Fanconi anemia (FA) is a rare recessive disorder characterized by genome instability, congenital malformations, progressive bone marrow failure, and predisposition to hematologic malignancies and solid tumors1. At the cellular level, hypersensitivity to DNA interstrand crosslinks (ICLs) is the defining feature in FA2. Mutations in thirteen distinct FA genes3 have been shown to interfere with the DNA-replication dependent repair of lesions involving crosslinked DNA at stalled replication forks4. Depletion of SLX4, which interacts with multiple nucleases and has been recently identified as a Holliday junction resolvase5–7, results in increased sensitivity of the cells to DNA crosslinking agents. Here we report the identification of biallelic SLX4 mutations in two patients with typical clinical features of FA and show that the cellular defects in the patients’ cells are complemented by wild-type SLX4, demonstrating that biallelic mutations in SLX4/FANCP cause a new subtype of Fanconi anemia, FA-P.
SLX4 is a multidomain scaffold protein interacting with three distinct nucleases SLX1, ERCC4/XPF-ERCC1, and MUS81-EME15–7. While the SLX4-SLX1 interaction is largely responsible for the Holliday junction resolvase activity seen in the complex, SLX4 can also stimulate the activity of ERCC4/XPF and MUS81 nucleases, both of which have been previously implicated in the processing of interstrand crosslinks (ICLs)8. The finding that depletion of SLX4 leads to increased sensitivity to cross-linking agents and camptothecin5–7 prompted us to investigate SLX4 as a candidate gene for Fanconi anemia1.
So far, mutations in thirteen genes are responsible for FA. Eight of the FA proteins (FANCA/B/C/E/F/G/L/M) form a core complex, a nuclear E3 ubiquitin ligase2 which ubiquitinates FANCI and FANCD29,10. These two activated proteins subsequently localize as an FANCI/FANCD2 (ID) complex to chromatin and direct repair4 in part through interaction with the newly identified nuclease FAN111–14. Cells with mutations in the FA core complex (except for FANCM) lack monoubiquitination of FANCD2. The other FA proteins are FANCJ/BRIP1, a helicase, and homologous recombination (HR) effectors, FANCN/PALB2 and FANCD1/BRCA2. Recently, RAD51C, also involved in HR repair, has been found to be mutated in three patients with an FA-like disorder15. Cells mutated in FANCJ/N/D1 and RAD51C have normal FANCD2 monoubiquitination and these genes are thought to work downstream of the ID complex.
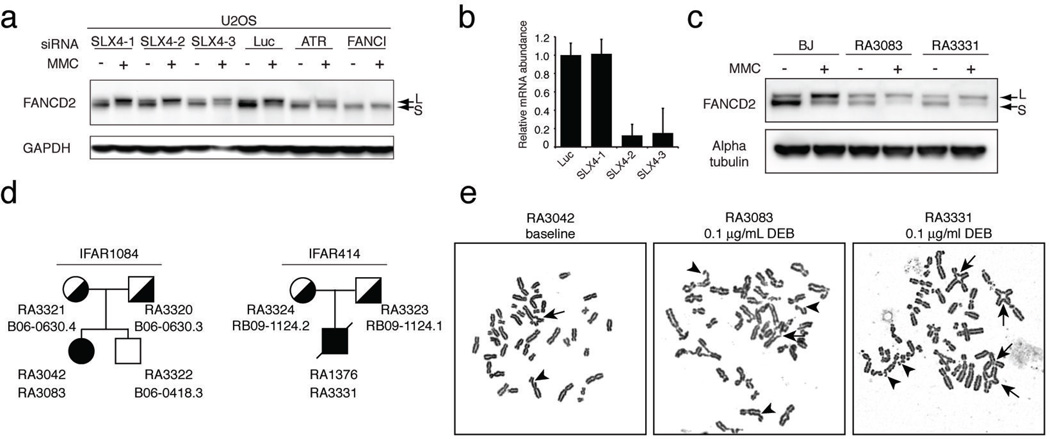
As depletion of SLX4 in a U2OS cell line does not affect FANCD2 ubiquitination (Figure 1A and B), we sequenced SLX4 in the families from the International Fanconi Anemia Registry16 with unassigned FA complementation groups and normal FANCD2 modification (Figure 1C) and identified two families carrying germline mutations, IFAR1084 and IFAR414 (Figure 1D). Phenotypes of the two patients are summarized in Table 1. The lymphoblastoid cell line (LCL) (RA3042) and fibroblasts (RA3083) from the patient 1084/1 showed increased genomic instability (Figure 1E and Table 2) and increased sensitivity to Mitomycin C (MMC) (Figure S1A). The 414/1 patient’s LCL (RA 1376) was not sensitive to MMC suggestive of reversion (Figure S1B); however, his skin fibroblasts (RA 3331) displayed a high degree of DEB induced chromosomal instability (Figure 1E and Table 2) and sensitivity to MMC. No UV sensitivity was observed in fibroblasts from either of the patients (Figure S1C and D). Fibroblasts from the patient 414/1 (RA3331), but interestingly not patient 1084/1 (RA3083), were sensitive to camptothecin (CPT), a topoisomerase I inhibitor (Figure S1E and F).
Figure 1.
Characterization of cell lines from FA patients with SLX4 mutations. A. Western blot analysis with an anti-FANCD2 antibody of U2OS cells transfected with the indicated siRNAs and treated with 1 µM MMC for 24 hours. L (long) indicates a monoubiquitinated form and S (short) indicates the non-monoubiquitinated form of FANCD2. B. RT-qPCR in U2OS cells transfected with various siRNAs against SLX4 used in experiment shown in A. Error bars indicate standard deviation of three replicates. C. Western blot analysis with an anti-FANCD2 antibody of BJ, RA3083 and RA3331 fibroblasts. Cells were left untreated or were treated with 1 µM MMC for 24 hours. D. Pedigrees of the two families described in this study showing accession numbers for cell lines (RA) and peripheral blood samples (B, RB). The two probands are indicated with filled symbols. Mutation carriers are indicated by half-filled symbols. E. Examples of metaphases of the LCL RA3042 (no drug treatment) and fibroblast RA3083 cell lines from the patient 1084/1 and the fibroblasts RA3331 from patient 414/1 (treatment with diepoxybutane (DEB)).
Table 1.
Characteristics of patients with Fanconi anemia and mutations in SLX4
| Individual | Maternal allele | Paternal allele | Ethnicity | Phenotypic and hematologic abnormalities |
|---|---|---|---|---|
| 1084/1 | c.1163+2T>A p.R317_F387del* |
c.1163+2T>A p.R317_F387del* |
South Indian |
15 year-old female, short stature (height -2.1SD, 1st percentile); vitiligo; presented at 9 years of age with isolated thrombocytopenia; |
| 414/1 | c.2013+225_3147 del4890insCC p.L672VfsX119** |
c.514delC p.L172FfsX22*** |
American of European descent |
bilateral absent thumbs and right radial aplasia, undescended left testicle, pelvic kidney, malformed auricle and short stature; squamous cell carcinoma of the tongue at 21 years of age; platelets 35,000 cells/µl, Hb 10 g/dL, MCV 105.5 fL; died at 22 years from complications of metastatic disease |
the predicted protein has an internal deletion from amino acids 317 to 387.
the predicted protein has 671 N-terminal amino acids of SLX4 followed by 119 non-SLX4 amino acids due to a frameshift.
the predicted protein has 172 N-terminal amino acids of SLX4 followed by 22 non-SLX4 amino acids due to a frameshift.
Table 2.
Chromosome breakage analysis in the indicated cell lines with and without diepoxybutane treatment (DEB).
| RA3042 (LCL) | RA3083 E6E7 | RA3331 E6E7 | BJ E6E7* | |||||
|---|---|---|---|---|---|---|---|---|
| DEB concentration (µg/ml) | 0 | 0.1 | 0 | 0.1 | 0 | 0.1 | 0 | 0.1 |
| # Metaphases | 56 | 29 | 53 | 32 | 50 | 31 | 63 | 51 |
| # Total breaks** | 41 | 221 | 8 | 140 | 7 | 217 | 8 | 8 |
| # Chromatid breaks | 29 | 123 | 6 | 92 | 7 | 123 | 6 | 8 |
| # Triradials | 5 | 44 | 1 | 16 | 0 | 36 | 1 | 0 |
| # Quadriradials | 1 | 5 | 0 | 8 | 0 | 11 | 0 | 0 |
| % Metaphases with breaks | 30 | 90 | 13 | 81 | 14 | 100 | 11 | 16 |
| # Breaks per metaphase | 0.73 | 7.6 | 0.15 | 4.4 | 0.14 | 7.0 | 0.11 | 0.16 |
BJ foreskin fibroblasts from a healthy donor were used as a normal control.
Total number of breaks included chromatid breaks and radial chromosomes.
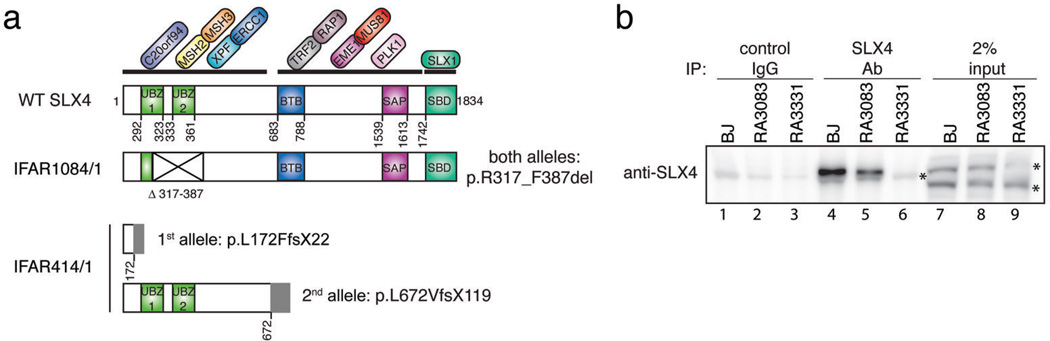
Sequencing of the cDNA from the 1084/1 patient’s cells revealed skipping of Exon 5 (Figure S2A), due to a homozygous point mutation in the canonical splice donor dinucleotide GT in intron 5 (c.1163+2T>A) in the genomic DNA (Figure S2B). Both parents were found to be heterozygous and an unaffected sibling was found to be negative for this mutation (Figure S2B). The predicted effect of this mutation is a 70 amino acid deletion of amino acids (aa) 317 to 387 of SLX4 (p.R317_F387del), leading to an in-frame deletion of the conserved Cys and Leu of the first UBZ domain and the whole second UBZ domain (Figure 2A, Figure S2C). Immunoprecipitation of SLX4 from the cell line RA3083 confirmed the presence of a slightly shorter protein product (Figure 2B, lane 5, Figure S2D)
Figure 2.
SLX4 is defective in two FA patients. A. Schematic of SLX4 (based on ref. 7) showing the domain architecture, the interacting proteins, and the predicted protein effect of SLX4 mutations in IFAR1084/1 and IFAR414/1 patients. B. Analysis of the mutant SLX4 protein in the patient’s cell lines. Cell extracts of primary BJ, RA3083, and RA3331 fibroblasts were subjected to immunoprecipitation using a control rabbit antibody (control IgG) or the SLX4 antibody. Asterisks indicate the crossreacting bands. Note that the antibody does not identify SLX4 in straight western (lanes 7 to 9).
In patient 414/1, a heterozygous frameshift mutation in exon 2 (c.514delC) was detected by sequencing of the full length RT-PCR product (Figure S3A) and confirmed in the genomic DNA of the patient and his father (Figure S3B). The predicted protein effect of this frame-shift mutation is a truncated protein with N-terminal 171aa of SLX4 followed by 22 non-SLX4 aa (p.L172FfsX22) (Figure 2A). The second allele of SLX4 in the second patient, identified as described in Online Methods, showed a large genomic deletion from intron 9 to exon 12 resulting in c.2013+225_3147delinsCC (Figure S3C, S3D, and S3E). The predicted effect of this mutation is a truncated protein with N-terminal 671aa of SLX4 followed by 119 non-SLX4 aa due to a frameshift (p.L672VfsX119) (Figure 2A). Consequently, immunoprecipitation with the antibody against SLX4 failed to identify the full-length protein in the patient’s fibroblasts RA3331 (Figure 2B, lane 6).
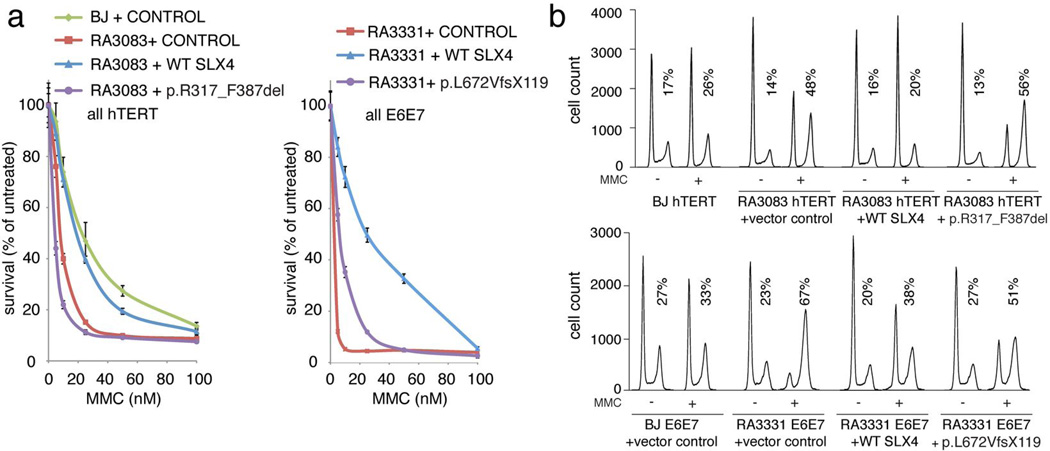
To prove that the mutations identified in SLX4 were causal for the FA phenotype of both patients, we introduced the WT or the mutant SLX4 cDNAs into the patients’ fibroblasts (RA3083 and RA3331) and performed functional complementation assays (Figure 3 and Figure S4). Expression of WT SLX4 in both cell lines almost fully rescued the MMC sensitivity (Figure 3A, Figure S4A, and S4B), the late S/G2 arrest with MMC treatment (Figure 3B, Figure S4C, S4D, and S4E), and the chromosomal instability after treatment with DEB (Figure S4F). Some residual MMC sensitivity, cell cycle arrest, and chromosomal breakage is most likely due to some cells losing expression of the SLX4 as evident by immunofluorescence analysis (data not shown). Introduction of the mutant proteins did not rescue the FA phenotypes of the patients’ cells although a slight improvement was noted in the various assays possibly due to overexpression of the mutant proteins, which might have residual function. These experiments demonstrate that biallelic SLX4 mutations cause a new subtype of Fanconi anemia, FA-P and FANCP becomes an alias for the SLX4 gene.
Figure 3.
Complementation of RA3083 and RA3331 cells with the SLX4 cDNA. A. Complementation of MMC sensitivity. Fibroblasts stably transduced with empty vector (control), or the vector expressing WT SLX4 or the mutant SLX4 cDNAs were exposed in triplicate to different levels of MMC ranging from 0 to 100 nM. After 8 days, cell number was determined using a coulter counter. Total cell numbers at each dose were divided by the number of cells in the untreated sample to arrive at percent survival. Error bars indicate standard deviation. B. Complementation of the cell cycle defect after MMC treatment. Indicated cells were treated with 100nM MMC and the cell cycle was analyzed 48 hours later. Untreated samples were analyzed in parallel. Expression levels of the exogenous proteins are shown in Figure S4A, S4B, and S4C. Quantification of the data is shown in Figure S4D and S4E.
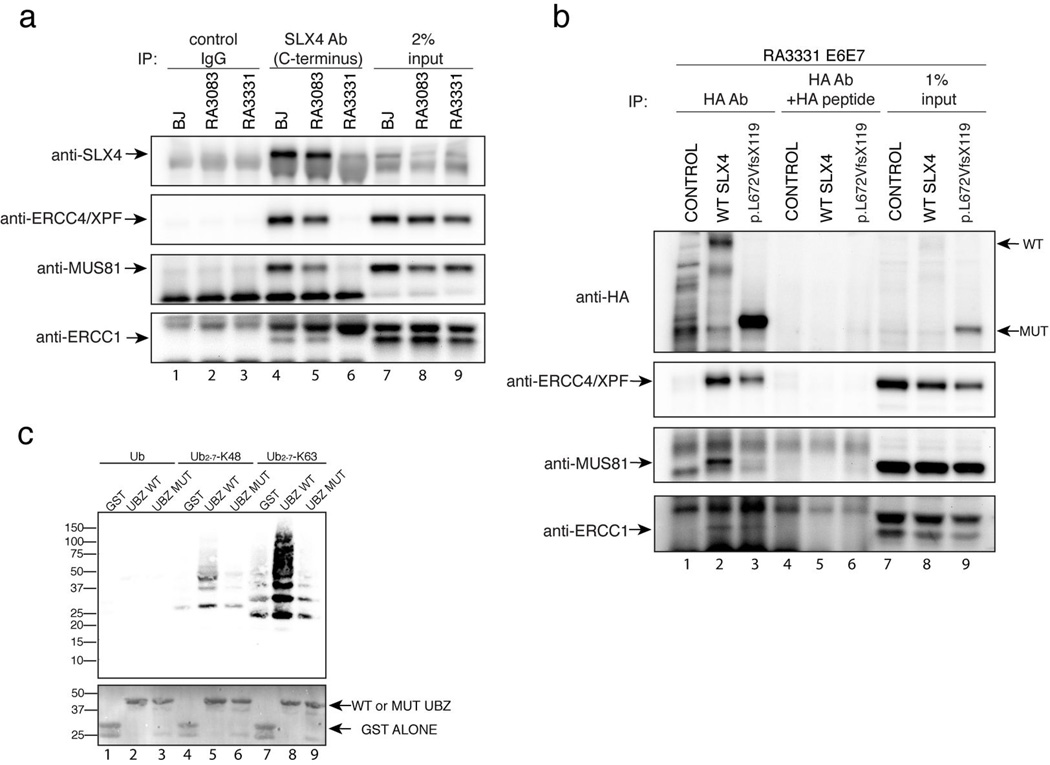
SLX4 interacts with multiple factors, two of which, ERCC4/XPF and MUS81 have been previously implicated in crosslink repair8. We therefore tested whether the mutant SLX4 proteins from both patients still interacted with the ERCC4/XPF and MUS81 complexes. We found that ERCC4/XPF, MUS81, and ERCC1 co-immunoprecipitate with endogenous mutant SLX4 (p.R317_F387del) from RA3083 fibroblasts (Figure 4A, lane 5 and Figure S5A, lane 4), although the levels of the mutant SLX4 protein were consistently lower in multiple experiments leading to diminished immunoprecipitation of the interacting factors. The second patient’s SLX4 p.L672VfsX119 overexpressed in RA3331 fibroblasts showed diminished but present interaction with ERCC4/XPF and ERCC1, but not with MUS81 (Figure 4B, lane 3). This was consistent with the previous findings that MUS81 interacts with aa 684–1834 fragment of the SLX4 protein7 which are deleted in the p.L672VfsX119 mutant protein. Immunoprecipitation with an antibody recognizing the N terminus of SLX4 from RA3331 cells showed greatly diminished interaction with ERCC4/XPF, ERCC1, and MUS81 (Figure S5B, lane 6).
Figure 4.
Interaction of mutant forms of SLX4 with its partners and with ubiquitin. A. Analysis of SLX4 interacting partners in SLX4 mutant cells. Cell extracts of primary fibroblasts (BJ, RA3083, and RA3331) were subjected to immunoprecipitation using the SLX4 antibody. Interacting proteins were identified by immunoblotting with the indicated antibodies. B. Analysis of SLX4 interacting partners in RA3331 cells. Cell extracts of RA3331 E6E7 cells expressing HA tagged control vector (CONTROL), WT SLX4, or the p.L672VfsX119 SLX4 were subjected to immunoprecipitation using HA antibody or HA antibody in the presence of 30 µg of HA peptide. Interacting proteins were identified by immunoblotting with the indicated antibodies. C. Interaction of the UBZ domains with ubiquitin. GST-purified GST control, WT UBZ domains (SLX4aa 251-402) and UBZ domains with four cysteines mutated to alanines (SLX4aa251-402_C296A_C299A_C336A_C339A) were incubated with the indicated forms of ubiquitin, purified by binding to GST beads, separated on a PAGE gel and immunobloted with anti-ubiquitin antibody. The bottom panel shows Ponceau staining of the GST proteins.
As UBZ domains are known to interact with ubiquitin17, we hypothesized that the absence of the tandem UBZ domains in the mutant SLX4 from patient 1084/1 might disrupt the binding of the SLX4 complex to ubiquitin chains of repair proteins at the sites of DNA damage, as shown for the tandem UBZ domains of RAP8018. We therefore performed in vitro ubiquitin binding assays (Figure 4C) that showed binding of the isolated UBZ domains of SLX4 to the K63 chains of ubiquitin (Figure 4C, lane 8). When the two conserved cysteines from each UBZ domain were mutated to alanines (Figure S2C) the binding was reduced to background levels seen with GST alone (Figure 4C, compare lane 7 and 9), suggesting the possibility that SLX4 may localize to the sites of damage through binding to K63 ubiquitinated substrates. As SLX4 would localize other proteins including ERCC4/XPF, MUS81 and SLX1 to sites of DNA damage, the SLX4 deficient patient cell lines described here are important tools to understand which interactions of SLX4 are essential for the repair of cross-linked DNA and ultimately to define the importance of the SLX4/FANCP function in the FA pathway. Patient phenotypes also provide an important clue. FA patients with mutations in FANCN/PALB2 or FANCD1/BRCA2 genes, which are essential for homologous recombination, have very early onset of childhood solid tumors and AML19,20. FA patients with SLX4/FANCP mutations show a milder phenotype more akin to that seen in FA patients with mutations in the FA core or the ID complex components. This suggests that the Holliday junction resolution, an integral step of homologous recombination, might not be the essential function of SLX4 in the somatic compartment during crosslink repair and that the repair depends on the other nucleases, ERCC4/XPF and MUS81 that interact with SLX4.
SLX4/FANCP represents a second protein (besides FANCM) that is conserved in lower eukaryotes, which do not have any other FA pathway components. Yeast SLX4, like human SLX4 interacts with orthologs of ERCC4/XPF and SLX1 and the work in this model organism will provide insight into the function of the FA pathway in human cells. Since germ-line mutations in three FA genes (FANCD1/BRCA1, FANCN/PALB2, FANCJ/BRIP1) and RAD51C, mutated in an FA-like disorder, are associated with a high risk of developing familial breast and ovarian cancers21–24, SLX4 should also be sequenced in patients from pedigrees where no other predisposing mutations could be identified.
SLX4 Reference Sequences
NCBI Reference Sequence: NM_032444.2, NCBI Reference Sequence: NP_115820.2
Methods
Subjects
Cell lines and genomic DNA samples were derived from individuals with Fanconi anemia and their family members registered in the International Fanconi Anemia Registry after obtaining informed written consent. The Institutional Review Board of The Rockefeller University approved the studies.
Cell Culture
U2OS cells were grown in Dulbecco Modified Eagle medium (DMEM) supplemented with 10% (v/v) FBS, 100 units of penicillin per ml, and 0.1 mg streptomycin per ml (all from Invitrogen). Lymphoblast cell lines were grown in RPMI supplemented as above except with 20% FBS. Fibroblasts were grown as above except with 15% FBS and non-essential amino acids. Fibroblasts were grown in 3% oxygen. BJ cells are normal foreskin fibroblasts and were obtained from ATCC. Patients’ fibroblasts were immortalized using a catalytic subunit of telomerase (hTERT) or were transformed using HPVE6 and E7 proteins.
cDNA and Genomic Sequencing
RNA was extracted from the cell lines using Qiagen RNeasy Plus Mini Kit. First strand cDNAs were then synthesized using the Invitrogen SuperScript™ III Reverse Transcriptase kit. SLX4 RT-PCR was done using primers 621 and 624 (see Supplementary Table 1) and Invitrogen Platinum® Pfx DNA Polymerase kit. PCR products were cleaned up using USB Exo-sapit kit before set up for sequencing, done by Genewiz, Inc. (NJ). Primers 623, 627, 628, 629, 630, 633, 634,639, 640, and 641 were used to sequence the full length SLX4 (see Supplementary Table 1). Genomic PCR was done using Qiagen Taq Polymerase with primers shown in Supplementary Table 1. The PCR products were cleaned up using USB Exo-sapit kit before set up for sequencing. In most cases, the PCR primers were also suitable for sequencing.
Identification of the second allele in IFAR414/1
In order to identify the second allele in RA 3331 cell line, full genomic sequencing of the coding exons was done on the patient’s and the maternal DNA. A genomic deletion was found likely, based on informative polymorphic markers in exon 12: c.3162G>A (p.S1054S, rs76488917), c.3365C>T (p.P1122L, rs714181). Amplification of parts of the cDNA followed by the detailed sequence analysis revealed retention of IVS9 through c.2013+224, with deletion of c.2013+225_3147 and insertion of two cytosine nucleotides in its place: hence c.2013+225_3147del4890insCC (Figure S2D). To confirm this result, a genomic PCR assay was set up using primers flanking the deletion. The resulting wild-type amplicon (6821bp) failed to amplify under the PCR conditions we used but the deleted allele, resulted in a 1931bp amplicon from the patient‘s and the mother’s DNA, but not the father’s DNA (Figure S3E). Direct sequencing of the mutant amplicon confirms the result observed in the cDNA (Figure S3C). The sequencing result appears homozygous since the wild type allele does not amplify.
Plasmids
The WT SLX4 cDNA was a kind gift from the Harper lab. Mutant alleles were amplified and recombined into pDONR22325. pDONR223 derivatives were recombined into pHAGE vectors or pDEST15 using LR clonase (Invitrogen). HPV16 E6E7 genes (gift from Howley lab) were subcloned into pMSCVneo (Clontech) and used to transform the primary cells. pWZLhTERT26 was used to immortalize RA3083 and BJ cell lines.
Antibodies
The following antibodies were used: FANCD2 (Novus NB100-182), GFP (Roche 11814460001), HA (Covance MMS-101R), SLX4 (Bethyl A302-269A and A302-270A) ERCC4/XPF (Neomarkers Ab-1), ERCC1 (Neomarkers Ab-2) Mus81 (Abcam, MTA30 2G10/3), Ubiquitin (Millipore, MAB1510) and an antibody raised against aa 251-402 of SLX4 (Bethyl).
Mutagenesis
Mutagenesis was performed using multisite mutagenesis kit (Agilent) using primers shown in Supplementary Table 2.
RNAi
siRNA transfections were performed using LipofectamineRNAiMAX as suggested by the manufacturer with the final siRNA concentration of 50 nM. siRNAs (Invitrogen) are shown in Supplementary Table 3.
RT-qPCR
Superscript III reverse transcriptase followed by Platinum cybergreen super mix (Invitrogen) were used according to the manufacturer’s instructions. Actin was used as control.
Ubiquitin binding assay
5 µg purified GST SLX4_aa251-402, GST SLX4_aa251-402 ZNF1andZNF2 mutants or GST alone as a control were added to 7.5 µl GST-beads and 1 µg mono-Ub, Ub-K48 or Ub-K63 (Boston Biochem) in 100 µl binding buffer (50 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Triton, 2 mM NEM, 200 µg/ml BSA) and incubated for 3 hours at 4°C. After washing 5 times with binding buffer, the samples were boiled and loaded on a Bis-Tris gel (Invitrogen) and immunoblotted with anti-ubiquitin antibody (Millipore, MAB1510).
Cell cycle studies
For cell cycle analysis, cells were left untreated or were treated with 100 nM MMC and were grown for 48 hours. Collected cells were resuspended in 300 µl PBS. While vortexing, 700 µl of ice cold 100% (v/v) ethanol were added drop-wise and the suspension was stored at −20°C at least overnight. 30 min before FACS, cells were spun down, resuspended in propidium iodine (PI) mix (1 ml PBS, 10 µl RNase [of stock solution of 20 mg/ml], 10 µl PI [of stock solution of 1 mg/ml]), and analyzed using FACSCalibur (Becton Dickinson). Cell cycle analysis was performed using the FlowJo software (Tree Star, Inc).
Breakage analysis
Cells were treated with 0.1 µg DEB per mL of media for 72 hours, arrested with colcemid (0.17 ug per mL of media) for 20 minutes (LCL) or 2 hours for fibroblasts, harvested, incubated for 10 min at 37°C in 0.075 M KCl, and fixed in the freshly prepared methanol:glacial acidic acid (3:1 vol/vol). Cells were stored at 4°C and when needed dropped onto wet slides and air-dried at 40°C for 60 minutes before staining with Karyomax Giemsa (Invitrogen) Gurr Buffer for 3 minutes. After rinsing with fresh Gurr Buffer followed by distilled water, the slides were fully dried 40°C for 60 minutes and scanned using the Metasystems Metafer application.
Immunoprecipitations
For immunoprecipitations, cells were lysed in MCLB (50 mM Tris, 150 mM NaCl and 0.5% NP-40) supplemented with protease Inhibitors (Roche), and phosphatase inhibitors (Calbiochem). 1 or 2 mg protein extract was incubated with 5 µg of the indicated antibody and 10 µl of Protein A/G PLUS-Agarose (Santa Cruz). Following five washes in lysis buffer, the immunoprecipitates were eluted in tris-Glycine SDS sample buffer and size-fractionated on Novex 3–8% Tris-Acetate gel (Invitrogen).
Mitomycin C sensitivity assay
Cells were plated in a 6-well plate in triplicate at a density of 2.5×104 cells per well. 24 hours later, MMC was added at final concentrations from 0 to 100 nM. After 8 days in culture, cell numbers were determined using a Z2 Coulter Counter (Beckman Coulter). The cell number after MMC treatment was normalized to the cell number in the untreated sample to arrive at the % survival.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the patients and their families for their participation in this study. We thank the Harper lab for reagents, Emily Foley for advice, and Johan de Winter for communicating unpublished results. H.H. is supported by the Deutsche Forschungsgemeinschaft SPP1230 and the BMBF networks for bone marrow failure syndrome (bmfs) and FoneFA. A.S. is supported by the Burroughs Wellcome Fund Career Award for Medical Scientists and is a Rita Allen Foundation and an Irma T. Hirschl Scholar.
Footnotes
AUTHOR CONTRIBUTIONS
The study was designed by A.S., Y.K., and F.P.L. Patient recruitment and sample collection was done by A.D.A., F.P.L., and A.S. Characterization with respect to Fanconi anemia subgroups was performed by A.S., F.P.L., H.H., and A.D.A. Mutation analysis and functional studies were performed by A.S., Y.K., F.P.L., and R.D. The manuscript was written by A.S. with help from other authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auerbach AD, Wolman SR. Susceptibility of Fanconi's anaemia fibroblasts to chromosome damage by carcinogens. Nature. 1976;261:494–496. doi: 10.1038/261494a0. [DOI] [PubMed] [Google Scholar]
- 3.Moldovan GL, D'Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knipscheer P, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fekairi S, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munoz IM, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 7.Svendsen JM, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Higuera I, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 10.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kratz K, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 12.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 13.MacKay C, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smogorzewska A, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaz F, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 16.Kutler DI, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101:1249–1256. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst) 2009;8:544–556. doi: 10.1016/j.dnarep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Sobhian B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howlett NG, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 20.Xia B, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 21.Meindl A, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42:410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 22.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seal S, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 24.Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 25.Lamesch P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.