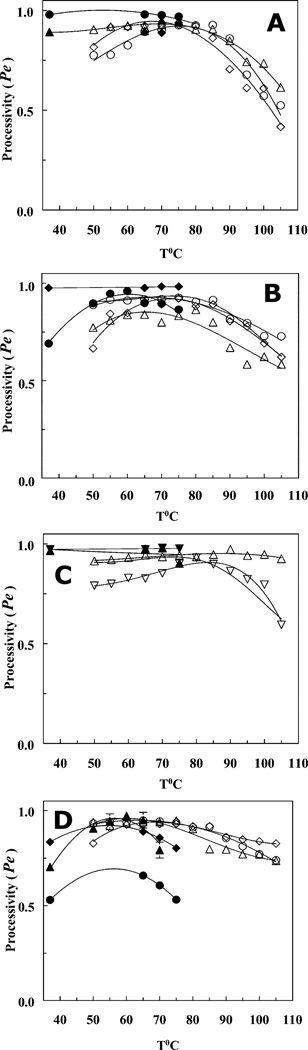
Dependencies of DNA polymerase processivity on temperature. Panel A, (○- PTJ1, ●- PTJ2) –Taq polymerase; (△- PTJ1, ▲- PTJ2) – Stoffel Fragment; (◇- PTJ1, ◆- PTJ2) – Klentaq. Panel B, (○- PTJ1,●- PTJ2) – N-TopoTaq in potassium glutamate; (△- PTJ1) – N-TopoTaq in NaCl; (◇- PTJ1, ◆- PTJ2) – TaqTopoC3. Panel C, Taq chimeras with C1 (△- PTJ1, ▲- PTJ2) and C2 (▽- PTJ1, ▼- PTJ2) tails. Panel D, Mka polB av19 (○- PTJ1, ●- PTJ2); Pfu DNA polymerase (△- PTJ1, ▲- PTJ2), and PfuC2 (◇- PTJ1, ◆- PTJ2). All kinetic measurements were carried out in triplicate, and the error bars represent standard deviations for each experimental value.
As shown in Fig. 3 (panels G–I), the rate of primer extension for PTJ2 with TaqTopoC1 virtually does not change in the 75°–105°C range. In addition, it turned out, that more than 96% of the substrate can be elongated if longer incubation with the enzyme is allowed. This implies that the melting rates of PTJ2 are much lower than the rates of primer extension at all temperatures of the assays. Therefore, the substrate melting does not affect measurement of the initial rates, particularly when the reactions get started by adding duplex substrates to the reaction mixtures containing DNA polymerases equilibrated at the temperatures of the assay (see Materials and Methods). However, this was not the case with PTJ1 (Fig. 3D), as melting of this substrate (Tm 73.2 ± 0.4 °C) prevents its efficient extension at high temperatures.
The temperature profiles of the polymerization rates of N-TopoTaq and TaqTopoC3 (Fig. 3B) above 70°C are very similar, and they do not differ significantly from those for the catalytic Stoffel Fragment, Taq polymerase, or KlenTaq (Fig. 3A). Therefore, it appears that the additional domains of these polymerases are not essential for the high temperature DNA synthesis by Taq polymerase catalytic domain. Also, because primer extension by Taq polymerase and its fragments is performed in 50 mM KCl (higher salt concentrations inhibit these polymerases), and the reaction with the chimeras are carried out in the presence of 0.25 M salts and 1 M betaine, one may conclude that heat inactivation of the Taq polymerase catalytic domain is not sensitive to the salt concentration. Accordingly, only minor differences are observed between the curves for N-TopoTaq in the presence of 0.25 M NaCl and 0.25 M K-Glu (Fig. 3B), although the absolute values of the initial rates are higher in K-Glu.
An effective high-temperature DNA synthesis (up to 105°C) by chimeric polymerases was observed only for C1- and C2-constructs (Fig. 3C, D). The effect of the C1 and C2 domains is seen in cases of chimeras containing either Taq or Pfu catalytic domains, which suggests that other thermostable polymerases could be adopted for DNA synthesis at extremely high temperatures, as well.
The profiles at lower temperatures (37–70°C) in Figure 3 show decreased activities for Taq polymerase, KlenTaq, TaqTopoC2, and TaqTopoC3 with respect to the Stoffel Fragment, suggesting that there are domain interactions that come forward at lower temperatures and interfere with DNA synthesis.
The data in Figure 3 and 4 allow for evaluation of apparent rates of substrate binding, (Vbapp; see Supporting Information for details) shown in Fig. 5 A–D. Figure 5 thus demonstrates that the drop in the, rates of substrate binding rather than the decrease in processivity (Pe) (Fig. 4 A–D) is the major contributor to the slowing down of the polymerization by the DNA polymerases at high temperatures.
Analysis of substrate binding by DNA polymerases at high temperatures
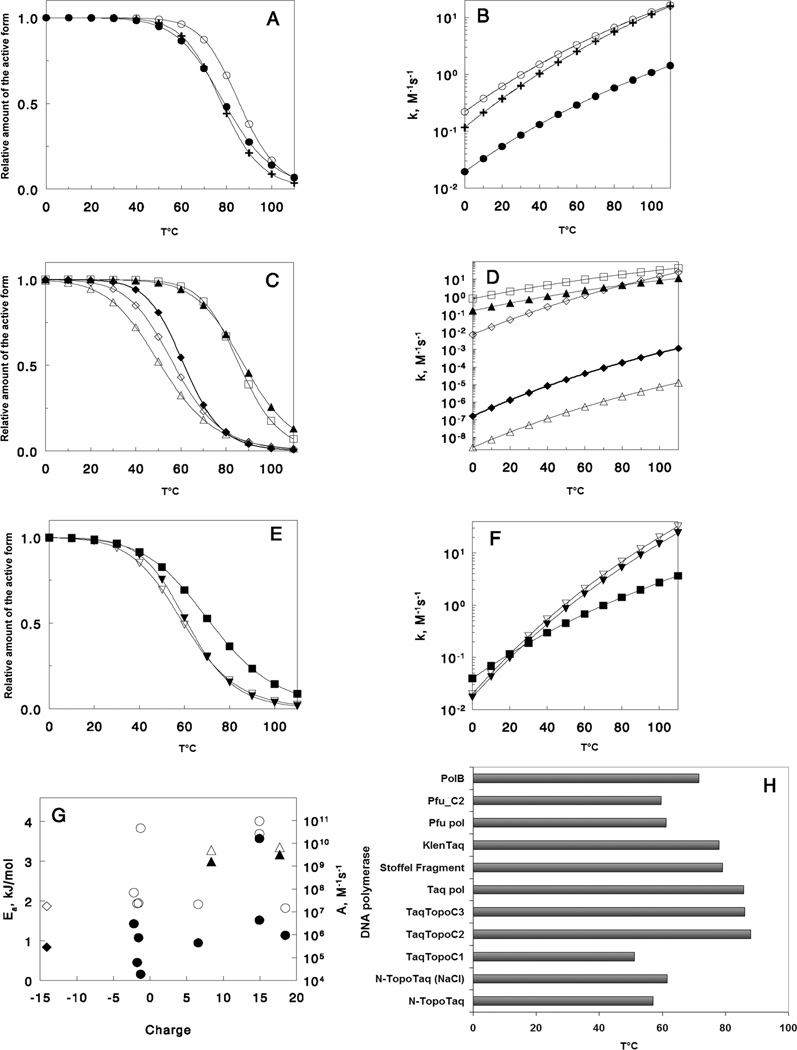
The analysis of the substrate binding was carried out using a general model that includes temperature - dependent reversible isomerization of DNA polymerases (see Supporting information for details). Accordingly, the dependencies of Vbapp on temperature for all polymerases shown in Figures 5 (A–D), can be approximated by theoretical bell-shaped curves predicted by this model. Table 2 summarizes values for the pre-exponential factor and activation energies of substrate binding to DNA polymerases and the enthalpy and entropy changes of the conformational transitions (see Supporting Information).
Table 2.
Values of the pre-exponential factor, activation energy for duplex substrate binding to DNA polymerases and values of the enthalpy and entropy changes for high-temperature reversible conformational transition of DNA polymerases that eliminates binding of substrate.
| DNA polymerase |
A M−1•s−1 |
Ea kJ•mol−1 |
ΔS‡ J•K−1 |
ΔH° kJ•mol−1 |
ΔS° J•K−1 |
|---|---|---|---|---|---|
| N-TopoTaqa | 1.63E+10 | 3.69 | −2.89 | 4.99 | 15.10 |
| N-TopoTaqb | 4.40E+06 | 4.01 | −6.79 | 6.41 | 19.16 |
| TaqTopoC1a | 1.86E+04 | 3.83 | −9.39 | 4.17 | 12.85 |
| TaqTopoC2a | 4.49E+05 | 1.92 | −7.88 | 5.70 | 15.78 |
| TaqTopoC3a | 9.47E+05 | 1.82 | −7.52 | 7.02 | 19.53 |
| Taq polc | 7.44E+05 | 1.95 | −7.64 | 7.13 | 19.87 |
| Stoffel Fragmentc | 6.09E+04 | 1.94 | −8.83 | 5.42 | 15.39 |
| KlenTaqc | 3.06E+06 | 2.21 | −6.97 | 6.58 | 18.76 |
| Pfu pola | 1.61E+09 | 3.28 | −3.99 | 5.09 | 15.21 |
| PfuC2a | 3.17E+09 | 3.35 | −3.67 | 4.40 | 13.23 |
| Mka PolBa | 2.83E+05 | 2.05 | −8.10 | 3.83 | 11.11 |
250 mM potassium glutamate
250 mM sodium chloride
50 mM potassium chloride
Fusion of various HhH domains with DNA polymerases causes relatively small changes in parameters of a DNA binding reaction; e.g., values of activation energies for the formation of complexes with PTJ are almost the same for Stoffel fragment, Taq polymerase, and the chimeric TaqTopoC2 and TaqTopoC3. This suggests that the additional TopoV domains at the C-termini of the Taq polymerase catalytic domain do not interfere with the initial binding of the duplex substrates to the polymerase active site. Accordingly, it implies that during initial substrate binding with the native Taq polymerase, the N-terminal exonuclease domain has a remote position with respect to the catalytic domain, as it was revealed by structural analysis (17). The slightly higher activation energy of the KlenTaq polymerase than those of the Taq polymerase and Stoffel fragment could indicate interference of the truncated exonuclease domain with the substrate binding during the reaction. In contrast, both N-TopoTaq and TaqTopoC1 possess much higher activation energy values. This could be a result of the localization of the TopoV C3 domain to be in close proximity to the polymerase active site, which would result in partial blockage of the substrate binding. Thus, the efficient binding of the substrate likely requires a conformational change such as removing the interfering domains. These findings support our earlier suggestion (6) that in the TaqTopoC1 construct, the C3 domain could be in close proximity to the catalytic site because of the length of the C-terminal tail of the chimera.
It is interesting that in PfuC2 DNA polymerase the C2 domain is positioned on the substrate binding side but it does not alter the activation energy of the substrate binding, which is already much higher than that for the Taq polymerase catalytic domain. It is possible that the high activation energy of the binding reflects conformational changes of Pfu catalytic domain similar to those in bacteriophage RB69 polB (18, 19). However, this is not necessarily a general property of polB catalytic domains, as Ea of M. kandleri DNA polymerase is very similar to that of Taq Stoffel fragment (Table 2).
Surprisingly, the empirical pre-exponential factor of the Arrhenius equation (see Supporting Information) was found to have variations in a very wide range between different DNA polymerases (Table 2). The pre-exponential factor can be interpreted as a product of collision frequency and a steric factor of the reaction (20). As the values of collision frequencies should be similar for the reactions of all studied DNA polymerases with the PTJs, the differences could be related to efficiency of the collisions or the steric factor.
In Figure 6, both the values of Ea and the pre-exponential factors are plotted against the calculated charge of the enzymes at pH 6.5 (estimated pH of the reaction in Tris-HCl buffer at 75°C). Although several groups of DNA polymerases have similar activation energy values or empirical pre-exponential factors, those values do not correlate with the total charge of the molecules (Fig. 6G). Therefore, the mechanism of electrostatic enhancement of diffusion-controlled association (21) suggesting the reduction of Ea for binding substrates as a result of charge interactions does not play a significant role in binding of PTJs to DNA polymerases in our case. Also, for charged molecules, the theory of diffusion-controlled association predicts that the electrostatic enhancement and, consequently, the rate of substrate binding would depend on total ionic strength of the solution. However, in the presence of NaCl, the pre-exponent factor for N-TopoTaq was almost four orders of magnitude lower than in potassium glutamate of about the same ionic strength, with only slightly higher Ea (Table 2). This result is in agreement with our previous finding that chloride ions can specifically block positive charges on HhH domains of TopoV (6) that interact with DNA, rather than decrease the rate of DNA binding by changing the total charge distribution of the molecules.
Figure 6.
Theoretical values calculated from the parameters of Scheme II. Accumulation of the inactive conformations with temperature increase and dependencies of DNA binding rate constant on temperature for Taq polymerase and its fragments (A, B), chimeric polymerases with Taq catalytic domain (C, D), and polB DNA polymerases (E, F): -○- – Taq polymerase; -●- – Stoffel Fragment; -+ - – Klentaq; -△- – TaqTopoC1; -▲- – TaqTopoC2; -□- – TaqTopoC3; -◇- – N-TopoTaq in potassium glutamate; -◆- – N-TopoTaq in NaCl; -▽- – Pfu DNA polymerase; -▼- – PfuC2; -■- – Mka polB AV19. G, dependencies of pre-exponential factor (filled symbols) and the activation energy (hollow symbols) of the Arrhenius equation for PTJ binding to DNA polymerases (6) on total charge of the protein molecules calculated at 75°C: (○, ●) – polymerases with Taq catalytical domain; (△, ▲) – polymerases with Pfu catalytical domain; (◇, ◆) – M.K. polB av19. Panel H displays the temperatures of half-inactivation for DNA polymerases (curves on panels A, C, and E).
In terms of transition state theory, the steric factor, which is included in the pre-exponential factor, can be related to the entropy of activation of the substrate binding. As all studied polymerases have strong negative values of entropy of activation, ΔS‡ (Table 2), it appears that the formation of polymerase-PTJ complexes renders structures of the proteins and PTJ more compact than in the solution before the interaction.
The calculated rate constants are presented in Figures 6B, D, and 6F. As compared to Taq polymerase and KlenTaq, the Stoffel Fragment has ~10 times lower rate constants of association with the PTJ due to low steric factor of the complex formation (Fig. 6B). These observations suggest an important role of the Taq polymerase 5’-exonuclease domain for providing efficient DNA association with the polymerase site, e.g., by offering additional DNA binding groups for favorable orientation of the substrate. Also, TaqTopoC2 and TaqTopoC3 have rate constants similar to that of the Taq polymerase, apparently due to the very close values of the pre-exponential factors and activation energies (Table 2). Therefore, Topo V HhH domains could substitute for the original Taq polymerase 5’ - exo- domain to provide proper orientation of the DNA substrate for faster binding to the polymerase site.
Fusion of C3 domain to the N- terminal of the Taq polymerase catalytic domain resulted in big changes to both the pre-exponential factor and the activation energy, with respect to the Stoffel Fragment alone (Table 2; N-TopoTaq without added NaCl). Nevertheless, the rate constants for N-TopoTaq and the Stoffel Fragment behaved very similarly (compare curves on Fig. 6B vs. Fig. 6D), since ~ 1,000 times increase of the pre-exponential factor in the Arrhenius equation for N-TopoTaq compensated the increase of the activation energy. In contrast, the addition of 0.25 M NaCl, which significantly reduces interaction of DNA with HhH domains (6), decreases the pre-exponential factor and the corresponding rate constant of binding ~10,000 times (Fig. 6D), as compared to the Stoffel fragment.
The extremely low pre-exponential factor found for TaqTopoC1, evidently, originates from poor steric conditions for DNA binding. In combination with very high activation energy (as in the case of N-TopoTaq; Table 2), it produces binding rate constants about 106 times lower than those of the Stoffel Fragment.
PolB DNA polymerases demonstrate the temperature profile of the binding rate constants comparable to that of Taq polymerase (Fig. 6F). Interestingly, fusion of the C2 domain to the C- terminal of the catalytic domain of Pfu DNA polymerase does not produce any significant differences neither in the pre-exponential factor, nor in the activation energy. It appears that the interaction of DNA with the HhH domains during the initial substrate binding does not improve the orientation of the substrate with respect to the polymerase DNA-binding site, or such interaction occurs only after the binding of DNA to the polymerase site. The binding parameters of DNA to M. kandleri PolB were similar to those of TaqTopoC2 (Table 2; Fig. 6F).
The reversible conformational transition of DNA polymerases makes the PTJ substrate binding to the polymerase catalytic domain at high temperatures inefficient. This transformation is controlled by the values of changes for enthalpy and entropy. As both the enthalpy and entropy changes are positive, it suggests that the polymerases adopt less compact and less stable conformations. However, since the changes of enthalpy and entropy have the opposite effect on the transformation, they determine a variety of the dependencies of the equilibrium (Scheme 2; Supporting Information) on temperature. Figure 6 (A, C, and E) shows the calculated relative amount of the substrate binding polymerase conformations on temperature. Surprisingly, such transitions occur at rather low temperatures: for the majority of the polymerases in this study the temperature of half-transformation is below 80°C (Fig. 6H). Moreover, the Pfu catalytic domain shows much lower transformation temperature than that of the Taq polymerase. Both C2 and C3 domains on the C-termini of the Stoffel Fragment, as well as the whole Taq exo domain of the original Taq polymerase seem to stabilize the active conformation. However, the transformation into an inactive form occurs at very low temperatures in the case of N-TopoTaq and TaqTopoC1. As this effect is not observed for Taq polymerase, its fragments, or other Taq hybrids, it can be attributed to the interference of the C3 domain with PTJ binding in these chimeras.
DISCUSSION
In light of this work, previous data on the increase of thermostability of the hybrids (6) become even more intriguing, as they demonstrate direct interactions of the polymerase domains and HhH domains. Our modeling of TopoV HhH domains in chimeras (6), which is in agreement with the crystal structure of several TopoV HhH domains (22), suggests that the HhH repeats B-L are folded into individual structures of similar shape connected with each other by flexible linker sequences. Therefore, the thermo-stabilizing effect in chimeras could occur either because of wrapping the flexible HhH tail around the polymerase domain, thereby restricting its conformational mobility, or through some specific contacts between the polymerase domain and C3 domain, which has its own stable structure (8). Comparison of the thermostabilities of chimeras (Fig. 2) points to a mechanism of thermostabilization that involves specific interactions of the polymerase catalytic domains with the HhH C3 domain. This conclusion is supported by the fact that the incorporation of the C3 domain alone suffices to produce the highest increase of chimera’s thermostability, while truncations of the C3 domain (Fig. 1) have the strongest effect on the stability of the hybrids at high temperatures. The specific interdomain contacts in chimeras are likely produced by electrostatic interactions (e.g., between positively charged Topo V domain L, and negatively charged N-termini in catalytic fragments of polA or thumb domains in polB), since chloride ions, which are known to impair charge-directed interactions of C3 domain, eliminate the gained stabilization.
The ability to increase the polymerase thermal stability is not limited to the C3 domain since other hybrid DNA polymerases with the C1Δ, C2 and C2Δ HhH domains are all more thermostable than the Stoffel fragment alone (Fig. 1 and 2). It is interesting in this regard that the exonuclease domain of the entire Taq polymerase, which also contains a single copy of the HhH fold, does not contribute to the thermostability of the polymerase domain. Unlike the highly temperature-resistant HhH domains of Topo V, the 5’-exo Taq polymerase domain melts separately at a lower temperature than the Taq polymerase catalytic domain (89°C and 97–100°C, respectively) (23), thus it cannot stabilize the catalytic domain at high temperatures.
Studies of the polymerase reaction at very high temperatures (> 80°C) reveal several effects of HhH domains on DNA synthesis. The most interesting information was obtained while analyzing the effect of temperature on elongation rates that, under the selected reaction conditions, reflects rates of binding of the DNA substrates to DNA polymerases and formation of the productive complexes for DNA elongation. We found that a decrease in the rate of DNA synthesis by DNA polymerases at high temperatures could be attributed to the reversible high temperature isomerization of the DNA polymerase catalytic domains. Another important parameter of the binding reaction is the rate of formation of the productive complexes upon association to a DNA substrate with the fraction of DNA polymerase molecules, which are present in active conformations. Despite the highly negative charge of DNA substrates, the mechanism of electrostatic enhancement of diffusion-controlled association (21) does not play a significant role in the initial binding of the studied polymerases to DNA. Rather, position of the additional charged domain is important for the mechanism of binding. For example, the calculated charge on a TaqTopoC3 molecule at 75°C is higher than that of N-TopoTaq (+18.46 vs +14.95). However, in 0.25 M K-Glu, the pre-exponential factor for N-TopoTaq is ~17,000 times higher than that for TaqTopoC3, suggesting that every collision of DNA with N-TopoTaq has 17,000 more chances to form the productive complex than collision with TaqTopoC3, if DNA and N-TopoTaq molecules have sufficient energies. Nonetheless, the activation energy for the binding reaction of DNA with N-TopoTaq is much higher than that of the binding reaction for TaqTopoC3 (Table 2), so that significantly smaller fraction of collisions with properly oriented N-TopoTaq molecules resulted in DNA binding. Moreover, the interaction of the N-TopoTaq HhH domains with the catalytic polymerase domain in the absence of the bound substrate decreases the number of N-TopoTaq molecules in a conformation suitable for DNA binding (Fig. 4A, C). As a result, overall rate of DNA synthesis by N-TopoTaq is much lower than synthesis by TaqTopoC3.
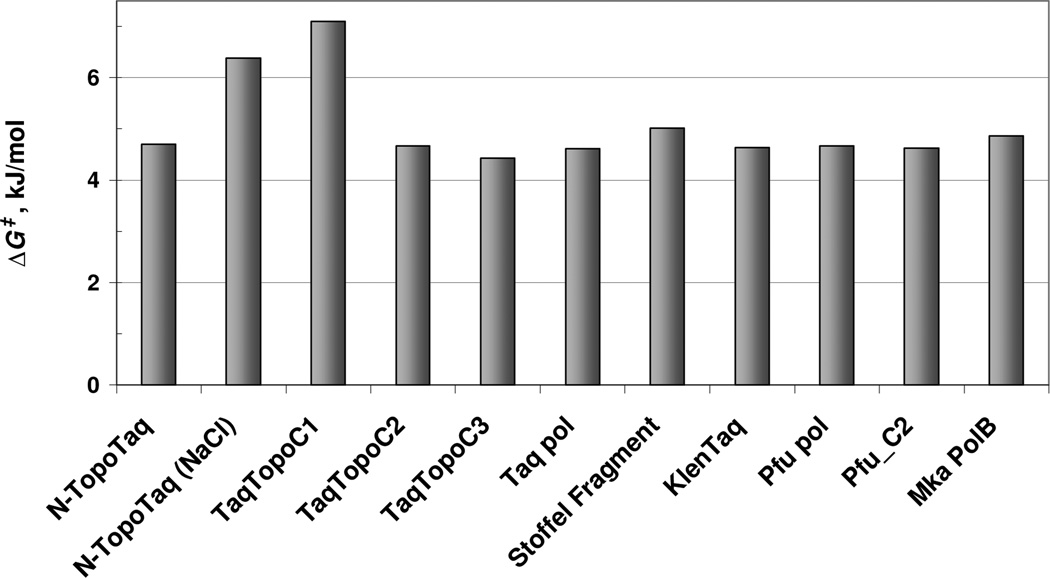
Data in Table 2 show that for the majority of the DNA polymerases of this study, the activation energy increases alongside the pre-exponential factor, pointing to a linear correlation between Ea and logA. In this case the Eyring equation for the rate constant (20) suggests constant value of the Gibbs activation energy among various DNA. Figure 7 shows that the values of the Gibbs energy of activation are very similar for DNA polymerases of various classes and with different HhH domains. Therefore, in the reaction of binding DNA and forming the productive complex, E + S → ES‡ → ES0, the transition states ES‡ of diverse DNA polymerases should have similar energies/geometries. Evidently, the latter is a basic property of intact DNA polymerase domains, which is not changed in the majority of the hybrid enzymes. Two exceptions, N-TaqTopo (NaCl) and TopoTaqC1, show that the transition state can be affected by unfavorable positioning of HhH domains.
Figure 7.
Values of the Gibbs energy of activation for binding PTJ1 to various DNA polymerases.
Moreover, the values of the Gibbs activation energy do not depend on the overall charges of the DNA polymerases suggesting that the transition state is not affected by the charge of DNA polymerases either, and the binding of the substrate to the active site is not controlled by diffusion.
The HhH tails in the chimeras can stabilize the enzyme complexes with the DNA substrate, as is noted by the processivity plots (Fig. 4). It is interesting that all TopoV HhH-Taq chimeras used in this study demonstrate similar low levels of stabilization, except for TaqTopoC1, which significantly exceeds the others. As all polymerase complexes with a primer template junction substrate are different with respect to salt resistance (6), this suggests that salt induced dissociation and heat induced dissociation of the same complexes have different mechanisms and involve different steps.
HhH domains are widely present in enzymes that act on nucleic acids and can be catalytically involved with base excision repair (7, 24). Little is known about functional interaction of HhH domains with other domains of DNA processing proteins. An important role of HhH domains in contact between the human polλ and PCNA has been reported (25). Using artificial hybrid proteins (6), we demonstrated other types of interdomain interactions involving the HhH motifs. In a recent paper (10), Salas and co-workers reported the successful fusion of HhH domains from the M. kandleri Topo V with phi29 DNA polymerase, which brings about a general improvement of amplification reactions carried out by the hybrid enzyme. Although the authors do not analyze the mechanism of domain interaction in the chimera, they assume that the major cause for the DNA synthesis enhancement is the improved binding of the hybrid polymerase to the DNA template. However, it is also possible that the reported improvement results from the direct interaction of the HhH domains with the polymerase.
Supplementary Material
ACKNOWLEDGMENT
We thank Vladimir Slesarev for help with manuscript preparation.
Research was supported by NIH grant 2R44CA101566 (to A. Slesarev).
ABBREVIATIONS
- Mka polB
Methanopyrus kandleri DNA polymerase of family B
- Bst DNA polymerase
Bacillus stearothermophilus DNA polymerase of family A
- LF Bst polymerase
Large Fragment of Bacillus stearothermophilus DNA polymerase of family A
- Pfu polB
Pyrococcus furiosus DNA polymerase of family B
- HhH domain
Helix-hairpin-Helix DNA binding domain
- PTJ
primer-template junction
- Vbapp
apparent rate of substrate binding
- Pe
processivity equivalence parameter
- p0
microscopic processivity parameter
Footnotes
Supporting Information Available
Detailed information such as a general model for substrate binding by DNA polymerases and kinetic schemes used to described effects of temperature on duplex binding and processivity of DNA synthesis is available free of cherge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Pavlov AR, Pavlova NV, Kozyavkin SA, Slesarev AI. Recent developments in the optimization of thermostable DNA polymerases for efficient applications. Trends Biotechnol. 2004;22:253–260. doi: 10.1016/j.tibtech.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Baar C, d'Abbadie M, Vaisman A, Arana ME, Hofreiter M, Woodgate R, Kunkel TA, Holliger P. Molecular breeding of polymerases for resistance to environmental inhibitors. Nucleic Acids Res. 2011;39:e51. doi: 10.1093/nar/gkq1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun S, Geng L, Shamoo Y. Structure and enzymatic properties of a chimeric bacteriophage RB69 DNA polymerase and single-stranded DNA binding protein with increased processivity. Proteins. 2006;65:231–238. doi: 10.1002/prot.21088. [DOI] [PubMed] [Google Scholar]
- 4.Pavlov AR, Pavlova NV, Kozyavkin SA, Slesarev AI. In: DNA Amplification: Current Technologies and Applications. Demidov VV, Broude NE, editors. Norwich U.K: Horizon Bioscience; 2004. pp. 3–20. [Google Scholar]
- 5.Pavlov AR, Pavlova NV, Kozyavkin SA, Slesarev AI. In: DNA Sequencing II: Optimizing Preparation and Cleanup. Kieleczawa J, editor. Sudbury, MA: Jones and Bartlett Publishers; 2006. pp. 241–258. [Google Scholar]
- 6.Pavlov AR, Belova GI, Kozyavkin SA, Slesarev AI. Helix-hairpin-helix motifs confer salt resistance and processivity on chimeric DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13510–13515. doi: 10.1073/pnas.202127199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belova GI, Prasad R, Kozyavkin SA, Lake JA, Wilson SH, Slesarev AI. A type IB topoisomerase with DNA repair activities. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6015–6020. doi: 10.1073/pnas.111040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belova GI, Prasad R, Nazimov IV, Wilson SH, Slesarev AI. The Domain Organization and properties of individual domains of DNA topoisomerase V, a type 1B topoisomerase with DNA repair activities. J. Biol. Chem. 2002;277:4959–4965. doi: 10.1074/jbc.M110131200. [DOI] [PubMed] [Google Scholar]
- 9.Slesarev AI, Stetter KO, Lake JA, Gellert M, Krah R, Kozyavkin SA. DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote. Nature. 1993;364:735–737. doi: 10.1038/364735a0. [DOI] [PubMed] [Google Scholar]
- 10.de Vega M, Lazaro JM, Mencia M, Blanco L, Salas M. Improvement of φ 29 DNA polymerase amplification performance by fusion of DNA binding motifs. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16506–16511. doi: 10.1073/pnas.1011428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairfield FR, Newport JW, Dolejsi MK, von Hippel PH. On the processivity of DNA replication. J. Biomol. Struct. Dyn. 1983;1:715–727. doi: 10.1080/07391102.1983.10507477. [DOI] [PubMed] [Google Scholar]
- 12.von Hippel PH, Yager TD. Transcript elongation and termination are competitive kinetic processes. Proc. Natl. Acad. Sci. U.S.A. 1991;88:2307–2311. doi: 10.1073/pnas.88.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slesarev AI, Mezhevaya KV, Makarova KS, Polushin NN, Shcherbinina OV, Shakhova VV, Belova GI, Aravind L, Natale DA, Rogozin IB, Tatusov RL, Wolf YI, Stetter KO, Malykh AG, Koonin EV, Kozyavkin SA. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4644–4649. doi: 10.1073/pnas.032671499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malykh AG, Polushin NN, Slesarev AI, Kozyavkin SA. In: Bacterial Artificial Chromosomes. Zhao S, Stodolsky M, editors. Totowa, NJ: Humana Press; 2004. pp. 221–229. [Google Scholar]
- 15.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Boeker EA. Initial rates. A new plot. Biochem J. 1982;203:117–123. doi: 10.1042/bj2030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eom SH, Wang J, Steitz TA. Structure of Taq polymerase with DNA at the polymerase active site. Nature. 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 19.Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 20.IUPAC. Compendium of Chemical Terminology (Gold Book) Online corrected version, International Union of Pure and Applied Chemistry. 1997 (2010). [Google Scholar]
- 21.Vijayakumar M, Wong K-Y, Schreiber G, Fersht AR, Szabo A, Zhou H-X. Electrostatic enhancement of diffusion-controlled protein-protein association: comparison of theory and experiment on barnase and barstar. J. Mol. Biol. 1998;278:1015–1024. doi: 10.1006/jmbi.1998.1747. [DOI] [PubMed] [Google Scholar]
- 22.Taneja B, Patel A, Slesarev A, Mondragon A. Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J. 2006;25:398–408. doi: 10.1038/sj.emboj.7600922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karantzeni I, Ruiz C, Liu C-C, LiCata VJ. Comparative thermal denaturation of Thermus aquaticus and Escherichia coli type 1 DNA polymerases. Biochem. J. 2003;374:785–792. doi: 10.1042/BJ20030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullen GP, Wilson SH. DNA polymerase beta in abasic site repair: a structurally conserved helix-hairpin-helix motif in lesion detection by base excision repair enzymes. Biochemistry. 1997;36:4713–4717. doi: 10.1021/bi962363a. [DOI] [PubMed] [Google Scholar]
- 25.Maga G, Blanca G, Shevelev I, Frouin I, Ramadan K, Spadari S, Villani G, Hubscher U. The human DNA polymerase lambda interacts with PCNA through a domain important for DNA primer binding and the interaction is inhibited by p21/WAF1/CIP1. FASEB J. 2004;18:1743–1745. doi: 10.1096/fj.04-2268fje. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.