Abstract
The winter-annual habit of Arabidopsis thaliana requires active alleles of FLOWERING LOCUS C (FLC), which encodes a potent flowering repressor, and FRIGIDA (FRI), an activator of FLC. FLC activation by FRI is accompanied by an increase in specific histone modifications, such as tri-methylation of histone H3 at lysine 4 (H3K4me3), and requires three H3K4 methyltransferases, the Drosophila Trithorax-class ARABIDOPSIS TRITHORAX1 (ATX1) and ATX2, and yeast Set1-class ATX-RELATED7/SET DOMAIN GROUP25 (ATXR7/SDG25). However, lesions in all of these genes failed to suppress the enhanced FLC expression caused by FRI completely, suggesting that another H3K4 methyltransferase may participate in the FLC activation. Here, we show that ATXR3/SDG2, which is a member of a novel class of H3K4 methyltransferases, also contributes to FLC activation. An ATXR3 lesion suppressed the enhanced FLC expression and delayed flowering caused by an active allele of FRI in non-vernalized plants. The decrease in FLC expression in atxr3 mutants was accompanied by reduced H3K4me3 levels at FLC chromatin. We also found that the rapid flowering of atxr3 was epistatic to that of atxr7, suggesting that ATXR3 functions in FLC activation in sequence with ATXR7. Our results indicate that the novel-class H3K4 methyltransferase, ATXR3, is a transcriptional activator that plays a role in the FLC activation and establishing the winter-annual habit. In addition, ATXR3 also contributes to the activation of other FLC clade members, such as FLOWERING LOCUS M/MADS AFFECTING FLOWERING1 (FLM/MAF1) and MAF5, at least partially explaining the ATXR3 function in delayed flowering caused by non-inductive photoperiods.
Keywords: ARABIDOPSIS TRITHORAX, Flowering, FLOWERING LOCUS C, Histone methylation, Winter-annual Arabidopsis
Introduction
Organisms are adapted to a wide range of environmental conditions by variation in genotype and phenotype. A well-studied example of such natural variation is the existence of both summer-annual and winter-annual habits in Arabidopsis thaliana. Summer-annual accessions flower rapidly and produce their offspring (i.e. seeds) in the same growing season in which they germinate. The summer-annual habit is likely to be advantageous in locations with a favorable growing environment that is sufficiently long and free of competition to complete the life cycle. Winter-annual accessions become established in the autumn season, overwinter, and then flower in the following spring. Winter-annual accessions typically have a strong requirement to undergo vernalization to prevent flowering in the autumn season in which they become established, thus ensuring spring flowering. Vernalization is the acquisition of competence to flower that results from exposure to the prolonged cold of winter (Chouard 1960). The winter-annual habit is advantageous in locations in which the autumn and early spring seasons provide the most favorable conditions for growth.
In Arabidopsis, the vernalization requirement/winter-annual habit is typically established by active alleles of FRIGIDA (FRI) and FLOWERING LOCUS C (FLC) (Napp-Zinn 1979, Koornneef et al. 1994, Lee et al. 1994). FLC encodes a potent flowering repressor, and FRI is required for FLC to be transcribed to a level that effectively suppresses flowering (Michaels and Amasino 1999, Sheldon et al. 1999, Johanson et al. 2000).
Most commonly used ‘lab strains’ of Arabidopsis lack active FRI and/or FLC alleles, and exhibit rapid-flowering behavior in inductive long days. For instance, the Columbia (Col) wild type possesses a null allele of fri (Johanson et al. 2000) and, when FRI is introgressed into Col, it increases FLC transcription and causes a delay of flowering in non-vernalized plants (Lee and Amasino 1995). FLC activation due to the presence of FRI is accompanied by and requires histone modifications, such as tri-methylation of histone H3 at lysine 4 (H3K4me3) (Kim et al. 2005, Kim and Michaels 2006, Martin-Trillo et al. 2006, Choi et al. 2011, Jiang et al. 2011).
In eukaryotic cells, post-translational covalent modifications of core histones are involved in establishing the level of transcriptional activity of genes (reviewed in Li et al. 2007, Liu et al. 2010). A number of the histone modifications, including H3K4me2/me3, are associated with transcriptional activation, and there are also a range of histone modifications, such as H3K27me3, which are associated with transcriptional repression. Histone methylations at specific lysine residues are catalyzed by SET domain-containing proteins (Rea et al. 2000). The Arabidopsis genome encodes at least 47 proteins with a SET domain (Baumbusch et al. 2001, Springer et al. 2003, Ng et al. 2007). Based on the similarity to known H3K4-specific methyltransferases such as a Drosophila Trithorax (TRX) and yeast (Saccharomyces cerevisiae) Set1, five ARABIDOPSIS TRITHORAX (ATX1–ATX5) and two ATX-RELATED (ATXR3/SDG2 and ATXR7/SDG25) proteins have been proposed as H3K4-specific methyltransferases (Ng et al. 2007). Moreover, based on the similarities in amino acid sequence of the SET domains and the domain structures of the proteins, ATX1–ATX5 are likely to be TRX-class H3K4 methyltransferases, and ATXR7 appears to be the only ortholog of yeast Set1 (Alvarez-Venegas and Avramova 2001, Alvarez-Venegas et al. 2003, Avramova 2009). ATXR3 does not appear to belong to either the canonical TRX- or Set1-class methyltransferases, although the domain structure is similar to that of ATXR7 (Springer et al. 2003).
Among the putative Arabidopsis H3K4 methyltransferases, ATX1, ATX2 and ATXR7 have been shown to be involved in FLC activation and histone methylation (Pien et al. 2008, Saleh et al. 2008a, Berr et al. 2009, Jiang et al. 2009, Tamada et al. 2009). Mutations in these ATX genes partially suppress the delayed flowering caused by FRI (Pien et al. 2008, Tamada et al. 2009). atx1 atxr7 double mutants in particular exhibit decreased FLC expression and reduced levels of H3K4 mono-, di- and tri-methylation in the presence of FRI, indicating that these ATXs are required for FLC activation, presumably by catalyzing H3K4 methylation (Tamada et al. 2009). However, we found that atx1 atx2 atxr7 does not completely suppress FRI-mediated increased FLC expression; thus, it is possible that other H3K4 methyltransferase(s) are required for FLC activation.
It was recently revealed that ATXR3/SDG2 is a major H3K4 tri-methyltransferase in Arabidopsis and is involved in many aspects of its development (Berr et al. 2010, Guo et al. 2010). For example, ATXR3 functions in the activation of FLC and flowering repression in the Col background (Guo et al. 2010). However, in the Col background, FLC expression is low due to lack of FRI, and FLC has a minor effect in flowering repression. It is thus of interest to analyze the ATXR3 function in FLC activation caused by an active allele of FRI. Furthermore, FLC activation cannot completely explain the role of ATXR3 function in flowering repression in non-inductive short days; thus, there must be another flowering repressor gene(s) that is activated by ATXR3.
Here, we show that a lesion in ATXR3 suppresses the delayed flowering as well as the increased FLC expression caused by FRI. Levels of H3K4me3 were reduced around the transcription start site of FLC in the FRI atxr3 mutant. In addition, rapid flowering of atxr3 is epistatic to that of atxr7. These results indicate that ATXR3 influences transcriptional activation of FLC by catalyzing H3K4 tri-methylation of FLC chromatin in sequence with ATXR7 to establish the winter-annual habit of Arabidopsis. Loss of ATXR3 also results in reduced expression levels of other FLC clade members including FLOWERING LOCUS M/MADS AFFECTING FLOWERING1 (FLM/MAF1) and MAF5, which at least partially accounts for the rapid flowering of atxr3 in short days.
Results
A lesion in ATXR3 results in more rapid flowering than the wild type only in short days in the absence of FRI
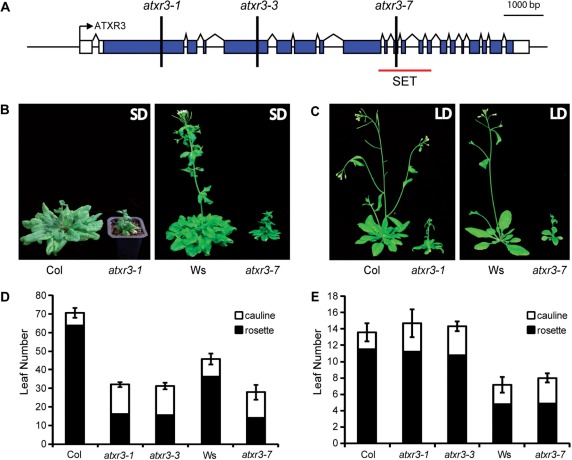
To investigate the role(s) of ATXR3/SDG2 in Arabidopsis flowering, we characterized the phenotypes of three homozygous insertion mutants: atxr3-1/sdg2-1 and atxr3-3/sdg2-3 in the Col background (Tamada et al. 2009, Berr et al. 2010) (which were named sdg2-2 and sdg2-1, respectively, by Guo et al. 2010) and atxr3-7 in the Wassilewskija (Ws) background (Fig. 1A). The three insertions are in the second, fifth and 11th exon, and are predicted to prevent production of a protein with a functional SET domain. Loss of full-length ATXR3 transcripts has been previously confirmed for atxr3-1 and atxr3-3 (Berr et al. 2010, Guo et al. 2010).
Fig. 1.
ATXR3 gene structure and phenotypes of atxr3 mutants. (A) ATXR3 gene structure and T-DNA insertion sites. An arrow, boxes and chevron lines indicate the transcription start site, exons and introns, respectively. Filled boxes represent the open reading frame regions, and open boxes indicate the untranslated regions. atxr3-1 and atxr3-3 are in the Col background, and atxr3-7 is in the Ws background. (B and C) Representative plants of Col, atxr3-1, Ws and atxr3-7 grown in short days (B, left panel, 13 weeks old; right panel, 12 weeks old) or in long days (C, left panel, 39 d old; right panel, 28 d old). atxr3-1 and atxr3-3 mutants have essentially identical phenotypes (data not shown). (D and E) Primary leaf number at flowering of Col, atxr3-1, atxr3-3, Ws and atxr3-7 grown in short days (D) or in long days (E). Closed and open bars indicate rosette and cauline leaves, respectively. The averages of the results from at least nine plants are shown. Error bars indicate the SE.
All three (atxr3-1, atxr3-3 and atxr3-7) alleles result in a similar pleiotropic phenotype that includes small plant size, as recently described (Guo et al. 2010), and increased cauline leaf number compared with the corresponding wild types (Fig. 1B–E). Guo et al. (2010) reported that atxr3/sdg2 flowers more rapidly than the Col wild type in both long and short days. In our growth conditions, all three atxr3 mutants flowered after forming fewer leaves than the corresponding wild types in short days (Fig. 1B, D); however, they exhibited flowering behavior similar to that of the wild types in long days (Fig. 1C, E). The growing temperature in our laboratory (21°C) is similar to that in the report by Guo et al. (2010) (22°C); thus, the discrepancies in the flowering time in long days is likely to be due to other differences in growth conditions such as light intensity.
In contrast to the lower number of leaves relative to the wild type that atxr3 produces in short days, the actual number of days to flowering was greater in the atxr3 mutants than in the wild types in both long and short days because the leaf initiation rate of atxr3 mutants is substantially slower than that of the wild type (Supplementary Fig. S1A–C). Therefore, from a developmental perspective (leaf number), an atxr3 lesion accelerates flowering in short days, but the overall time to flowering is increased due to the substantially slower growth of the mutant. In another study, the leaf initiation rate of atxr3 was reported to be comparable with that in the wild type before flowering (Berr et al. 2010). The difference in leaf initiation rate before flowering may be due to differences in growth conditions: we grew the plants in growth chambers, whereas a glasshouse was used in the study by Berr et al. (2010).
ATXR3 belongs to a unique class of SET-domain proteins
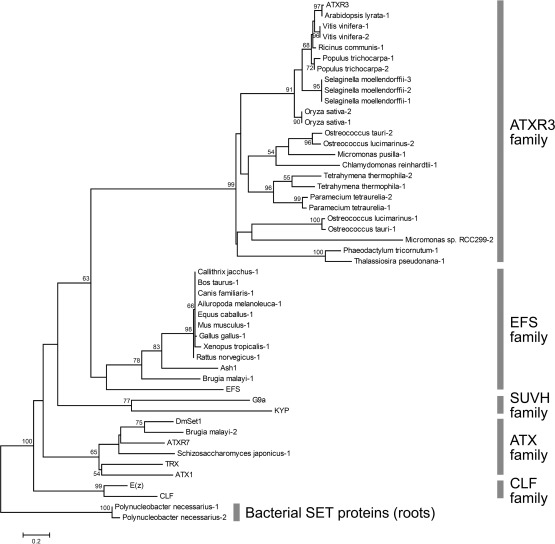
We compared ATXR3 and proteins with a related amino acid sequence from a range of organisms in both bikont (Alveolata, Stramenopiles and Viridiplantae) and unikont (Fungi and Metazoa) groups (the sequences were obtained from BLASTp searches using ATXR3 as a query; Supplementary Table S1) with known SET-domain proteins from Arabidopsis [ATX1, ATXR7, EARLY FLOWERING IN SHORT DAYS (EFS), CURLY LEAF (CLF) and KRYPTONITE (KYP)] and Drosophila {TRX, Set1 (DmSet1), Absent, small or homeotic discs 1 (Ash1), Enhancer of zeste [E(z)] and G9a}. As presented in the phylogenetic tree constructed from the SET domain sequences of these proteins (Fig. 2), ATXR3 and related proteins from bikonts formed a distinct clade, separate from those of unikonts and other known SET-domain proteins. Thus, ATXR3 appears to belong to a novel class of SET-domain proteins for which there was likely to be a founding member in the common ancestor of Alveolata, Stramenopiles and Viridiplantae. Alternatively, one cannot exclude the possibility that ATXR3-like proteins were present in both the bikont and unikont groups, but were lost in the unikonts.
Fig. 2.
Phylogenic relationships among ATXR3 and known histone methyltransferases. An ML tree was drawn using the SET domain sequences of known histone methyltransferases from Arabidopsis and Drosophila, bacterial SET proteins (Polynucleobacter necessarius-1, 2) and proteins that share similar amino acid sequences with ATXR3 (other proteins shown as Organism name-number) with PhyML (Guindon and Gascuel 2003, Guindon et al. 2010). KYP and G9a belong to the SUV39 HOMOLOG (SUVH) H3K9 methyltransferase family, and E(z) and CLF are in the H3K27 methyltransferase family. Numbers next to branches indicate bootstrap statistical support values for branches (1,000 replicates). Bootstrap values >50% are presented. The scale is the estimated branch length indicating the number of substitutions per site.
The atxr3 mutant suppresses FRI-dependent delayed flowering
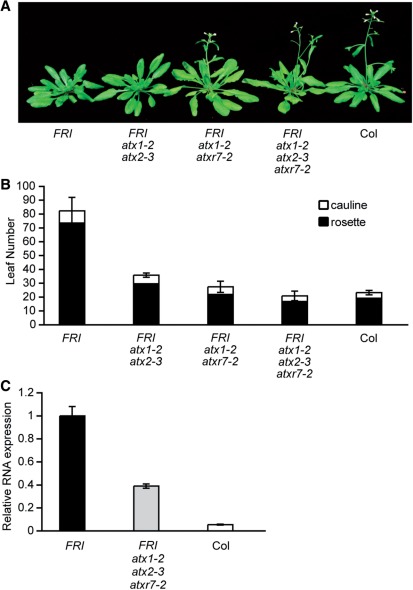
The Arabidopsis winter-annual habit/vernalization requirement is conferred by the presence of active alleles of both FRI and FLC (Napp-Zinn 1979, Koornneef et al. 1994, Lee et al. 1994). FRI-mediated FLC activation is accompanied by an increase of H3K4me3 at FLC chromatin (Kim et al. 2005, Kim and Michaels 2006, Martin-Trillo et al. 2006, Choi et al. 2011, Jiang et al. 2011). To date, among a number of known/putative H3K4-specific methyltransferases, ATX1, ATX2 and ATXR7 are known to be required for FRI-mediated FLC activation (Pien et al. 2008, Tamada et al. 2009). However, the FLC mRNA levels in the FRI atx1 atx2 atxr7 triple mutants are not as low as those in the Col wild type (i.e. a fri mutant) (Fig. 3), indicating that an additional H3K4 methyltransferase(s) could be involved in FLC activation. To evaluate the possible involvement of ATXR3 in FLC activation, an active FRI locus was introgressed into the atxr3 mutant and the flowering time was analyzed. The atxr3 mutation significantly suppressed the FRI-induced delayed flowering (Fig. 4A, B). Thus, ATXR3 is also required for FRI- and FLC-dependent delayed flowering.
Fig. 3.
Phenotypes of the atx1 atx2 atxr7 triple mutant. (A) Representative plants of FRI-Col, FRI atx1-2 atx2-3, FRI atx1-2 atxr7-2, FRI atx1-2 atx2-3 atxr7-2 and Col (7 weeks old) grown in long days. At this stage, half of the FRI atx1-2 atx2-3 plants started to flower, but none of the FRI-Col plants had flowered. (B) Primary leaf number at flowering of FRI-Col, Col and atx multiple mutants grown in long days. Closed and open bars indicate rosette and cauline leaves, respectively. Error bars indicate the SD. (C) Real-time PCR was performed to analyze expression levels of FLC in FRI-Col, FRI atx1-2 atx2-3 atxr7-2 and Col. RNA was isolated from whole 10-day-old seedlings grown in continuous light. The averages of the results from three different biological replicates are shown. Each experiment was normalized to ACTIN7 (ACT7) expression. Error bars indicate the SE.
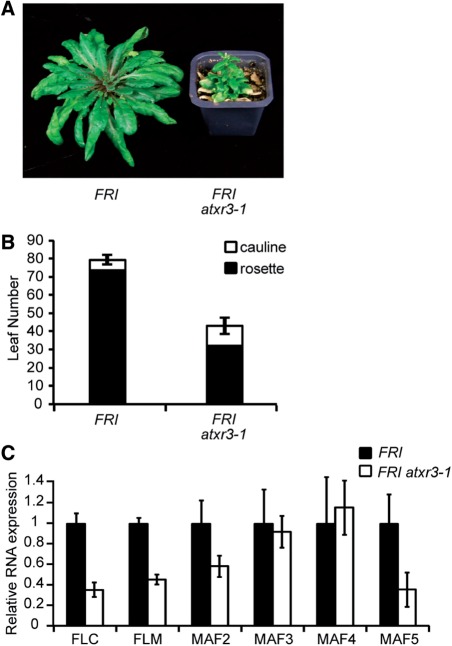
Fig. 4.
Phenotypes of atxr3 in a FRI-Col background. (A) Representative plants of FRI-Col and FRI atxr3-1 (12 weeks old) grown in long days. (B) Primary leaf number at flowering of FRI-Col and FRI atxr3-1 grown in long days. Closed and open bars indicate rosette and cauline leaves, respectively. The averages of the results from at least 12 plants are shown. Error bars indicate the SE. (C) Real-time PCR was performed to analyze the expression of FLC and the FLC-clade genes in FRI-Col (filled bars) and FRI atxr3-1 (open bars). Almost fully expanded rosette leaves that look vital and completely green were sampled from several 6-week-old plants. Plants had not flowered yet at this stage. The averages of the results from four different biological replicates are shown. The data from each experiment were normalized to ACT7 expression. Error bars indicate the SE.
ATXR3 is required for proper expression of FLC and FLC clade genes
To assess whether ATXR3 is involved in setting the level of FLC expression in the presence of FRI, FLC mRNA levels were analyzed in FRI atxr3 and FRI-Col (Col into which FRI has been introgressed; Lee and Amasino 1995). FLC mRNA levels in FRI atxr3 were lower compared with those in the wild type FRI-Col (P < 0.05; Fig. 4C). Thus, the earlier developmental stage when flowering occurs in FRI atxr3 relative to FRI-Col is likely to be due, at least in part, to a reduction of FLC expression.
A reduction of FLC expression alone has only a minor effect in flowering without an active FRI allele (Michaels and Amasino 2001); thus, rapid flowering of atxr3 in short days in the fri (i.e. Col and Ws) backgrounds (Fig. 1; Guo et al. 2010) cannot be explained by only reduced FLC expression. Most of the histone modifiers that play a role in the FLC activation are also involved in the transcriptional activation of other FLC clade members, such as FLM/MAF1 and/or MAF2-5 (Kim et al. 2005, Cao et al. 2008, Xu et al. 2008, Gu et al. 2009, Schmitz et al. 2009, Tamada et al. 2009, Xu et al. 2009). In fact, FRI atxr3-1 exhibited reduced levels of FLM and MAF5 transcripts relative to FRI-Col wild type (P < 0.05; Fig. 4C). However, no significant change was observed for MAF2, MAF3 and MAF4 expression in the mutant (P > 0.05; Fig. 4C). FLM is known to be required for the photoperiod-dependent repression of flowering (Ratcliffe et al. 2001, Ratcliffe et al. 2003, Scortecci et al. 2003). Reduced expression of FLM and MAF5, in addition to FLC, at least partially accounts for the attenuation of the photoperiod-dependent delay of flowering in atxr3.
Loss of ATXR3 affects histone modifications at FLC chromatin
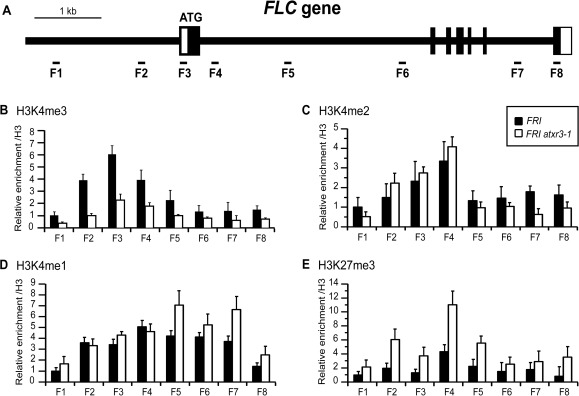
As discussed above, we have established that ATXR3 is required for proper levels of FLC expression in the presence of FRI (Fig. 4). ATXR3 encodes a member of a novel class of SET domain proteins (Fig. 2) which is a major H3K4-specific methyltransferase (Berr et al. 2010, Guo et al. 2010). Although ATXR3 catalyzes all mono-, di- and tri-methylation of H3K4 in vitro, only global reduction of tri-methylation (H3K4me3) was observed in atxr3 in vivo (Berr et al. 2010, Guo et al. 2010). The effects of an atxr3 lesion on global levels of H3K4me1/me2 are different in the reports by Guo et al. (2010) and Berr et al. (2010): Guo et al. observed increased levels of global H3K4me2 in atxr3/sdg2, while Berr et al. obtained slightly decreased H3K4me2 and increased H3K4me1 levels in the mutant. It was of interest for us to evaluate the effect of loss of ATXR3 function on H3K4 methylations at FLC in a FRI background using chromatin immunoprecipitation (ChIP) with anti-H3K4me3, H3K4me2 or H3K4me1 antibodies (Fig. 5A).
Fig. 5.
Histone methylation statuses at FLC in FRI-Col and the FRI atxr3-1 mutant. (A) Schematic model of primer positions at the FLC locus. (B–E) Levels of H3K4me3 (B), H3K4me2 (C), H3K4me1 (D) and H3K27me3 (E) at FLC. The x-axis and y-axis indicate the primer number described in A and relative levels of modifications, respectively. Relatively young rosette leaves were sampled from several 4-week-old plants. Plants had not flowered at this stage. The averages of the results from at least two different biological replicates are shown. The data from each experiment were normalized to either ACT7 (H3K4 methylations) or AGAMOUS (AG) (H3K27me3) and then were further normalized to the total histone H3 ChIP. Error bars indicate the SE.
Levels of an active mark, H3K4me3, were enriched particularly around the transcriptional start site of the active FLC locus in FRI-Col (Fig. 5B; similar results have been reported in He et al. 2004, Kim et al. 2005, Oh et al. 2008, Tamada et al. 2009). However, loss of ATXR3 function resulted in reduced H3K4me3 levels across the entire FLC genomic region (Fig. 5B), and the most significant reduction was around the transcriptional start site relative to FRI-Col wild type (P < 0.05; Fig. 5B: F2-4). The reduced H3K4me3 levels at FLC in FRI atxr3-1 correlate with the reduction of FLC transcription in the mutant (Fig. 4C). In FRI-Col wild type, the pattern of H3K4me2 is similar to that of H3K4me3 (Fig. 5C). However, unlike the case for H3K4me3, the atxr3 mutation did not result in a reduction of H3K4me2 around the transcriptional start site (Fig. 5C: F2-4), although there was a slight reduction of H3K4me2 in the transcribed region of FLC (Fig. 5C: F5-8). Finally, there was no significant reduction of the H3K4me1 level across the FLC region in the atxr3 mutant (Fig. 5D); in fact, there is a slight increase in H3K4me1 in the transcribed region of FLC (Fig. 5D: F5-7). Overall, the effects of an atxr3 lesion on the H3K4 methylation levels at FLC are similar to those on global H3K4 methylation observed by Berr et al. (2010).
Another chromatin modification, H3K27me3, is involved in the transcriptional repression of FLC. H3K27me3 is highly enriched at FLC when its expression is perturbed (see the review in this special issue; Buzas et al. 2012). For example, loss of the ATXR7 function causes reduced FLC mRNA levels and increased H3K27me3 levels at FLC (Tamada et al. 2009). However, another H3K4 methyltransferase, ATX1, is the only known exception; both FLC expression and the H3K27me3 levels at FLC are lower in atx1 than in the wild type (Pien et al. 2008). Therefore, it is of interest to examine whether a novel class H3K4 methyltransferase, ATXR3, functions like ATX1 or ATXR7 for H3K27me3. In FRI atxr3-1, we observed an increased level of H3K27me3 relative to FRI-Col wild type across the entire FLC locus with ChIP (Fig. 5E). This indicates that ATXR3 functions like ATXR7 for H3K27me3, and ATXR3-mediated H3K4 tri-methylation and the concomitant transcription, which is enhanced by H3K4me3, acts to antagonize H3K27 tri-methylation at FLC (Buzas et al. 2011, Schmitges et al. 2011).
In short days atxr3 is epistatic to atxr7 for flowering time whereas atxr7 is epistatic to atxr3 with respect to growth rate
Although the SET domain of ATXR3 is diverged from those of other known H3K4 methyltransferases (Fig. 2), the overall protein structure of ATXR3 is similar to that of ATXR7: both ATXR3 and ATXR7 share GYF domains in the N-terminus (Springer et al. 2003). In addition, like ATXR3, ATXR7 is required for repression of flowering both in short days and in the presence of FRI (Tamada et al. 2009). To investigate the genetic interaction between ATXR3 and ATXR7 in flowering, an atxr3-1 atxr7-1 double mutant was created and analyzed for flowering time in short days relative to the wild type and the single mutants. Consistent with our previous findings, the atxr7-1 single mutant flowered after forming ∼50 leaves in short days (Fig. 6A, B). Interestingly, the atxr3-1 atxr7-1 double mutant flowered after forming ∼30 leaves, which is similar to the leaf number at flowering of the atxr3-1 single mutant (Fig. 6B). Thus, a lesion in ATXR7 does not cause any further developmental acceleration of flowering when combined with a lesion in ATXR3, indicating that the role of ATXR7 is largely dependent on ATXR3 with respect to short day flowering.
Fig. 6.
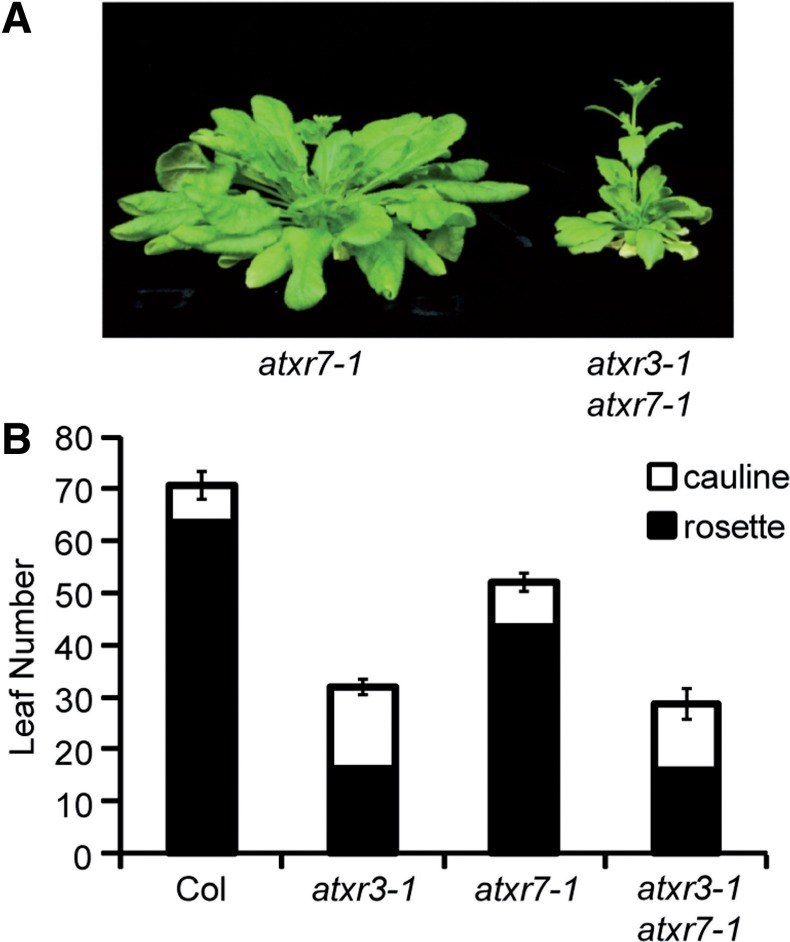
Phenotypes of the atxr3 atxr7 double mutant. (A) Representative atxr7-1 and atxr3-1 atxr7-1 plants (9 weeks old) grown in short days. (B) Primary leaf number at flowering of Col, atxr3-1, atxr7-1 and atxr3-1 atxr7-1 grown in short days. Closed and open bars indicate rosette and cauline leaves, respectively. The averages of the results from at least nine plants are shown. Error bars indicate the SE.
We also monitored the days to the appearance of a visible inflorescence of the atxr3-1 atxr7-1 double mutant and each single mutant. As mentioned earlier, atxr3-1 exhibited an extended time to flower due to the slower growth rate relative to wild-type Col (Supplementary Fig. S1A–C). The atxr7-1 single mutant has an essentially wild-type growth rate and thus required less time to flower than the atxr3-1 single mutant (Supplementary Fig. S1B, C). Notably, in the atxr3 atxr7 double mutant, the extended time to flower of atxr3 was suppressed; the double mutant had a growth rate similar to that of the atxr7-1 single mutant (Supplementary Fig. S1B, C). Therefore, atxr7 is epistatic to atxr3 with respect to the growth rate, unlike the epistatic relationship for the developmental acceleration of flowering described above.
Discussion
The level of gene expression is often correlated with the levels of the H3K4me3 enrichment around transcription and translation start sites. It has been previously shown that certain Arabidopsis H3K4 methyltransferases, ATX1 and ATX2 in the TRX class and ATXR7 in the Set1 class, function in the FLC activation and flowering repression caused by FRI (Pien et al. 2008, Tamada et al. 2009). In the work reported herein, we have shown that ATXR3/SDG2, which is a member of a novel class of H3K4-specific methyltransferases (Fig. 2; Berr et al. 2010, Guo et al. 2010), also plays a role in FRI-dependent FLC activation. An ATXR3 lesion suppresses the FLC activation and the delayed flowering of the FRI-containing line FRI-Col in long days (Fig. 4). The reduced FLC expression in atxr3 mutants correlates with decreased H3K4me3 levels at the FLC locus (Fig. 5B). Collectively, three divergent types of H3K4-specific methyltransferases encoded by four genes (TRX-class ATX1 and ATX2, Set1-class ATXR7 and ATXR3) appear to be required for full levels of H3K4me3 at FLC chromatin and FLC expression in the presence of an active FRI allele, and thus for the winter-annual habit of Arabidopsis.
In contrast to H3K4me3, minor differences were found for the H3K4me1/me2 levels between FRI atxr3 and the wild type in the gene body of FLC: the H3K4me2 levels are reduced and the H3K4me1 levels are increased in FRI atxr3 compared with the wild type (Fig. 5C, D). These data suggest that H3K4me3, but not H3K4me1/me2, is involved in the FLC activation. We also found that, in FRI atxr3, the levels of the repressive epigenetic mark, H3K27me3, are increased compared with the FRI-Col wild type (Fig. 5E). Thus, ATXR3 appears to be part of an antagonistic system of methyltransferases that set the proper level of FLC expression by establishing and maintaining a balance between H3K4 and H3K27 methyltransferase activities (reviewed in Buzas et al. 2012). The antagonistic nature of H3K4 and H3K27 methylations at target loci has been observed in a range of eukaryotic systems (Papp and Muller 2006, Suganuma and Workman 2008, Schmitges et al. 2011).
Loss of the ATXR3 function also suppresses the delayed flowering in short days of lines that lack FRI (Fig. 1; Guo et al. 2010). This suppression of delayed flowering is associated with reduced expression of both FLC (Guo et al. 2010) and FLC clade members FLM and MAF5 (Fig. 4C). Thus, ATXR3 participates in the repression of flowering by maintaining the activated state of FLC as well as certain FLC-clade genes that are not affected by FRI. The expression of FLM and MAF5 was not increased in atxr3 in a previous microarray analysis (Guo et al. 2010). This discrepancy may be caused by the differences in the developmental stage of the plant material (we used almost fully expand rosette leaves compared with entire 12-day-old seedlings) and/or in methods [real-time reverse transcription–PCR (RT–PCR) vs. microarray].
Although ATXR3 and ATXR7 have divergent SET domains (Fig. 2), they have a similar domain organization, with GYF domains in the N-terminus (Springer et al. 2003), and share similar roles in flowering repression in the presence of FRI and in non-inductive short days (Figs. 1, 3; Berr et al. 2009, Tamada et al. 2009, Guo et al. 2010). It was therefore of interest to evaluate their possible relationship by characterizing the atxr3 atxr7 double mutant. The developmental acceleration of flowering is stronger in the atxr3 single mutant than in the atxr7 single mutant and, interestingly, in the atxr3 atxr7 double mutant there is not any additional flowering acceleration compared with the atxr3 single mutant (Fig. 6B). Thus, atxr3 is epistatic to atxr7 with respect to a developmental measure of flowering. This epistatic relationship indicates that the ability of ATXR7 to participate in activation of FLC and other flowering repression genes (such as FLC clade genes) is largely dependent on ATXR3.
How do ATXR3 and ATXR7 function together in FLC activation? One possibility is that they have different methyltransferase activities and together establish the H3K4me3 mark at FLC. As described above, the atxr3 lesion in a FRI background results in reduced H3K4me2/me3 levels and increased H3K4me1 levels at FLC (Fig. 5). In contrast, the levels of all mono-, di- and tri-methylations of H3K4 are decreased in FRI atxr7 relative to those in FRI-Col (Tamada et al. 2009). This difference may be accounted for by the ‘Phe/Tyr switch’ model; a tyrosine at a particular amino acid residue in the SET domain is considered to indicate that the protein has mono- and/or di-methyltransferase activities, whereas a phenylalanine indicates di- and/or tri-methyltransferase activities (Collins et al. 2005, Couture et al. 2008). ATXR3 has a phenylalanine in the key position and is thus predicted to have di- and tri-methyltransferase activities (Guo et al. 2010) and ATXR7 has a tyrosine and thus mono- and di-methyltransferase activities. In this case, the increased levels of H3K4me1 in FRI atxr3 are likely to be due to the attenuation of H3K4 di- and tri-methylation activities, and reduced levels of H3K4me3 in FRI atxr7 could result from decreased H3K4 mono- and di-methylation activities as such activities are required to produce the substrate for tri-methylation. Given that the FLC activation is associated with the H3K4me3 enrichment around the transcription and translation start sites, di- and tri-methyltransferase activity of ATXR3, together with mono- and di-methyltransferase activities of ATXR7, are required for full FLC expression. However, we should also mention that ATXR3 is reported to possess all mono-, di- and tri-methyltransferase activities in vitro (Guo et al. 2010). Further biochemical analyses for ATXR3 and ATXR7 will be interesting. Another possibility is that GYF domains in ATXR3 and ATXR7 together play a role in the expression of FLC and other flowering time genes (Springer et al. 2003). The GYF domain has been reported to interact with proline-rich sequences in proteins involved in splicing and mRNA export from the nucleus (reviewed in Kofler and Freund 2006). The GYF domains in ATXR3 and ATXR7 may function together in the splicing and mRNA export of the transcripts of FLC and other flowering time genes.
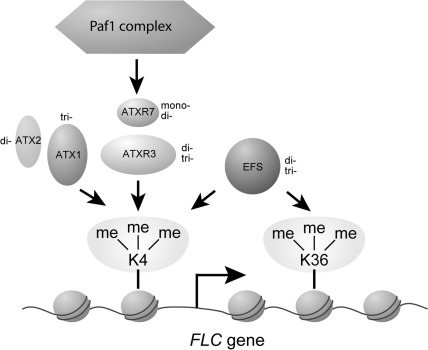
The epistatic relationship between atxr3 and atxr7 is reminiscent of that between early flowering7 (elf7) and atxr7 in which elf7 is epistatic to atxr7 (Tamada et al. 2009). ELF7 is a component of the RNA polymerase II-associated factor 1 complex (Paf1c), and this epistatic relationship is consistent with a model in which ELF7 may function in ATXR7 recruitment (Krogan et al. 2003, Tamada et al. 2009). This raises the possibility that there exists a functional link between ATXR3 and Paf1c. It is also interesting to note that the function of Paf1c and ATXR7 is largely independent of that of EFS, which appears to possess di- and tri-methyltransferase activities on H3K4 and H3K36 (Ko et al. 2010), based on the previous observation of transgressive phenotypes in atxr7 efs or elf7 efs double mutants (Tamada et al. 2009). Based on these epistatic relationships, ATXR3 function is also likely to be independent of EFS. In support of this model, it was recently shown that the genome-wide expression pattern in atxr3 is not correlated with that of atx1 or efs (Guo et al. 2010). From the results we and other groups have obtained, we propose a hypothetical model of FLC activation by the H3K4 methyltransferases and Paf1c (Fig. 7).
Fig. 7.
Model of a possible relationship between chromatin-modifying complexes that are involved in FLC activation. Our results indicate that the function of ATXR7 in flowering regulation is dependent on ATXR3. Relationships among ATXR7, ATX1, Paf1 complex and EFS were described previously (Tamada et al. 2009). Mono-, di- and tri- next to the factors indicate the predicted or examined activities of H3K4 methyltransferases. ATXR3 has phenylalanine and ATXR7 has tyrosine at the key residue for the methyltransferase activities, suggesting that ATXR7 catalyzes mono- and di-methylation of H3K4, and that ATXR3 catalyzes di- and tri-methylation to activate FLC. The activities of other H3K4 methyltransferases were described in previous studies (Pien et al. 2008, Saleh et al. 2008b, Guo et al. 2010, Ko et al. 2010). EFS contributes to H3K4 methylations at FLC most probably only in the presence of FRI (Kim et al. 2005, Zhao et al. 2005, Xu et al. 2008, Ko et al. 2010). K4, Lys4 of histone H3 (H3K4); K36, Lys36 of histone H3 (H3K36); me, methyl groups on lysine residues.
In addition to the epistasis in the flowering time regulation, we also observed the epistatic relationship in the growth rate regulation between ATXR3 and ATXR7: the slower growth rate in atxr3 vs. that in the wild type was suppressed by atxr7, suggesting that atxr7 is epistatic to atxr3 with respect to growth rate (Supplementary Fig. S1). If the gene(s) responsible for the slower growth rate is directly regulated by both ATXR3 and ATXR7 as in the case of FLC, ATXR3 and ATXR7 may have opposite roles in the transcription (i.e. ATXR7 or ATXR3 may have the unexpected function of transcriptional repression). Another possibility is that the gene(s) responsible may not be the direct target of ATXR3 and/or ATXR7. In fact, both Berr et al. (2010) and Guo et al. (2010) found a significant number of genes (two out of five genes and 22 out of 30 genes, respectively) with the microarray and individual ChIP analyses, for which changes in the mRNA levels are not correlated with those in the H3K4me3 levels. These genes are thought to be the indirect targets of ATXR3 (Berr et al. 2010, Guo et al. 2010). Isolation of the responsible gene(s) will be necessary to understand the epistasis in the growth rate regulation between ATXR3 and ATXR7.
Materials and Methods
Plant materials and growth conditions
FRI-Col is the Col genetic background into which an active FRI locus of accession San feliu-2 was introgressed (Lee and Amasino 1995). Seeds of atxr3-1 (WiscDsLox361D10) and atxr3-3 (SALK_021008) were obtained from the Arabidopsis Biological Resource Center (ABRC) (Alonso et al. 2003, Woody et al. 2007). atxr3-7 (FLAG_224A03/G01) seeds were obtained from the National Institute for Agricultural Research (INRA) (Samson et al. 2002).
Plants were grown at 21°C in long days (16 h light/8 h dark), short days (8 h light/16 h dark) or continuous light from cool-white fluorescent tubes [photosynthetic photon flux density (PPFD) approximately 60–70 μmol m−2 s−1]. Seeds were incubated in water at 4°C for 2 d and were then directly sown on the soil surface (Sun-Gro; MetroMix360), or incubated on agar-solidified medium containing 0.65 g l−1 Peters Excel 15-5-15 fertilizer (Grace Sierra) at 4°C for 2 d, and were transferred on the same agar medium to either long days, short days or continuous light.
Phylogenic analysis and multiple alignments
BLASTp searches were performed using the entire or the SET domain sequence of ATXR3 as a query on the National Center for Biotechnology Information (NCBI) website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). SET domain sequences of known histone methyltransferases, bacterial SET proteins (Polynucleobacter necessarius-1, 2) that were used for the roots, and the proteins from the BLASTp searches (Supplementary Table S1) were obtained using the CDD program on the NCBI website (Marchler-Bauer et al. 2009). Protein models from organisms in Alveolata, Stramenopiles and Viridiplantae that lack a SET domain or that are too short to include in the phylogenic analyses were omitted from the analyses. The SET domain sequences were then aligned using the linsi program on the mafft alignment application (Katoh et al. 2005; http://mafft.cbrc.jp/alignment/software/) and the alignment of sequences was refined manually using MEGA5 (Tamura et al. 2011; http://www.megasoftware.net/). From the alignment, a maximum likelihood (ML) tree was obtained with PhyML (Guindon and Gascuel 2003, Guindon et al. 2010; http://atgc.lirmm.fr/phyml/) using a Neighbor–Joining (NJ) tree and 200 random trees as initial trees with eight substitution categories, Best of NNI and SPR moves, and a bootstrap analysis (1,000 replicates). We adopted the default settings for the other parameters.
Molecular analyses
Total RNAs were isolated using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. First-strand cDNAs were synthesized from 3 μg of total RNA by using M-MLV Reverse Transcriptase (Promega) according to the manufacturer's recommendations. Real-time PCR was performed on a 7500 Fast Real-time PCR system (Applied Biosystems) using a DyNAmo HS SYBR Green qPCR kit (Finnzymes) according to the manufacturer's protocol. The PCR conditions were one cycle of 15 min at 95°C, and 45 cycles of 30 s at 95°C, 30 s at 58°C and 30 s at 72°C, followed by the dissociation stage as recommended by the manufacturer. Primers used to amplify the cDNAs are listed in Supplementary Table S2. All real-time RT–PCR data shown herein are the averages of the results from at least three different biological replicates, and were normalized to ACT7 expression.
ChIP was performed as previously described (Johnson et al. 2002, Gendrel et al. 2005). Antibodies used in this study were purchased from Millipore: histone H3 (06-755), H3K4me3 (07-473), H3K4me2 (07-030), H3K4me1 (07-436) and H3K27me3 (07-449). Real-time PCR was performed on a StepOnePlus Real-time PCR system (Applied Biosystems) using SYBR premix Ex Taq (TAKARA BIO INC.) according to the manufacturer's protocol. The PCR conditions were one cycle of 30 s at 95°C, and 45 cycles of 5 s at 95°C, 15 s at 55°C and 30 s at 70°C, followed by the dissociation stage as recommended by the manufacturer. Primers used to detect the enrichment are listed in Supplementary Table S2. All real-time ChIP-PCR data shown herein are the averages of the results from at least two different biological replicates, and were normalized to the level of either ACT7 or AG enrichment.
Accession numbers
Accession numbers of genes in this article are as follows: At2g31650 (ATX1), At1g05830 (ATX2), At3g61740 (ATX3), At4g27910 (ATX4), At5g53430 (ATX5), At4g15180 (ATXR3/SDG2), At5g42400 (ATXR7/SDG25), At4g00650 (FRI), At1g77300 (EFS), At5g10140 (FLC), At1g77080 (FLM), At5g65050 (MAF2), At5g65060 (MAF3), At5g65070 (MAF4), At5g65080 (MAF5), At5g09810 (ACT7), At4g18960 (AG), At2g23380 (CLF) and At5g13960 (KYP). Accession numbers of the genes used in the phylogenic analysis are described in Supplementary Table S1. Stock numbers of the T-DNA insertion lines described in this manuscript are as follows: WiscDsLox361D10 (atxr3-1), SALK_021008 (atxr3-3), FLAG_224A03/G01 (atxr3-7) and SALK_149692 (atxr7-1).
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the University of Wisconsin; the National Institutes of Health [grant No. 1R01GM079525]; the GRL Program of the Ministry of Education, Science, and Technology/Korea Foundation for International Cooperation of Science and Technology [to R.A.]; the Japanese Ministry of Education, Culture, Sports, Science and Technology [ID:22870036], NINS program for cross-disciplinary study [to Y.T.].
Supplementary Material
Acknowledgments
We thank Xiaoyu Zhang, Mark R. Doyle, Young-Min Jeong, Joohyun Lee, Issei Ohshima, Kenji Fukushima and Mitsuyasu Hasebe for helpful advice and discussion, and Seung chul Woo, Noriko Masuda, Asuka Murata and Yoona Jang for assisting in our experiments. We also thank other members of the Amasino Laboratory, the Donna E. Fernandez Laboratory and the Sebastian Y. Bednarek Laboratory at the University of Wisconsin as well as the Hasebe Laboratory at the National Institute for Basic Biology (NIBB) for helpful discussions. We acknowledge Hiroyo Nishide and the Data Integration and Analysis Facility at NIBB for providing the efficient programs to construct the phylogenic trees, and ABRC and INRA for providing the Arabidopsis T-DNA insertion lines.
Glossary
Abbreviations
- ABRC
Arabidopsis Biological Resource Center
- ACT
ACTIN
- AG
AGAMOUS
- Ash1
Absent, small or homeotic discs 1
- ATX
ARABIDOPSIS TRITHORAX
- ATXR
ATX-RELATED
- ChIP
chromatin immunoprecipitation
- CLF
CURLY LEAF
- Col
Columbia
- ELF7
EARLY FLOWERING7
- EFS
EARLY FLOWERING IN SHORT DAYS
- E(z)
Enhancer of zeste
- FLC
FLOWERING LOCUS C
- FLM
FLOWERING LOCUS M
- FRI
FRIGIDA
- INRA
National Institute for Agricultural Research
- KYP
KRYPTONITE
- MAF
MADS AFFECTING FLOWERING
- ML
maximum likelihood
- NCBI
National Center for Biotechnology Information
- NIBB
National Institute for Basic Biology
- NJ
Neighbor–Joining
- Paf1c
RNA polymerase II-associated factor 1 complex
- RT–PCR
reverse transcription–PCR
- SDG
SET DOMAIN GROUP
- SUVH
SUV39 HOMOLOG
- TRX
Trithorax
- Ws
Wassilewskija.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Avramova Z. Two Arabidopsis homologs of the animal trithorax genes: a new structural domain is a signature feature of the trithorax gene family. Gene. 2001;271:215–221. doi: 10.1016/s0378-1119(01)00524-8. [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 2003;13:627–637. doi: 10.1016/s0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- Avramova Z. Evolution and pleiotropy of TRITHORAX function in Arabidopsis. Int. J. Dev. Biol. 2009;53:371–381. doi: 10.1387/ijdb.082664za. [DOI] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, et al. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, McCallum EJ, Menard R, Meyer D, Fuchs J, Dong A, et al. Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell. 2010;22:3232–3248. doi: 10.1105/tpc.110.079962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, Xu L, Gao J, Cognat V, Steinmetz A, Dong A, et al. SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 2009;151:1476–1485. doi: 10.1104/pp.109.143941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzas DM, Robertson M, Finnegan EJ, Helliwell CA. Transcription-dependence of histone H3 lysine 27 trimethylation at the Arabidopsis polycomb target gene FLC. Plant J. 2011;65:872–881. doi: 10.1111/j.1365-313X.2010.04471.x. [DOI] [PubMed] [Google Scholar]
- Buzas DM, Tamada Y, Kurata T. FLC: a hidden polycomb response element shows up in silence. Plant Cell Physiol. 2012;53, doi: 10.1093/pcp/pcr163. 10.1093/pcp/PCR163. [DOI] [PubMed] [Google Scholar]
- Cao Y, Dai Y, Cui S, Ma L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell. 2008;20:2586–2602. doi: 10.1105/tpc.108.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Kim J, Hwang HJ, Kim S, Park C, Kim SY, et al. The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell. 2011;23:289–303. doi: 10.1105/tpc.110.075911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouard P. Vernalization and its relations to dormancy. Annu. Rev. Plant Physiol. 1960;11:191–238. [Google Scholar]
- Collins RE, Tachibana M, Tamaru H, Smith KM, Jia D, Zhang X, et al. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J. Biol. Chem. 2005;280:5563–5570. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture JF, Dirk LM, Brunzelle JS, Houtz RL, Trievel RC. Structural origins for the product specificity of SET domain protein methyltransferases. Proc. Natl Acad. Sci. USA. 2008;105:20659–20664. doi: 10.1073/pnas.0806712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- Gu X, Jiang D, Wang Y, Bachmair A, He Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 2009;57:522–533. doi: 10.1111/j.1365-313X.2008.03709.x. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guo L, Yu Y, Law JA, Zhang X. SET DOMAIN GROUP2 is the major histone H3 lysine 4 trimethyltransferase in Arabidopsis. Proc. Natl Acad. Sci. USA. 2010;107:18557–18562. doi: 10.1073/pnas.1010478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Doyle MR, Amasino RM. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Gu X, He Y. Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell. 2009;21:1733–1746. doi: 10.1105/tpc.109.067967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Kong NC, Gu X, Li Z, He Y. Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet. 2011;7:e1001330. doi: 10.1371/journal.pgen.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, He Y, Jacob Y, Noh YS, Michaels S, Amasino R. Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell. 2005;17:3301–3310. doi: 10.1105/tpc.105.034645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Michaels SD. SUPPRESSOR OF FRI 4 encodes a nuclear-localized protein that is required for delayed flowering in winter-annual Arabidopsis. Development. 2006;133:4699–4707. doi: 10.1242/dev.02684. [DOI] [PubMed] [Google Scholar]
- Ko JH, Mitina I, Tamada Y, Hyun Y, Choi Y, Amasino RM, et al. Growth habit determination by the balance of histone methylation activities in Arabidopsis. EMBO J. 2010;29:3208–3215. doi: 10.1038/emboj.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler MM, Freund C. The GYF domain. FEBS J. 2006;273:245–256. doi: 10.1111/j.1742-4658.2005.05078.x. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Blankestijn-de Vries H, Hanhart CJ, Soppe W, Peeters T. The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J. 1994;6:911–919. [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Lee I, Amasino RM. Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 1995;108:157–162. doi: 10.1104/pp.108.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Michaels SD, Masshardt AS, Amasino RM. The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 1994;6:903–909. [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Liu C, Lu F, Cui X, Cao X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Trillo M, Lazaro A, Poethig RS, Gomez-Mena C, Pineiro MA, Martinez-Zapater JM, et al. EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development. 2006;133:1241–1252. doi: 10.1242/dev.02301. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napp-Zinn K. On the genetical basis of vernalization requirement in Arabidopsis thaliana (L.) Heynh. In: Champagnat P, Jaques R, editors. La Physiologie de la Floraison. Paris: Colloques Internationaux du Centre National de la Recherche Scientifique; 1979. pp. 217–220. [Google Scholar]
- Ng DW, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S, Hall TC. Plant SET domain-containing proteins: structure, function and regulation. Biochim. Biophys. Acta. 2007;1769:316–329. doi: 10.1016/j.bbaexp.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Park S, van Nocker S. Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 2008;4:e1000077. doi: 10.1371/journal.pgen.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Fleury D, Mylne JS, Crevillen P, Inze D, Avramova Z, et al. ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell. 2008;20:580–588. doi: 10.1105/tpc.108.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell. 2003;15:1159–1169. doi: 10.1105/tpc.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 2001;126:122–132. doi: 10.1104/pp.126.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z. Dynamic and stable histone H3 methylation patterns at the Arabidopsis FLC and AP1 loci. Gene. 2008a;423:43–47. doi: 10.1016/j.gene.2008.06.022. [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Yilmaz M, Le O, Hou G, Sadder M, et al. The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell. 2008b;20:568–579. doi: 10.1105/tpc.107.056614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, et al. FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 2002;30:94–97. doi: 10.1093/nar/30.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Tamada Y, Doyle MR, Zhang X, Amasino RM. Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol. 2009;149:1196–1204. doi: 10.1104/pp.108.131508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci K, Michaels SD, Amasino RM. Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol. Biol. 2003;52:915–922. doi: 10.1023/a:1025426920923. [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, et al. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, Chandler VL, et al. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 2003;132:907–925. doi: 10.1104/pp.102.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Crosstalk among histone modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- Tamada Y, Yun JY, Woo SC, Amasino RM. ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell. 2009;21:3257–3269. doi: 10.1105/tpc.109.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ. The WiscDsLox T-DNA collection: an arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant Res. 2007;120:157–165. doi: 10.1007/s10265-006-0048-x. [DOI] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, et al. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 2008;28:1348–1360. doi: 10.1128/MCB.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Menard R, Berr A, Fuchs J, Cognat V, Meyer D, et al. The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 2009;57:279–288. doi: 10.1111/j.1365-313X.2008.03684.x. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 2005;7:1256–1260. doi: 10.1038/ncb1329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.