Abstract
Tumors have been a highlight in the research of nanomedicine for decades. Despite all the efforts in the decoration of the nano systems, tumor specific targeting is still an issue due to the heterogeneous nature of tumors. Hypoxia is frequently observed in solid tumors. The consequent acidification of tumor extracellular matrices may bring new insight to tumor targeting. In this review, we present the polymeric nano systems that target tumor extracellular pH (pHe).
Keywords: Tumor, extracellular pH, Targeting, nanoparticles, drug delivery, pH sensitive polymers
As a self-derived disease, the consistent differences between normal tissue and tumor are often inadequate to facilitate the development of effective therapy. No significant change in mortal rate among patients with advanced cancer are observed over more than half a century [1, 2]. Among all the thriving new technology in cancer treatment, chemotherapy remains the most widely used method. However, toxic agents that can kill cancer cells can also damage normal cells. Hence there are numerous side/adverse effects, but limited treatment outcomes. Further research revealed a difference of functional vasculature between tumor and normal tissue. Nano system formed from various polymeric carriers brought new promise to treatments. Studies of anti-cancer nanoparticles thrive in the area of enhanced permeability and retention (EPR) effect, and ligand/receptor facilitated internalization [3, 4]. However, the heterogeneity among cancer cell populations as well as the heterogeneous up-regulation/expression of receptors/antigens on cancer cell membranes limits the clinical application of nanoparticles decorated with one kind of ligands [5, 6].
The functional vasculature in a tumor area is often mal-developed and insufficient to provide enough nutrition to fast dividing cells. The resulting lack of oxygen and nutrients triggers an alteration of metabolism in tumor cells as an adaption. The anaerobic condition leads to a production of lactic acid, resulting in an acidic pH in many solid tumors. Although acidic environment causes trouble in drug permeability and facilitates tumor invasion in some cases, it also brings opportunity for anti-cancer nano systems. In this review, we present the polymeric nano systems that target tumor pHe.
1. Tumor Vasculature and Angiogenesis
As fast growing masses, tumors require an extra supply of oxygen and other nutrition. This requirement of supply triggers the formation of new vasculature. In a growing tumor, the origin of vessels includes the original host vessels that run through the tumor tissue, and the neovasculature formed as a result of tumor angiogenesis factors [7-13]. The preexisting host vessels per unit tumor mass do not increase over time, but the shape of venules is often deformed, elongated, and often dilated [7, 9]. As tumors grow, some original vessels are crushed, while remaining vessels seem to be able to adopt the change and resist the destruction brought about by tumor growth. However, tumor arterioles often lack spontaneous vasomotion, which is typical in normal vessels [7-11].
Although the vessels vary in different tumor types, sometimes even within one tumor mass, the new vessels formed in accelerated growth exhibit abnormalities both in structure and in function. For example, structurally, the vessel wall is incomplete, lack pericytes and biological receptors; the vessels are often elongated and exhibit an arteriovenous shunt; the vascular density is chaotic and the intercapillary space is expanded. Functionally, the vessels are more fragile; the speed and direction of blood flow is unstable; the vascular permeability is increased, which may result in hemoconcentration and high interstitial fluid pressure [7, 14]. When increased vascular permeability is combined with the often malfunctioned lymphatic drainage, it results in the EPR effect, which is used in most antitumor nano systems [15-17].
Therefore blocking neovascularization and starving tumors to death was considered beneficial to cancer patients. The vascular endothelial growth factor (VEGF) and its receptor VEGFR2 are among the most investigated. Clinically, bevacizumab (monoclonal anti-VEGF antibody), sunitinib and sorafenib (second-generation multitargeted receptor tyrosine kinase inhibitors) have given numerous patients prolonged lifespans [18-24]. However, the increased span is often limited to months. Recent research revealed what seems to be contradictory – treatment with VEGF inhibitors may trigger metastasis [18, 25, 26], which is the primary cause of mortality in cancer patients [27]. To further understand the situation, we need to investigate the hypoxia and corresponding metabolism in tumors.
2. Hypoxia and Metabolism under Hypoxia
A growing tumor mass needs to meet the bioenergetic and biosynthetic demands, despite the fluctuating nutrition and oxygen supply. In blood, healthy tissue, and typical hypoxic solid tumors, the oxygen concentration ranges from 10-12.5%, 3-6% and 1-2% respectively [28, 29]. Hypoxia can trigger the activation of different genes, which can in turn change the metabolism in the cells [30]. For example, the hypoxia-inducible factor 1 (HIF-1) is up-regulated in most malignant tumors and over 60% of metastases [29, 31]. It activates the glycolytic or tumor metabolic phenotype [32, 33], which results in the acidification of tumor extracellular environments. HIF consists of two subunits – α- and β-subunits. Three α-subunit isoforms are discovered so far, with HIF-1α being the most studied [34-36]. For all three isoforms, the post-translational regulation of stability is similar in mechanism. Under normoxic conditions (normal pO2 in tissues), proline in the oxygen-dependent degradation domain (ODDD) and asparagine in the C-terminal activation domain (C-TAD) can be hydroxylated by the oxygen sensor prolyl hydroxylase domain (PHD) dioxygenases and factor inhibiting HIF (FIH) respectively. However, under hypoxia, PHD proteins and FIH are inactive and the stability of HIF-α is preserved [36-39]. HIF-α then translocates into the nucleus, heterodimerizes with HIF-β and binds with hypoxia-response elements (HRE) in the promoter or enhancer regions of DNA. Thus a series of downstream changes are triggered, and the metabolic balance is altered [36, 40-42].
The expression of glycolytic enzymes and glucose transporters (GLUT1 and GLUT3) are up-regulated by HIF [42]. As a result, glucose molecules are more efficiently caught and converted to pyruvate, which helps the cancer cells to survive and proliferate under a limited oxygen and nutrient supply. The resulting pyruvate, instead of going through a tricarboxylic acid (TCA) cycle, is converted into lactic acid directly. However, under aerobic metabolism, one molecule of glucose can produce 38 adenosine triphosphate (ATP) molecules maximum; whereas in anaerobic metabolism, one molecule of glucose can produce only 2 ATP molecules [33, 36]. The phenomenon of preference for using the truncated pathway – Warburg effect – was observed on cancer cells even in the presence of oxygen [33, 36], but it is still not totally clear why cancer cells use this pathway which is 19-fold less efficient in ATP production. One explanation is believed to have to do with the decrease of the downstream products that prevent/prohibit and the increase of the ones that facilitate tumorigenesis, and harnessing glucose metabolism in cancer cells may be of therapeutic benefits [33, 36, 43-44].
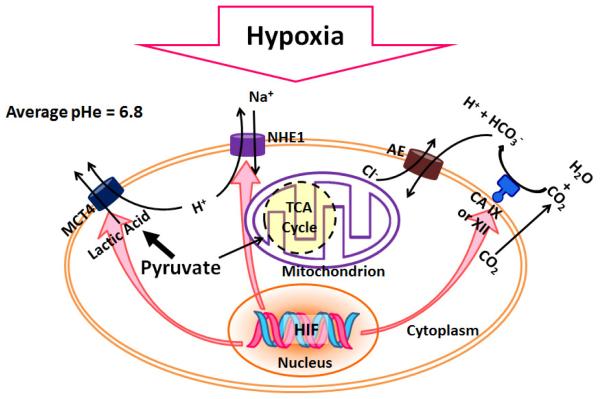
Aerobic glycolysis is one primary metabolic change often observed in cancer cells, where above 90% of pyruvate (which is produced from glucose) is converted to lactate, which is eventually transported outside the cell membrane [33, 45]. H+ ions are formed during glycolysis, ATP hydrolysis, and glutaminolysis and also transported out the cell. Large amount of H+ ions are produced as a result of high rate of glycolysis and lactic acid production. They would normally be washed out by blood and the interstitial pH remains unchanged [46]. However, the blood flow rate in tumors is often decreased as the result of abnormalities in vasculature. For example, in Grade I astrocytoma’s blood flow can be as low as 0.03 ml/g/min, compared with 0.25 – 0.78 ml/g/min in normal gray matter. Therefore the over-produced H+ ions are accumulated in the tumor interstice [47]. The low tumor pHe is then a result of over-production of lactic acid and carbonic acid. Some isoforms of H+/lactate monocarboylate transporters (MCT) are up-regulated under hypoxia and can pump lactate outside the cells [48]. HIF also induces ecoenzyme carbonic anhydrase (CA) IX or XII, which present in the cell membrane and convert carbon dioxide into carbonic acid when carbon dioxide molecules diffuse out from the membrane. HCO3− is subsequently taken up by the cells and H+ is left to add on the acidity of tumor extracellular fluid [49]. The expression and activity of Na+/H+ exchanger 1 (NHE1) is also enhanced under hypoxia. NHE1 can substitute one molecule of extracellular Na+ with one molecule of intracellular H+. NHEs play an important role in maintaining the intracellular pH of cells [50, 51]. The decreased tumor pHe can facilitate the invasion of tumor, slow down the uptake of basic anticancer drugs (such as doxorubicin (DOX)) [52, 53]. Thus combined with the altered metabolism, the tumor cells usually become more resistant to chemotherapy. However, it also offers an opportunity for anticancer nano-treatment. An illustration of the influence of hypoxia on tumor pHe is shown in Fig. 1.
Fig. 1.
Hypoxia and decreased pHe. Under hypoxia, HIF maintains the pHi by changes including up-regulation of NHE1, MCT4 (which exports H+ and lactate), CA IX or XII which converts CO2 to carbonic acid. The dissociated weak base HCO3- influxes through AE (anion exchangers) and H+ is left outside. The overall results is the decreased pHe.
3. pH Measurement and Estimates of Tumor pHe
pH electrodes are the most frequently used measurement in tumor pH estimates [54]. The measurements by probe tips in μm to mm range mainly reflect the tumor pHe. However, insertion of the probe into the tumor tissue may cause damage to cells and surrounding capillaries [55]. Thus, the resulting estimates may be influenced. Finer probe tips were also designed, but with a loss of sensitivity [56].
31P-nuclear magnetic resonance (NMR) is a non-invasive spectroscopy based on the pH-dependent chemical shift of phosphates, H2PO4- and HPO42- (H2PO4- = HPO42- + H+) [57-61]. However, these inorganic phosphates are mainly distributed intracellularly. Therefore, the measurements largely reflect intracellular pH. By changing phosphates into organic phosphates, the distribution can be altered into the entire water phase or extracellular water phase [62-65]. Dimethyl methylphosphonate (DMMP) and 3-aminopropylphosphonate (3-APP) are non-toxic and chemically inert. DMMP can distribute through the entire water phase, while 3-APP is extracellular. Employment of DMMP and 3-APP in 31P-NMR can measure not only the compartmental volume, but also pHe.
Tumor pHe estimates are generally lower than those of normal tissue (Fig. 2). Volk et al. [66] examined 268 human xenograft samples of 30 tumor cell lines on rnu/rnu rats. Glass electrodes inserted into a 25 gauge bevelled needle were used in the measurement, with a length of the sensing portion of 250 μm. The resulting average xenograft pHe is 6.84, ranging from 6.71 to 7.01, with a variation within single tumor of 0.3 to 0.8 units, in contrast to the physiological pH of arterial blood of 7.4. Studies on human patient reveal a similar trend. A separate study [67] on 67 tumor nodules in 58 patients results in an average pHe of 7.01 (5.66-7.78). Average pHe of examined adenocarcinomas, soft tissue sarcomas, squamous cell carcinomas and malignant melanomas are 6.93 ± 0.08 (5 .66-7.78), 7.01 ± 0.21 (6.25-7.4), 7.16 ± 0. 08 (6.2-7.6), and 7.36 ± 0.1 (6.98-7.77). In contrast, the pHe of normal tissue are estimated to be 7.33 ± 0.03 (7.35 ± 0. 07) [68, 69]. However, both increase and decrease of pHe values with tumor volumes are observed. The confusing results may be due to necrosis in large tumor masses, which increases the environmental pH [67, 70]. (Fig. 2)
Fig. 2.
pHe of different solid tumor xenografts, with an average of 6.84 (Fig. 1 in [66]. Reprinted by permission from Macmillan Publishers Ltd: [Br. J. Cancer] [66], copyright (2011).)
4. Modulation of Tumor pHe
From the descriptions above, we expect that not all the tumor types have a pHe value that is low enough for effective targeting. However, one advantage of tumor pHe targeting is the feasibility of manipulating the pHe values by simple administration of glucose. The manipulation gives pHe targeting an advantage over general ligand targeting in that it is hard to further up-regulate desired cell membrane receptors in vivo for enhanced targeting.
The results of a study on human xenografts on rnu/rnu rats [66] shows that 4 hours after administration of glucose (2.5 times increase of blood glucose), the average xenograft pHe drops to 6.43, ranging from 6.12 to 6.78, while the pH of arterial blood remained 7.4 and the mean pH in normal tissues is 6.97. For the four main histopathological tumor kinds – breast, lung and gastrointestinal carcinomas, and sarcomas, pH response to glucose administration doesn’t show a major difference. Inorganic phosphates and m-iodobenzylguanidine (MIBG) can further decrease pHe under hyperglycemia, while insulin doesn’t have the same effect [71].
Further study was conducted on cancer patients [68]. Glucose is orally administrated to fasting patients. According to the response, patients are sorted into three categories --- normal (transient hyperglycemic), asymptomatic Type II diabetics (persistent hyperglycemic), and hypoglycemic responders. Maximum reduction in pHe was obtained within 90 minutes. Normal glucose responders seemed to have the most benefit in terms of pHe drop with a maximum ΔpH of −0.4. One thing to be noted is that from this set of study, over half (56%) of cancer patients were normal responders. It would be great if the time course of pHe response could be more detailed to make it more predictable and a better guidance for clinical applications.
5. Tumor pHe Targeting Nano-system
There are mainly two types of pH sensitive polymer carriers. One is the polymerization of monomers which have pKa values around tumor pHe, or attachment of small molecules with similar pKa values to an existing polymer. Thus change in pH causes the protonation/deprotonation of the pH sensitive block, decreases the stability of the nano systems and triggers the release of the drug loaded in the systems. Another includes chemical bonds/ligand-receptor binding that are less stable at tumor pHe. pH decrease results in either a decrease in stability and release of loaded drug, or the exposure of targeting ligand.
5.1. Protonation/Deprotonation of Polymer
Most pH sensitive block in this kind of polymeric carrier contains nitrogen/amine, such as polyhistidine and poly(β-amino ester), and is hydrophobic at physiological pH (higher pH). When they are linked with a hydrophilic block (usually PEG (poly(ethylene glycol)), micelle structures are formed with the hydrophobic pH sensitive block in the core, loaded with antitumor drug. Decrease in pH causes the increase of hydrophilicity in the core due to the protonation on nitrogen. The micelle structure is then destabilized and loaded drug released. One exception is polysulfonamide, which is negatively charged/deprotonated at physiological pH and neutralized at tumor pHe. Therefore polysulfonamide is a very promising carrier in shielding polycations, which are more toxic to the organisms.
5.1.1 pH Sensitive Block Changing from Anionic to Neutral When pH Decreases
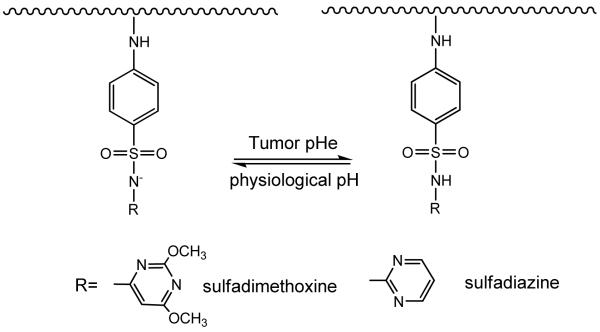
Sulfonamide is an antibacterial drug. The pH sensitivity brought about by the secondary amine (-NH-) linked to the sulfone group can be used in tumor pHe targeting (Structure 1). When polymerized/oligomerized, the resulting oligo-sulfonamides (OSAs) possess different proton buffering capacities [72]. These oligo-sulfonamides can also be conjugated with PEG (OSA-PEG) without the loss of pH sensitivity [73, 74]. For example, oligosulfadimethoxine-PEG possesses a pKa of around 6.8 [75], which is ideal for tumor pHe targeting. Above pH 6.8, such as under physiological pH 7.4, oligosulfadimethoxine-PEG is negatively charged and can form complex with polycations. The complex can protect the polycation from unselective targeting. When pH drops, the polymer is protonated, resulting in the loss of negative charge hence the ionic interaction between the polymer and the polycations. The polycations can thus be released from the complex to execute their activities (Fig. 3).
Structure 1.
Protonation and diprotonation of polysulfonamide
Fig. 3.
Schematic illustration of shielding (under physiological pH) and dishielding (under tumor pHe) of cationic ligand.
One application of the polymer is in gene therapy studies. Condensation of plasmid DNA by polycations, such as polyethyleneimine (PEI), has been used for decades for gene therapy. Main disadvantages, however, are the toxicity and the non-selectivity brought by the positive charges on the polycations. Shielding the complex with OSA-PEG showed promise in reducing the toxicity while preserving the transfection efficiency [73]. The DNA/PEI/OSA-PEG complex size was observed to be ~300 nm at pH 7.4, with a low cytotoxicity and transfection efficiency at pH 7.4 (physiological pH). The high cytotoxicity and transfection efficiency at pH 6.6 indicates the detachment of the OSA-PEG, and the release of DNA/PEI complex from the nanoparticles. Targeting at lower pH (6.6 in this case) suggests the targeting ability to tumor pHe.
It can also facilitate ligand targeting systems. One disadvantage of macromolecules is the slow cell entrance [76]. Ligand conjugated nanoparticles can enhance the entrance by receptor facilitated endocytosis [77, 78], which depends on the increased amount of receptors presented in tumor cell surface. However, the kinds and amount of up-regulated receptors are not consistent among tumor cells [79-82]. One universal solution is to conjugate cationic cell penetrating peptides (CCPs) [83-85], such as trans-activating transcriptional activator (TAT) from Human Immunodeficiency Virus 1 (HIV-1) [86-88], which have broad spectra of cell entrance. The apparent disadvantage is the non-selectivity by cell penetrating peptides. However, the positive charge on CCPs produces an opportunity to shield them by polysulfonamides (OSA-PEG) [74], or biodegradable polysulfonamide with disulfide bond [89]. The mechanism is similar to that of DNA/PEI/OSA-PEG complex (Fig. 3). In short, OSA-PEG can shield TAT molecules electrostatically at pH 7.4, and expose (deshield) TAT below pH 6.8. The zeta potential of the micelle was observed to be close to zero above pH 6.8, and as high as 6.0 mV from pH 6.6 to 6.0. The result is in consistence with the low cell internalization of TAT micelles shielded with OSA-PEG at pH 7.4 and the high cell internalization at pH 6.6, which indicates the deshielding. An optimized polysulfonamide thus is feasible to be developed into an in vivo shielding/deshielding system.
Sulfadimethoxine can also be conjugated to succinylated pullulan acetate (PA), and self-assembled PA/SDM hydrogel nanoparticles are formed in aqueous media. The system showed decreased stability below pH 7. When loaded with anticancer drug, the release of drug was faster at pH 6.8 and 6.4, compared to the release rate at pH 7.4 [90, 91]. The system can be potentially developed to deliver anticancer drug to tumor interstitial space.
5.1.2 pH Sensitive Block Changing from Neutral to Cationic When pH Decreases
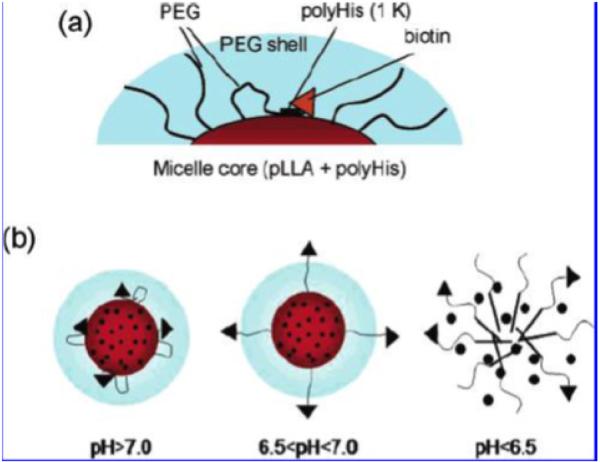
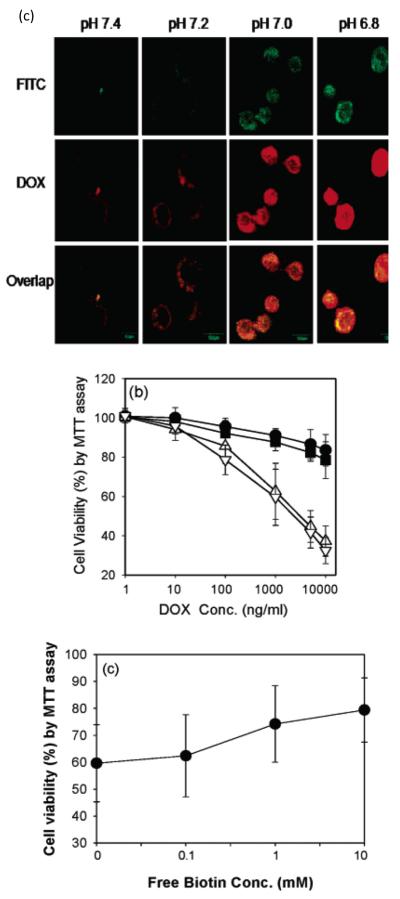
The imidazole ring on histidine has a pKb of around 6.5, which comes from the lone pairs of electrons on the unsaturated nitrogen (Structure 2). The imidazole ring is less charged above pH 7, and more charged below pH 7. After polymerization, the chain is hydrophobic above pH 7, and hydrophilic below pH 7. Therefore, polyHis (polyhistidine) or other histidine modified polymers are often used as endosomal pH targeting drug carrier [92-95]. However, it can also be used in tumor pHe targeting with delicate design, where a low molecular weight polyhistidine chain is sandwiched between the ligand and the micelle. At physiological pH (above 7), the hydrophobicity of the short chain will shrink and drag the attached ligand close to the core of the micelle. The ligand is thus hidden and will not facilitate the internalization of the micelle. At tumor pHe, the hydrophilicity of the short chain will force the attached ligand to move away from the hydrophobic core. The ligand is thus exposed and will facilitate the internalization of the micelle at tumor pHe. Biotin was used in a proof-of-concept study [96]. A mixed micelle system of polyHis-b-PEG and PLLA (poly(L-lactic acid))-b-PEG-b-polyHis (Mw 1000)-biotin was fabricated. The micelle was stable above pH 7.2. When avidin was added to the micelle solution under different pH values, the turbidity increased when pH dropped below pH 7. This was explained as the binding of the exposed biotin with avidin, which is a tetrameric protein that can bind with four biotin molecules (Fig. 4 (a) and (b)). Higher cellular uptake and cytotoxicity was observed at lower pH values, but cell viability is higher when cells were co-treated with higher concentration of free biotins because of the competition between free biotin and biotin-liganded micelles (Fig. 4 (c)). When TAT was used to replace biotin, the internalization of the micelle at pH 7 is 30 times greater than that at pH 7.4 [97]. This indicates the exposure of the cell penetrating peptide and the facilitated cell entrance at tumor pHe. Further in vivo study also proved the effectiveness of the system on drug resistant tumor models [97].
Structure 2.
Protonation and diprotonation of imidazole ring
Fig. 4.
Illustration of polyhistidine micelles (a) Schematic illustration of micelles in physiological pH, where targeting ligands (biotin molecules) are hidden, because of the hydrophobicity of the polyHis (1k) chain that drags the targeting ligands close to the core and prevents them from exposing to the surface. (b) Schemic illustration of micelles approaching tumor pHe (6.5<pH<7), where targeting ligands begin to expose, due to the increase of hydrophilicity of polyHis (1k) chain that pushes the chain and the ligands out. Dimicellization occurs at even lower pH (pH<6.5). (c) In vitro results, where the upper one is ther confocal images of treated MCF-7 cells. DOX is loaded in FITC labeled micelles and the treatments are done at different pH values. The highest internalization of both the micelles and DOX molecules are observed at the lowest experimental pH (pH = 6.8). The middle figure is the cytotoxicity result at different pH values, i.e. pH 7.4 (●), 7.2 (■), 7.0 (△), and 6.8 (▽). Again, higher cytotoxicity is obtained at lower pH values. The lower one shows the competitive interactions of free biotin on the cytotoxicity of the test micelles (DOX content 1000 μg/mL). The cell viability increases as the concentration of free biotin increases. (Figs. 2 and 4 from [96]. Reprinted with permission from [96] E.S. Lee, K. Na and Y.H. Bae, Nano Lett., 5 (2005) 325. Copyright 2011 American Chemical Society.)
In a separate study, poly(lactide-co-glycolide) (PLGA)-b-PEG-b-PLGA copolymer is capped by N-Boc-histidine on each end. The modification didn’t impact the biodegradability and biocompatibility of the polymer. DOX was incorporated in the micelle. The accumulative DOX release at 12 hours at pH 6.2 doubled that at pH 7.4. Cellular uptake of DOX was studied on human breast cancer cell MDA-MB-435. The higher uptake under pH 6.2 was attributed to both DOX released at extracellular pH and internalized micelles [98].
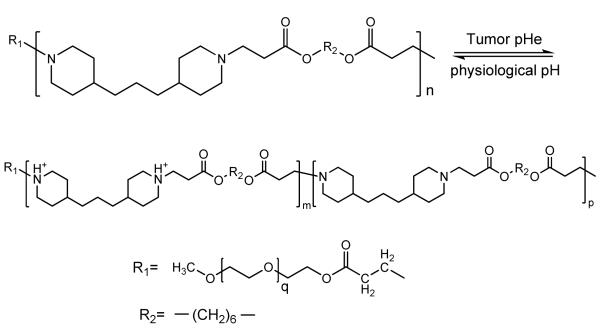
mPEG-b-poly(β-amino ester) (PAE) block copolymer was synthesized and the property was similar to that of PEG-b-polyHis (Structure 3). PAE is considered to be non-cytotoxic and biodegradable [116, 117]. The tertiary diamine moieties stay uncharged at physiological pH and form hydrophobic cores that can carry hydrophobic drug molecules; micelles at physiological pH are also more stable. As pH value goes down, the stability of the cores decreases as a result of gradual protonation of the tertiary diamine moieties. When pH reaches 6.4, a sharp transition of micellization/demicellization occurs [101]. With the destruction of the micelle structure, carried drug molecules are also released. Thus, cellular uptake of released drug, hence the decrease in cell viability, was also greater under pH 6.4 [99]. However, a composition of PAE-b-PEG to PLGA-b-PEG can increase the stability of the system and increase the internalization of intact micelles [100].
Structure 3.
Protonation and diprotonation of poly(β-amino ester)
Anticancer drug camptothecin (CPT) was incorporated in the pH sensitive micelle (pH-PM) by solvent evaporation. The incorporation helped maintain the active lactone form of CPT, and the sharp transition enabled the release of CPT at tumor pHe. The study on human breast cancer MDA-MB231 cell line revealed that at the CPT concentration of 100 μg/mL, the cell viability of drug loaded micelle group at pH 6.2 matched that of free drug treatment group (40% of cell viability), while the cell viability of drug loaded micelle group at pH 7.4 almost doubled that. In vivo accumulation in the tumor area was studied by incorporation of a fluorescence dye tetramethylrhodamine isothiocyanate (TRITC). The fluorescence intensity of TRITC-pH-PM in tumor was seven times higher than that in TRITC loaded pH insensitive micelle control group. This was a further support for efficacy study, where the tumor growth of the group treated by CPT-pH-PM (CPT 10 mg/kg) 32 days after injection was suppressed by 5.8%, compared to the 48.6% in the group treated by equal amount of CPT [101].
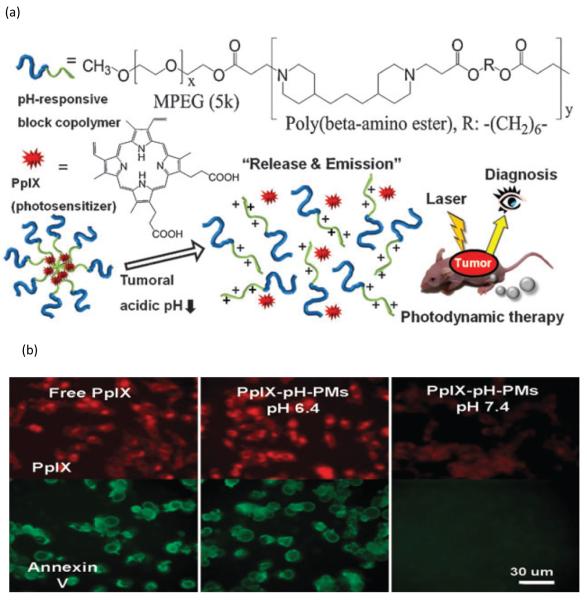
The same polymeric system was also used in tumor diagnosis and photodynamic therapy by encapsulation of protoporphyrin IX (PpIX). The result of in vivo study revealed a strong fluorescence at and a clear delineation around tumors one day post injection, and a 10 fold photon counts in tumor tissue 48 hours post injection for PpIX-pH-PM treatment group. Tumor growth in PpIX-pH-PM treatment group was almost completely suppressed 20 days post injection [102] (Fig. 5). In a separate study, DOX was also loaded in mPEG-PAE block copolymer micelles and the system studied both in vitro and in vivo. On B16F10 melanoma bearing mice, the pH sensitive system could suppress tumor growth significantly even at the dose of 1-2 mg DOX/kg [103].
Fig. 5.
Illustration of micelles containing PAE structure. (a) Scheme of diagnosis and photodynamic therapy. The demicellization at tumor acidic pH releases PpIX and enables both diagnosis and photodynamic therapy. (b) Cellular uptake of free PpIX is not affected by pH value. Difference in cell internalization at different pH values only occurred for micelle group (PpIX-pH-PMs). Higher cellular uptake is observed at low pH (pH 6.8). (Figs. 1 and 3 from [102]. Reproduced by permission of Royal Society of Chemistry.)
The system can also be used for tumor imaging. Both TRITC and black hole quencher (BHC) were encapsulated in the micelle in a quenched state. When the micelle dissociated at lower pH, the fluorescence was recovered. This attribute was used in an in vivo study on MDA-MB-231 tumor bearing mice. One day post injection, strong fluorescence signal was shown in the tumor tissue on mice administered with this nanoflash system, while only minimum fluorescence was observed from mice treated with pH insensitive PEG-b-PLLA particles [104]. However, from the above results, especially the one in [100], we can see one minor disadvantage is the low transition pH of PAE. The transition pH of 6.4 might be too low for some types of tumor. Also, higher cytotoxicity is achieved at pH 6.2 [100], which overlaps with the endosomal pH. Therefore, this system may be better described as a combination of tumor pHe and endosomal pH targeting.
5.1.3 Other Polymeric Carriers
Surfactant-like chitosan-g-poly(N-isopropylacrylamide) (PNIPAAm) was synthesized and nanoparticles were assembled with chitosan as the shell and PNIPAAm as the core. The pH sensitivity was obtained by careful adjustment of the mass ratio of PNIPAAm and chitosan. When the composition of the system was PNIPAAm and chitosan of 4:1 (w/w), the optimal sensitivity was achieved. CPT is then loaded in the nanoparticles and drug release was tested. The release at pH 6.8 was optimal at 37 oC, but decreased rapidly either below pH 6.5 or above pH 6.9. Cytotoxicity at pH 6.8 was also significantly higher than at pH 7.4. The mechanism is still under study, but the release is believed to be a combinational result of both the expansion of chitosan shell and the minification of PNIPAAm core [105]. It is also observed that graft ratio of chitosan to PNIPAAm can affect the swelling ratio of the hydrogels, and swelling ratios at different pH values [121]. PNIPAAm is also widely used in temperature sensitive systems [118, 119], which gives an opportunity of temperature- and pH-dual targeting. However, the toxicity of PNIPAAm [119, 120] limits the use of this system, and is an issue that needs to be solved in the future research. Tertiary amine can also be used to introduce the pH sensitivity. The pKa of the amine in 3-diethylaminopropyl group is 7.0-7.3. When it is linked onto chitosan, the resulting nanogel has an apparent pKa of 6.74. When DOX is loaded in the nanogel, the release at pH 6.8 almost triples that at pH 7.4 [105]. Poly(Nε-(3-diethylamino)propyl isothiocyanato-l-lysine) (poly(DEAP-Lys))-b-PEG-PLLA was synthesized as a pH sensitive carrier. At higher pH, the polymer could form flower-like micelle structure, with poly(DEAP-Lys) and PLLA forming the hydrophobic core and PEG curled in the outside like a petal [106].
5.2 Different Stability of Chemical Bonds at Different pH Values
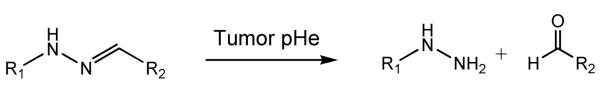
This “Smart” system differs from other systems in that the pH sensitivity is obtained by the cleavage of hydrazone bond, which is less stable when pH drops [107] (Structure 4). A longer PEG chain was linked onto the micelle/liposome via hydrazone bond [108]. Thus under physiological pH when the bond maintains intact, the longer PEG chain could shield the ligand (biotin or TAT peptide) that was chemically linked to the system either directly or via a shorter PEG chain. However, the longer PEG chain could be removed from the system due to the acidic hydrolysis of hydrazone bond under pH 5.0 – 6.0. Thus the ligand could be exposed and facilitate the internalization of the nano system. This transition could happen within 15-30 minutes. In vitro internalization study was conducted by incorporation of rhodamine in to the nanosystem. A significant enhance in fluorescence appeared after a brief incubation under pH 5.0 (20 −30 minutes), which suggested the cellular uptake of the nano system. Similarly, “Smart” pH-sensitive PEGylated TAT-modified liposomes were also able to accumulate in ischemic area and enhance transfection in vivo [109, 110].
Structure 4.
Hydrolysis of hydrazone bond under tumor pHe
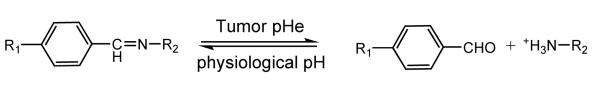
Benzoic imine bond can also be used for similar purpose [111]. In this system, PEGylation to polycation occurred via benzoic imine bond, which can hydrolyze under pH 6.8. This system differs from the above system in that the surface charge of micelle is switchable in response to environmental pH (Structure 5). This is attributed to the property that the imine bond can be reformed in aqueous solution, hence leading to a reshielding of polycation aggregates [112].
Structure 5.
Reversible reaction of benzoic imine under tumor pHe and physiological pH
5.3 Different pH-Dependent Ligand-Binding Strength of Ligand-Binding
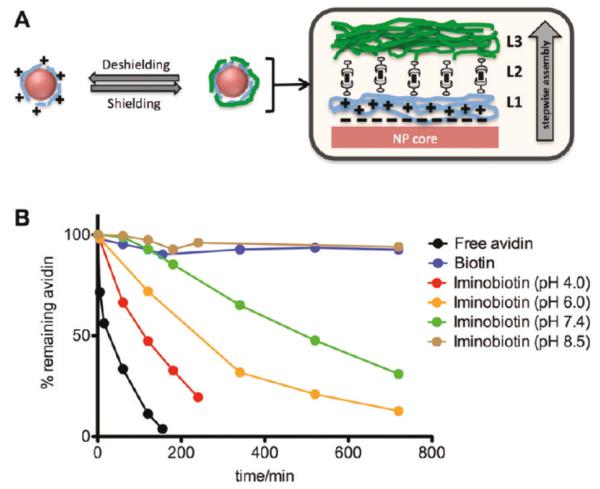
Besides pH sensitivity introduced by the monomer units and cleavage of chemical bonds, the difference in the strength of ligand-binding under different pHs can also be used to target tumor pHe. Iminobiotin-avidin bonds are stable at pH 8-12. When pH drops below 6, the affinity between iminobiotin and avidin decreased due to the protonation of iminobiotin, with a drop in binding constant of 7 magnitudes [113]. Avidin can immobilize iminobiotin-PEI and multilayer assembly formed in basic solution (pH = 12). When biotin is added or pH is dropped below 7, iminobiotin-PEI can be released from the assembly [114]. This attribute was used in the formation of a pH-sheddable layer in Lay-by-Layer (LbL) nanoparticles. Iminobiotin-PLL (poly-l-Lysine) coated nanoparticles and iminobiotin-PEG are complexed by neutravidin – the deglycosylated form of avidin, where iminobiotin – PLL coated nanoparticles are the inner layer, iminobiotin-PEG is the outer layer, and neutravidin is the layer in between. When the LbL nanoparticles encounter lower pH, the outer PEG layer is eroded because of the loss in affinity between nav and iminobiotin. The inner PLL layer is thus exposed and cell internalization facilitated. In vivo tumor targeting was tested with quantum dots (QD) incorporated nanoparticles. QD and nanoparticles constructed by biotin (PLLb) are used as control group for nanoparticles constructed by iminobiotin (PLLib). At 48h post injection, QD/PLLib/nav/PEG group had the highest fluorescence in tumor area, suggesting significantly higher nanoparticle uptake into tumor cells [115].
6. Conclusion
Above all, tumor pHe is a more universal way of tumor targeting. However, systems truly targeting tumor pHe are still limited, compared to tumor receptor targeting or tumor endosomal pH targeting, which might be a result of the narrow pH window that requires a sharp transition of the system. Also, data on the kinetics of change of the systems under reduced pH are limited. Addition of this information will further benefit the prediction of drug release and/or cellular internalization of the nano systems in vivo, and better serves the clinical applications. It would also be great if more information of the disposal/elimination of the pH sensitive polymers could be provided. In general, there is still vast space to investigate in tumor extracellular pH targeting area, the research in which we believe will be as fruitful as, if not more fruitful than, that in other tumor targeting areas.
Fig. 6.
Illustration of LbL system. (a) Schematic illustration of layer-by-layer nanoparticles, where L1 is PLL-iminobiotin, and L2 is neutravidin, and L3 is PEG-biotin. L2 helps to link L1 and L3. (b) Rate of loss of avidin-FITC from PEI-iminobiotin at different pH values. The behavior of the iminobiotin-avidin bond at different pH values can be used to predict that of the iminobiotin-neutravidin bond. The stability of the bond also decreases as pH drops. The bond dissociate fast in acidic environment, where pH is below 6. (Fig. 1 in [115] Reprinted with permission from [115] Z. Poon, D. Chang, X. Zhao and P.T. Hammond, ACS Nano., 5 (2011) 4284. Copyright 2011 American Chemical Society.)
Acknowledgment
This review work is supported by NIH CA 122356.
Reference
- [1].Jemal A, Siegel R, Xu J, Ward E. CA Cancer J. Clin. 2010;60:277. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- [2].Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. CA Cancer J. Clin. 2003;53:5. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- [3].Lee ES, Na K, Bae YH. J. Control. Release. 2003;91:103. doi: 10.1016/s0168-3659(03)00239-6. [DOI] [PubMed] [Google Scholar]
- [4].Lee ES, Na K, Bae YH. J. Control. Release. 2005;103:405. doi: 10.1016/j.jconrel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- [5].Park LS, Waldron PE, Friend D, Sassenfeld HM, Price V, Anderson D, Cosman D, Andrews RG, Bernstein ID, Urdal DL. Blood. 1989;74:56. [PubMed] [Google Scholar]
- [6].Li P, Yu X, Ge K, Melamed J, Roeder RG, Wang Z. Am. J. Pathol. 2002;161:1467. doi: 10.1016/S0002-9440(10)64422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Vaupel P, Kallinowski F, Okunieff P. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- [8].Folkman J. Cancer Res. 1974;34:2109. [PubMed] [Google Scholar]
- [9].Grunt TW, Lametschwandtner A, Staindl O. Microvasc. Res. 1985;29:371. doi: 10.1016/0026-2862(85)90026-3. [DOI] [PubMed] [Google Scholar]
- [10].Shubik P, Cancer J. Res. Clin. Oncol. 1982;103:211. doi: 10.1007/BF00409698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vaupel P, Kallinowski F. Biol. Syst. 1988;18:265. [Google Scholar]
- [12].Folkman J. Nature Medicine. 1995;1:27. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- [13].Ausprunk DH, Folkman J. Microvasc. Res. 1977;14:53. doi: 10.1016/0026-2862(77)90141-8. [DOI] [PubMed] [Google Scholar]
- [14].Reinhold HS, van den Berg-Blok A. CIBA Found. Symp. 1983;100:100. doi: 10.1002/9780470720813.ch7. [DOI] [PubMed] [Google Scholar]
- [15].Matsumura Y, Maeda H. Cancer Res. 1986;46:6387. [PubMed] [Google Scholar]
- [16].Duncan R, Sat YN. Ann. Oncol. 1988;9:39. [Google Scholar]
- [17].Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. J. Control. Release. 2000;65:271. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- [18].Loges S, Mazzone M, Hohensinner P, Carmeliet P. Cancer Cell. 2009;15:167. doi: 10.1016/j.ccr.2009.02.007. [DOI] [PubMed] [Google Scholar]
- [19].Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, Ferrara N, Fyfe G, Rogers B, Ross R, Kabbinavar F. N. Engl. J. Med. 2004;350:2335. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- [20].Miller K, Wang M, Gralow J, Dickler M, Cobleigh M, Perez EA, Shenkier T, Cella D, Davidson NE. N. Engl. J. Med. 2007;357:2666. doi: 10.1056/NEJMoa072113. [DOI] [PubMed] [Google Scholar]
- [21].Demetri GD, van Oosterom AT, Garrett CR, Blackstein ME, Shah MH, Verweij J, McArthur G, Judson IR, Heinrich MC, Morgan JA, Desai J, Fletcher CD, George S, Bello CL, Huang X, Baum CM, Casali PG. Lancet. 2006;368:1329. doi: 10.1016/S0140-6736(06)69446-4. [DOI] [PubMed] [Google Scholar]
- [22].Motzer RJ, Michaelson MD, Redman BG, Hudes GR, Wilding G, Figlin RA, Ginsberg MS, Kim ST, Baum CM, DePrimo SE, Li JZ, Bello CL, Theuer CP, George DJ, Rini BI. J. Clin. Oncol. 2006;24:16. doi: 10.1200/JCO.2005.02.2574. [DOI] [PubMed] [Google Scholar]
- [23].Abou-Alfa GK, Schwartz L, Ricci S, Amadori D, Santoro A, Figer A, De Greve J, Douillard JY, Lathia C, Schwartz B, Taylor I, Moscovici M, Saltz LB. J. Clin. Oncol. 2006;24:4293. doi: 10.1200/JCO.2005.01.3441. [DOI] [PubMed] [Google Scholar]
- [24].Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA, Rolland F, Demkow T, Hutson TE, Gore M, Freeman S, Schwartz B, Shan M, Simantov R, Bukowski RM, TARGET Study Group N. Engl. J. Med. 2007;356:125. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- [25].Pàez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Viñals F, Inoue M, Bergers G, Hanahan D, Casanovas O. Cancer Cell. 2009;15:220. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ebos JM, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS. Cancer Cell. 2009;15:232. doi: 10.1016/j.ccr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gupta GP, Massague J. Cell. 2006;127:679. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- [28].Atkuri KR, Herzenberg LA. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3756. doi: 10.1073/pnas.0409910102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ivanovic Z. J. Cell. Physiol. 2009;219:271. doi: 10.1002/jcp.21690. [DOI] [PubMed] [Google Scholar]
- [30].Jones RG, Thompson CB. Genes Dev. 2009;23:537. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. Am. J. Pathol. 2000;157:411. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Stubbs M, Bashford CL, Griffiths JR. Curr. Mol. Med. 2003;3:49. doi: 10.2174/1566524033361645. [DOI] [PubMed] [Google Scholar]
- [33].Gatenby RA, Gillies RJ. Nat. Rev. Cancer. 2004;4:891. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- [34].Brahimi-Horn MC, Pouysségur J. Int. Rev. Cytol. 2005;242:157. doi: 10.1016/S0074-7696(04)42004-X. [DOI] [PubMed] [Google Scholar]
- [35].Maynard MA, Qi H, Chung J, Lee EH, Kondo Y, Hara S, Conaway RC, Conaway JW, Ohh M. J. Biol. Chem. 2003;278:11032. doi: 10.1074/jbc.M208681200. [DOI] [PubMed] [Google Scholar]
- [36].Brahimi-Horn MC, Pouysségur J. Essays Biochem. 2007;43:165. doi: 10.1042/BSE0430165. [DOI] [PubMed] [Google Scholar]
- [37].Schofield CJ, Ratcliffe PJ. Biochem. Biophys. Res. Commun. 2005;338:617. doi: 10.1016/j.bbrc.2005.08.111. [DOI] [PubMed] [Google Scholar]
- [38].Kaelin WG., Jr. J. Am. Soc. Nephrol. 2003;14:2703. doi: 10.1097/01.asn.0000092803.69761.41. [DOI] [PubMed] [Google Scholar]
- [39].Peet D, Linke S. Novanis. Found. Symp. 2006;272:37. [PubMed] [Google Scholar]
- [40].Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Blood. 2005;105:659. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- [41].Schofìeld CJ, Ratcliffe PJ. Nat. Rev. Mol. Cell Biol. 2004;5:343. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- [42].Semenza GL. Nat. Rev. Cancer. 2003;3:721. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- [43].Fantin VR, St-Pierre J, Leder P. Cancer Cell. 2006;9:425. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- [44].Rajendran JG, Mankoff DA, O’Sullivan F, Peterson LM, Schwartz DL, Conrad EU, Spence AM, Muzi M, Farwell DG, Krohn KA. Clin. Cancer Res. 2004;10:2245. doi: 10.1158/1078-0432.ccr-0688-3. [DOI] [PubMed] [Google Scholar]
- [45].Fantin VR, St-Pierre J, Leder P. Cancer Cell. 2006;9:425. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- [46].Kallinowski F, Schienger KH, Runkel S, Kloes M, Stohrer M, Okunieff P, Vaupel P. Cancer Res. 1989;49:3759. [PubMed] [Google Scholar]
- [47].Beancy RP, Brooks DJ, Leenders KL, Thomas DG, Jones T, Halnan KE. J. Neurol. Ncurosurg. Psychol. 1985;48:310. doi: 10.1136/jnnp.48.4.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ullah MS, Davies AJ, Halestrap AP. J. Biol. Chem. 2006;281:9030. doi: 10.1074/jbc.M511397200. [DOI] [PubMed] [Google Scholar]
- [49].Svastová E, Hulíková A, Rafajová M, Zat’ovicová M, Gibadulinová A, Casini A, Cecchi A, Scozzafava A, Supuran CT, Pastorek J, Pastoreková S. FEBS Lett. 2004;577:439. doi: 10.1016/j.febslet.2004.10.043. [DOI] [PubMed] [Google Scholar]
- [50].Bobulescu IA, Di Sole F, Moe OW. Curr. Opin. Nephrol. Hypertens. 2005;14:485. doi: 10.1097/01.mnh.0000174146.52915.5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Shimoda LA, Fallon M, Pisarcik S, Wang J, Semenza GL. Am. J. Physiol. Lung Cell Mol. Physiol. 2006;291:L941. doi: 10.1152/ajplung.00528.2005. [DOI] [PubMed] [Google Scholar]
- [52].Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B, Gillies RJ. Cancer Res. 2006;66:5216. doi: 10.1158/0008-5472.CAN-05-4193. [DOI] [PubMed] [Google Scholar]
- [53].Adams DJ, Morgan LR. Curr. Med. Chem. 2011;18:1367. doi: 10.2174/092986711795029609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].van den Berg AP, Wike-Hooley JL, van den Berg-Blok AE, van der Zee J, Reinhold HS. Eur. J. Cancer Clin. Oncol. 1982;18:457. doi: 10.1016/0277-5379(82)90114-6. [DOI] [PubMed] [Google Scholar]
- [55].Calderwood SK, Dickson JA. Adv. Radiat. Biol. 1983;10:135. [Google Scholar]
- [56].Bicher HI, Hetzel FW, Sandhu TS, Frinak S, Vaupel P, O’Hara MD, O’Brien T. Radiology. 1980;137:523. doi: 10.1148/radiology.137.2.7433686. [DOI] [PubMed] [Google Scholar]
- [57].Ng TC, Evanochko WT, Hiramoto RN, Ghanta VK, Lilly MB, Lawson AJ, Corbett TH, Durant JR, Glickson JD. J. Magn. Reson. 1982;49:271. [Google Scholar]
- [58].Evanochko WT, Ng TC, Lilly MB, Lawson AJ, Corbett TH, Durant JR, Glickson JD. Proc. Nati. Acad. Sci. USA. 1983;80:334. doi: 10.1073/pnas.80.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Okunieff PG, Koutcher JA, Gerweck L, McFarland E, Hitzig B, Urano M, Brady T, Neuringer L, Suit HD. Int. J. Radiat. Oncol. Biol. Phys. 1986;1986(12):793. doi: 10.1016/0360-3016(86)90038-6. [DOI] [PubMed] [Google Scholar]
- [60].Oberhaensli RD, Hilton-Jones D, Bore PJ, Hands LJ, Rampling RP, Radda GK. Lancet. 1986;2:8. doi: 10.1016/s0140-6736(86)92558-4. [DOI] [PubMed] [Google Scholar]
- [61].Daly PF, Cohen JS. Cancer Res. 1989;49:770. [PubMed] [Google Scholar]
- [62].Barry JA, McGovern KA, Lien YH, Ashmore B, Gillies RJ. Biochemistry. 1993;32:4665. doi: 10.1021/bi00068a026. [DOI] [PubMed] [Google Scholar]
- [63].Clarke K, Anderson RE, Nédélec JF, Foster DO, Ally A. Magn. Reson. Med. 1994;32:181. doi: 10.1002/mrm.1910320206. [DOI] [PubMed] [Google Scholar]
- [64].Gillies RJ, Liu Z, Bhujwalla Z. Am. J. Physiol. 1994;267:C195. doi: 10.1152/ajpcell.1994.267.1.C195. [DOI] [PubMed] [Google Scholar]
- [65].Bhujwalla ZM, McCoy CL, Glicksonl JD, GiIfies RJ, Stubbs M. Br. J. Cancer. 1998;78:606. doi: 10.1038/bjc.1998.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Volk T, Jähde E, Fortmeyer HP, Glüsenkamp KH, Rajewsky MF. Br. J. Cancer. 1993;68:492. doi: 10.1038/bjc.1993.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Engin K, Leeper DB, Cater JR, Thistlethwaite AJ, Tupchong L, McFarlane JD. Int. J. Hyperthermia. 1995;11:211. doi: 10.3109/02656739509022457. [DOI] [PubMed] [Google Scholar]
- [68].Leeper DB, Engin K, Thistlethwaite AJ, Hitchon HD, Dover JD, Li DJ, Tupchong L. Int. J. Radiat. Oncol. Biol. Phys. 1994;28:935. doi: 10.1016/0360-3016(94)90114-7. [DOI] [PubMed] [Google Scholar]
- [69].Engin K, Leeper DB, Thistlethwaite AJ, Tupchong L, McFarlane JD. Int. J. Radiat. Oncol. Biol. Phys. 1994;29:125. doi: 10.1016/0360-3016(94)90234-8. [DOI] [PubMed] [Google Scholar]
- [70].Thistlethwaite AJ, Leeper DB, Moylan DJ, 3rd, Nerlinger RE. Int. J. Radiat. Oncol. Biol. Phys. 1985;11:1647. doi: 10.1016/0360-3016(85)90217-2. [DOI] [PubMed] [Google Scholar]
- [71].Jähde E, Volk T, Atema A, Smets LA, Glüsenkamp KH, Rajewsky MF. Cancer Res. 1992;52:6209. [PubMed] [Google Scholar]
- [72].Kang HC, Bae YH. Adv. Funct. Mater. 2007;17:1263. [Google Scholar]
- [73].Sethuraman VA, Na K, Bae YH. Biomacromolecules. 2006;7:64. doi: 10.1021/bm0503571. [DOI] [PubMed] [Google Scholar]
- [74].Sethuraman VA, Bae YH. J. Control. Release. 2007;118:216. doi: 10.1016/j.jconrel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Han SK, Na K, Bae YH. Colloids Surf. A Physicochem. Eng. Asp. 2003;214:49. [Google Scholar]
- [76].Jiang W, Kim BY, Rutka JT, Chan WC. Nat. Nanotechnol. 2008;3:145. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- [77].Allen TM. Nat. Rev. Cancer. 2002;2:750. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- [78].Cho K, Wang X, Nie S, Chen ZG, Shin DM. Clin. Cancer Res. 2008;14:1310. doi: 10.1158/1078-0432.CCR-07-1441. [DOI] [PubMed] [Google Scholar]
- [79].Schally AV, Nagy A. Eur. J. Endocrinol. 1999;141:1. doi: 10.1530/eje.0.1410001. [DOI] [PubMed] [Google Scholar]
- [80].Lappi DA. Semin. Cancer Biol. 1995;6:279. doi: 10.1006/scbi.1995.0036. [DOI] [PubMed] [Google Scholar]
- [81].Lu Y, Low PS. J. Control. Release. 2003;91:17. doi: 10.1016/s0168-3659(03)00215-3. [DOI] [PubMed] [Google Scholar]
- [82].Klijn JG, Berns PM, Schmitz PI, Foekens JA. Endocr. Rev. 1992;13:3. doi: 10.1210/edrv-13-1-3. [DOI] [PubMed] [Google Scholar]
- [83].Zorko M, Langel U. Adv. Drug Deliv. Rev. 2005;57:529. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- [84].Chugh A, Eudes F, Shim YS. IUBMB Life. 2010;62:183. doi: 10.1002/iub.297. [DOI] [PubMed] [Google Scholar]
- [85].Hällbrink M, Florén A, Elmquist A, Pooga M, Bartfai T, Langel U. Biochim. Biophys. Acta. 2001;1515:101. doi: 10.1016/s0005-2736(01)00398-4. [DOI] [PubMed] [Google Scholar]
- [86].Zhao M, Weissleder R. Med. Res. Rev. 2004;24:1. doi: 10.1002/med.10056. [DOI] [PubMed] [Google Scholar]
- [87].Brooks H, Lebleu B, Vivès E. Adv. Drug Deliv. Rev. 2005;57:559. doi: 10.1016/j.addr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- [88].Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Proc. Natl. Acad. Sci. U S A. 1994;91:664. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Sethuraman VA, Lee MC, Bae YH. Pharm. Res. 2008;25:657. doi: 10.1007/s11095-007-9480-4. [DOI] [PubMed] [Google Scholar]
- [90].Na K, Bae YH. Pharm. Res. 2002;19:681. doi: 10.1023/a:1015370532543. [DOI] [PubMed] [Google Scholar]
- [91].Na K, Lee ES, Bae YH. J. Control. Release. 2003;87:3. doi: 10.1016/s0168-3659(02)00345-0. [DOI] [PubMed] [Google Scholar]
- [92].Lee ES, Shin HJ, Na K, Bae YH. J. Control. Release. 2003;90:363. doi: 10.1016/s0168-3659(03)00205-0. [DOI] [PubMed] [Google Scholar]
- [93].Mohajer G, Lee ES, Bae YH. Pharm. Res. 2007;24:1618. doi: 10.1007/s11095-007-9277-5. [DOI] [PubMed] [Google Scholar]
- [94].Lee ES, Na K, Bae YH. J. Control. Release. 2005;103:405. doi: 10.1016/j.jconrel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- [95].Yin H, Lee ES, Kim D, Lee KH, Oh KT, Bae YH. J. Control. Release. 2008;126:130. doi: 10.1016/j.jconrel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Lee ES, Na K, Bae YH. Nano Lett. 2005;5:325. doi: 10.1021/nl0479987. [DOI] [PubMed] [Google Scholar]
- [97].Lee ES, Gao Z, Kim D, Park K, Kwon IC, Bae YH. J. Control. Release. 2008;129:228. doi: 10.1016/j.jconrel.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Chang G, Li C, Lu W, Ding J. Macromol. Biosci. 2010;10:1248. doi: 10.1002/mabi.201000117. [DOI] [PubMed] [Google Scholar]
- [99].Ko J, Park K, Kim YS, Kim MS, Han JK, Kim K, Park RW, Kim IS, Song HK, Lee DS, Kwon IC. J. Control. Release. 2007;123:109. doi: 10.1016/j.jconrel.2007.07.012. [DOI] [PubMed] [Google Scholar]
- [100].Zhao H, Duong HH, Yung LY. Macromol. Rapid Commun. 2010;31:1163. doi: 10.1002/marc.200900876. [DOI] [PubMed] [Google Scholar]
- [101].Min KH, Kim JH, Bae SM, Shin H, Kim MS, Park S, Lee H, Park RW, Kim IS, Kim K, Kwon IC, Jeong SY, Lee DS. J. Control. Release. 2010;144:259. doi: 10.1016/j.jconrel.2010.02.024. [DOI] [PubMed] [Google Scholar]
- [102].Koo H, Lee H, Lee S, Min KH, Kim MS, Lee DS, Choi Y, Kwon IC, Kim K, Jeong SY. Chem. Commun. (Camb) 2010;46:5668. doi: 10.1039/c0cc01413c. [DOI] [PubMed] [Google Scholar]
- [103].Ko J, Park K, Kim YS, Kim MS, Han JK, Kim K, Park RW, Kim IS, Song HK, Lee DS, Kwon IC. J. Control. Release. 2007;123:109. doi: 10.1016/j.jconrel.2007.07.012. [DOI] [PubMed] [Google Scholar]
- [104].Ko JY, Park S, Lee H, Koo H, Kim MS, Choi K, Kwon IC, Jeong SY, Kim K, Lee DS. Small. 2010;6:2539. doi: 10.1002/smll.201001252. [DOI] [PubMed] [Google Scholar]
- [105].Fan L, Wu H, Zhang H, Li F, Yang T, Gu Ch., Yang Q. Carbohydr. Polym. 2008;73:390. [Google Scholar]
- [106].Oh KT, Oh YT, Oh NM, Kim K, Lee DH, Lee ES. Int. J. Pharm. 2009;375:163. doi: 10.1016/j.ijpharm.2009.04.005. [DOI] [PubMed] [Google Scholar]
- [107].Zhou L, Yu L, Ding M, Li J, Tan H, Wang Z, Fu Q. Macromolecules. 2011;44:857. [Google Scholar]
- [108].Sawant RM, Hurley JP, Salmaso S, Kale A, Tolcheva E, Levchenko TS, Torchilin VP. Bioconjug. Chem. 2006;17:943. doi: 10.1021/bc060080h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Kale AA, Torchilin VP. “Smart” drug carriers: PEGylated TATp-modified pH-sensitive liposomes. J Liposome Res. 2007;17(3-4):197–203. doi: 10.1080/08982100701525035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kale AA, Torchilin VP. J. Drug Target. 2007;15:538. doi: 10.1080/10611860701498203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Wang C, Wang G, Wang Z, Zhang X. Chemistry. 2011;17:3322. doi: 10.1002/chem.201003502. [DOI] [PubMed] [Google Scholar]
- [112].Gu J, Cheng WP, Liu J, Lo SY, Smith D, Qu X, Yang Z. Biomacromolecules. 2008;9:255. doi: 10.1021/bm701084w. [DOI] [PubMed] [Google Scholar]
- [113].Hofmann K, Titus G, Montibeller JA, Finn FM. Biochemistry. 1982;21:978. doi: 10.1021/bi00534a024. [DOI] [PubMed] [Google Scholar]
- [114].Inoue H, Sato K, Anzai J. Biomacromolecules. 2005;6:27. doi: 10.1021/bm0495856. [DOI] [PubMed] [Google Scholar]
- [115].Poon Z, Chang D, Zhao X, Hammond PT. ACS Nano. 2011;5:4284. doi: 10.1021/nn200876f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Lynn DM, Langer R. J. Am. Chem. Soc. 2000;122:10761. [Google Scholar]
- [117].Nguyen MK, Lee DS. Macromol. Biosci. 2010;10:563. doi: 10.1002/mabi.200900402. [DOI] [PubMed] [Google Scholar]
- [118].Ruel-Gariépy E, Leroux JC. Eur. J. Pharm. Biopharm. 2004;58:409. doi: 10.1016/j.ejpb.2004.03.019. [DOI] [PubMed] [Google Scholar]
- [119].Bysell H, Månsson R, Hansson P, Malmsten M. Adv. Drug Deliv. Rev. 2011 doi: 10.1016/j.addr.2011.08.005. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- [120].Vihola H, Laukkanen A, Valtola L, Tenhu H, Hirvonen J. Biomaterials. 2005;26:3055. doi: 10.1016/j.biomaterials.2004.09.008. [DOI] [PubMed] [Google Scholar]
- [121].Alvarez-Lorenzo C, Concheiro A, Dubovik AS, Grinberg NV, Burova TV, Grinberg VY. J. Control. Release. 2005;102:629. doi: 10.1016/j.jconrel.2004.10.021. [DOI] [PubMed] [Google Scholar]