Abstract
Aims
A sequence variant, rs7025486[A], in DAB2IP on chromosome 9q33 has recently been associated with coronary heart disease (CHD). We sought to replicate this finding and to investigate associations with a panel of inflammatory and haemostatic biomarkers. We also sought to examine whether this variant, in combination with a chromosome 9p21 CHD variant (rs10757278) and the Framingham risk score (FRS), could improve the prediction of events compared with the FRS alone.
Methods and results
rs7025486 was genotyped in 1386 CHD cases and 3532 controls and was associated with CHD [odds ratio (OR) of 1.16, 95% confidence interval (CI) 1.05–1.29, P= 0.003]. Meta-analysis, using data from the original report and from genome-wide association studies in both the Wellcome Trust Case Control Consortium and the Cardiovascular Health Study, comprising 9968 cases and 20 048 controls, confirmed the association (OR of 1.10, 95% CI 1.06–1.14, P= 3.2 × 10−6). There was no association with a panel of CHD biomarkers, including any lipid, inflammation, or coagulation trait, nor with telomere length. Addition to the FRS of this variant plus rs10757278 on chromosome 9p21 improved the area under the receiver-operating characteristic curve (AROC) from 0.61 to 0.64 (P= 0.03) as well as improving the reclassification (net reclassification index = 11.1%, P= 0.007).
Conclusion
This study replicates a previous association of a variant in DAB2IP with CHD. Addition of multiple variants improves the performance of predictive models based upon classical cardiovascular risk factors.
Keywords: DAB2IP, Coronary heart disease, Genetic, Single-nucleotide polymorphism, Genomics
Introduction
A recent genome-wide association study (GWAS) found a sequence variant in DAB2IP (rs7025486[A]) to be strongly associated with the presence of abdominal aortic aneurysm (AAA) with a per-allele odds ratio (OR) of 1.21 (P= 4.6 × 10−10).1 The authors also found an association with early-onset myocardial infarction (MI) (OR 1.18, P= 3.1 × 10−5), MI at all ages (OR= 1.08, P= 0.0012), peripheral arterial disease (PAD) (OR= 1.14, P= 3.9 × 10−5), venous thrombo-embolism (VTE, OR 1.12, P= 0.0079), and pulmonary embolism (OR= 1.20, P= 0.00030) but not intracranial aneurysm or ischaemic stroke. This variant did not appear to act through classical intermediate phenotypes such as smoking, lipids, obesity, type 2 diabetes (T2DM), and hypertension.
DAB2IP, located on chromosome 9q33, is a GTPase-activating protein thought to play an important role in prostate cancer metastasis.2 A single-nucleotide polymorphism (SNP) in this gene has been associated with aggressive prostate cancer,3 whereas in vitro functional studies have demonstrated that loss of the protein leads to enhanced cell proliferation and reduced apoptosis, via the Pi3-Akt pathway.4 It is possible therefore, that this SNP exerts its effect by regulating cell senescence and proliferation. In 2007, three groups reported that another locus on the short arm of chromosome 9 at p21.3, tagged by the SNP rs10757278, was strongly associated with coronary heart disease (CHD).5–8 This SNP was also subsequently found to be associated with the risk of AAA and PAD but does not appear to act through classical intermediate phenotypes.5–7,9–11 There is mounting evidence to suggest that this SNP affects expression of the large non-coding RNA ANRIL, which in turn may affect expression of nearby genes such as the CDKN2A/B cluster which also play a role in cellular proliferation and senescence.12–14
We aimed to confirm the association of rs7025486[A] with CHD. In addition, associations with circulating biomarkers of cardiovascular disease (CVD), including lipids, inflammatory markers, and haemostatic markers, were investigated. Since shorter leucocyte telomere length (LTL) has been consistently shown to be associated with increased risk of CHD15,16 in both population-based case–control studies and patients with familial hypercholesterolaemia (FH), a monogenic form of premature CHD, we examined the relationship between this SNP and telomere length. Finally, whether or not this variant, in combination with the 9p21 locus and the Framingham risk score (FRS), could improve the prediction of CHD over and above the FRS alone in the Northwick Park Heart Study II (NPHS-II) prospective study of UK men was investigated.
Methods
Study groups
Table 1 shows the baseline characteristics of each of the studies used. Ethics approval was granted for all studies. Detailed descriptions of each study are provided elsewhere. NPHS-II is a prospective study of healthy middle-aged men (50–64 years) recruited from nine UK general practices.17 In the 2742 Caucasian men with genotype data, by December 2009, there had been 272 CHD events comprising 175 acute CHD events (42 fatal), 74 coronary artery revascularization procedures, and 23 silent MI. The HIFMECH study18 compares male survivors of a first MI aged <60 years (excluding patients with FH and insulin-dependent diabetes mellitus) and population-based individuals of the same age and region recruited from four centres in Europe: Stockholm (Sweden) and London (England) for the North and Marseille (France) and San Giovanni Rotondo (Italy) representing the south. In all, a total of 598 post-infarction patients and 653 controls were included in the study. The Simon Broome Study recruited 409 patients with definite FH (all with a total cholesterol concentration of >7.5 mmol/L) with 127 definite CHD events as defined in Neil et al.19 The UDACS20 consists of 1014 consecutive subjects recruited from the diabetes clinic at University College London Hospitals NHS Trust (UCLH) 2001–02 (629 men; 600 Caucasians with T2DM). All patients had diabetes according to the WHO criteria and analysis was restricted to the Caucasian subjects with T2DM to remove possible heterogeneity within the sample. Coronary heart disease was defined in Wootton et al.20 The coronary artery bypass graft (CABG) patients were drawn from the coronary artery surgery inflammation study and are described elsewhere.21 Briefly, all patients undergoing elective first-time CABG at the Middlesex Hospital, London, UK, between October 1999 and September 2000 were invited to participate. Subjects undergoing additional surgical procedures (such as valvular surgery or aneurysmectomy), subjects with evidence of a pre-existing inflammatory state or unstable coronary artery disease, and subjects who suffered potentially confounding infective post-operative complications or circulatory failure requiring inotropic support were excluded. The CABG group includes 439 people (20% women) having different ethnic origin (83% Caucasians, 8% Asians, 2% Afro-Caribbean, 2% of other ethnicity, and 5% of unknown origin). Non-Caucasian subjects were excluded from the genetic analysis.
Table 1.
Baseline characteristics of studies
| UDACS |
NPHS-II |
Simon Broome |
HIFMECH |
CABG | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHD−, n= 358 | CHD+, n = 135 | P-value | CHD−, n= 2406 | CHD+, n= 274 | P-value | CHD−, n= 214 | CHD+, n= 127 | P-value | CHD−, n= 554 | CHD+, n= 518 | P-value | CHD, n= 332 | |
| Age (years) | 66.2 (10.9) | 69.5 (9.9) | 0.003 | 56.0 (3.4) | 56.6 (3.5) | 0.01 | 44.6 (13.8) | 56.5 (10.4) | <0.001 | 51.5 (5.4) | 51.9 (5.4) | — | 64.9 (9.2) |
| % male | 56.4 (202) | 69.6 (94) | 0.008 | 100 (2406) | 100 (274) | — | 43.0 (92) | 66.9 (85) | <0.001 | 100 (554) | 100 (518) | — | 81.9 (272) |
| SBP (mmHg) | 142.0 (18.5) | 137.9 (20.5) | 0.03 | 136.8 (18.6) | 141.4 (19.4) | 0.0002 | 124.1 (16.3) | 130.8 (19.9) | 0.001 | 128.1 (14.5) | 127.7 (16.8) | 0.71 | |
| DBP (mmHg) | 79.5 (10.9) | 77.1 (10.5) | 0.03 | 84.4 (11.2) | 86.8 (11.5) | 0.0008 | 76.6 (9.9) | 78.5 (13.1) | 0.14 | 84.1 (8.5) | 81.6 (10.2) | <0.001 | |
| BMI (kg/m2) | 29.2 (5.3) | 29.5 (5.3) | 0.54 | 26.2 (3.4) | 26.6 (3.3) | 0.05 | 23.7 (4.0) | 25.2 (3.5) | 0.003 | 26.1 (3.2) | 27.0 (3.3) | <0.001 | 28.5 (4.5) |
| % ever smokers | 50.7 (173) | 59.9 (79) | 0.08 | 67.3 (1618) | 77.7 (213) | <0.001 | 38.5 (82) | 65.4 (83) | <0.001 | 61.9 (343) | 82.1 (425) | <0.001 | 78.0 (259) |
| Cholesterol (mmol/L) | 5.19 (1.06) | 4.68 (1.11) | <0.001 | 5.71 (1.01) | 6.07 (1.02) | <0.001 | 6.85 (1.28) | 6.28 (1.32) | <0.001 | 5.53 (0.98) | 5.39 (1.18) | 0.04 | 4.79 (1.10) |
| Triglyceride (mmol/L) | 1.94 (1.10) | 1.96 (1.08) | 0.85 | 1.77 (0.93) | 2.05 (1.07) | <0.001 | 1.24 (0.54) | 1.44 (0.67) | 0.003 | 1.44 (0.61) | 1.87 (0.76) | <0.001 | |
| CRP (g/L) | 1.70 (1.41) | 1.86 (1.59) | 0.27 | 2.46 (2.43) | 3.26 (3.33) | <0.001 | 1.19 (1.39) | 1.48 (1.72) | 0.11 | 1.24 (1.42) | 2.23 (2.53) | <0.001 | 2.25 (2.75) |
| Fibrinogen (g/L) | — | — | — | 2.70 (0.52) | 2.80 (0.49) | 0.002 | 2.77 (0.82) | 3.07 (1.00) | 0.003 | 3.41 (0.69) | 3.71 (0.92) | <0.001 | 3.63 (0.77) |
In silico analysis
The Cardiovascular Health Study is a longitudinal study of 3291 men and women, primarily of self-described European ancestry, who were free of clinical CVD at baseline, consented to the use of their DNA for cardiovascular research, and were successfully genotyped on the Illumina 370 CNV platform. There were 532 incident CHD events during follow-up. Data from the Cardiovascular Health Study GWAS of MI were accessed through a publicly available database (http://www.ncbi.nlm.nih.gov/gap). Data from the WTCCC of coronary disease22 were accessed, which included genotype data on 1988 CHD cases and 3004 controls from the National Blood Service and the 1958 Birth Cohort.
Genotyping
rs7025486 and rs10757274 were genotyped using TaqMan technology [Applied Biosciences (ABI), Warrington, UK]. Reactions were performed on 384-well microplates and analysed using ABI TaqMan 7900HT software (Applied Biosciences).
Measurement of telomere length
Leucocyte DNA was extracted by the salting-out method.15 Leucocyte telomere length was measured using a quantitative polymerase chain reaction (PCR)-based method,23 as adapted in our laboratory. The relative telomere length was calculated as the ratio of telomere repeats to single-copy gene (SCG) copies (T/S ratio). For each sample, the quantity of telomere repeats and that of SCG copies were determined in comparison to a reference sample in a telomere and an SCG quantitative PCR, respectively. All PCRs were performed in duplicate on the Rotor-Gene 6000 (Corbett Research Ltd, Cambridge, UK), and the raw data were analysed using the comparative quantification analysis (Rotor-Gene 6000 software, Corbett Research Ltd). The specificity of all amplifications was determined by melting curve analysis. Using the linear regression line between measures obtained by both the PCR-based method and the conventional terminal restriction fragment analysis for the same set of 32 samples, as previously described, the corresponding telomere length in base pairs (bp) was calculated from the T/S ratio measured in each subject.15
Statistical analysis
Allele frequencies between groups were compared using χ2 test, and we tested for departure from the Hardy–Weinberg equilibrium in controls. Mean values for continuous variables were compared between those with and without CHD using unpaired t-tests. Where necessary, variables were log-transformed before analysis and results are presented as geometric means with approximate standard deviations for these variables. Categorical variables were compared using χ2 tests. Genetic associations were tested using regression models using an additive (per-allele) model. Hazard ratios (HRs) were calculated in the prospective study (NPHS-II), using Cox's regression models with corresponding 95% confidence intervals (CIs). Odds ratios and their corresponding 95% CIs were calculated from case–control and cross-sectional studies using logistic regression models. Gene–disease associations were adjusted for age as a continuous outcome. In NPHS-II, analyses are stratified by general practice. For the conventional model, a score was derived based on age, triglycerides, cholesterol, smoking, and systolic blood pressure. We then fitted a model including both conventional factors and rs7025486 and rs10757274 genotypes and obtained a second score by weighting according to the β coefficients from the model. The area under the receiver-operating characteristic (ROC) curve was calculated for both scores and the difference between the conventional model and that incorporating genotypes was tested. We assessed the effect of adding the genetic variables on the ability of the score to assign participants to the correct risk category using the net reclassification index (NRI).24
For meta-analysis of CHD outcomes, study-specific ORs and standard errors were obtained and pooled generating a summary estimate and its 95% CI. There was no evidence of between-study heterogeneity (I2= 0%); therefore, a fixed-effects model was used. An additive model was used to confirm the association for additive effects found by Gretarsdottir et al.1 All analyses were carried out using Stata version 10 (Statcorp, TX, USA).
Results
The characteristics of the study groups are presented in Table 1, which show the expected differences in classical risk factors in cases compared with controls and in baseline characteristics in those who subsequently went on to develop CHD in the prospective NPHS-II study. In the UDACS study, many risk factors are higher in the CHD-negative group, which is indicative of higher proportion cases being treated with statins and blood pressure-lowering medication. For both SNPs, the genotype distributions were as expected from the Hardy–Weinberg principles with the risk-allele frequency for the DAB2IP SNP rs7025486 ranging from 0.222 to 0.294 and for the Chr9p SNP rs10757274 ranging from 0.482 to 0.608.
Association of rs7025486 with coronary heart disease
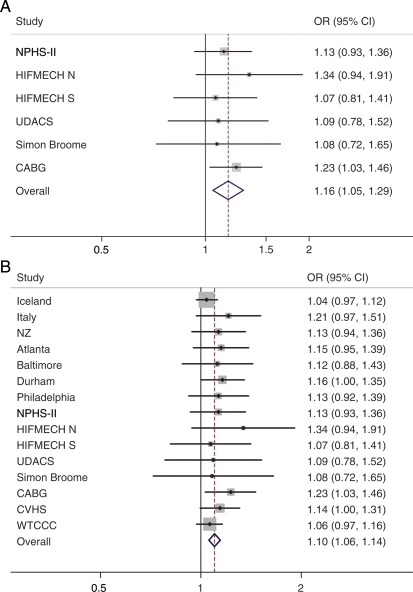
In each of the study groups, the rare allele was associated with 1.08–1.34 higher risk of CHD, although only one of these (CABG vs. healthy controls in NPHS-II) was nominally statistically significant using a P-value threshold of 0.05 (OR 1.2, 95% CI 1.03–1.46, P= 0.021). Pooled analysis of the results from each of the studies (Figure 1A) demonstrated that there was a consistent association between rs7025486 and CHD (OR 1.16, 95% CI 1.05–1.29, P= 0.003). Meta-analysis of data from the original report,1 the newly genotyped studies, and the WTCCC and CVHS GWAS data also demonstrated a consistent association (OR 1.1, 95% CI 1.06–1.14, P= 3.2 × 10−6), forest plot shown in Figure 1B.
Figure 1.
(A) Meta-analysis studies genotyped in this paper for association between rs7025486 and coronary heart disease. Combined odds ratio (95% confidence interval) is 1.16 (1.05–1.29); P= 0.003. Fixed-effects additive genotype model. (B) Meta-analysis of association between rs7025486 and coronary heart disease, including newly genotyped, previously published data and genome-wide association study data from the WTCCC and CVHS. Odds ratio 1.10 (95% confidence interval 1.06–1.14, P = 3.2 × 10−6). Fixed-effects additive genotype model.
Association of rs7025486 with intermediate traits
There was no significant association between this SNP and any of the cardiovascular biomarkers examined including lipid profiles and inflammatory markers (C-reactive protein and interleukin-6) or phenotypes such as stroke and diabetes. In addition, we found no association with a panel of haemostatic markers including fibrinogen, plasminogen activator inhibitor-1 (PAI-1), and a number of clotting factors, nor with the telomere length (see Supplementary material online, Tables 2a–d, Figure 4).
Conventional risk factors and coronary heart disease events
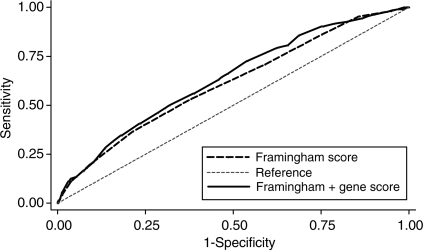
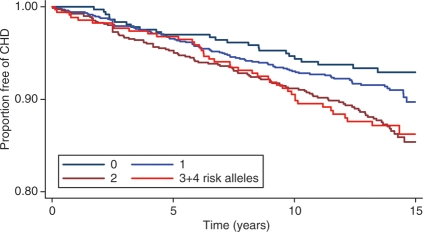
The baseline characteristics of the NPHS-II, stratified by subsequent CHD event, are presented in Table 1. The men who went on to develop CHD during follow-up (n= 274) were older, had higher plasma cholesterol, triglycerides, and blood pressure, and had lower HDL-cholesterol, and the prevalence of smoking was higher than in those who remained CHD free (n= 2406). On the basis of the measured variables included in the Framingham algorithm, the AROC given by this set of classical risk factors (CRFs) was 0.61 (95% CI 0.572–0.651) (Figure 2). We created a simple additive ‘gene score’ (GS) based upon carriage of rs7025486 and rs10757278, combined with CRFs, to assess whether or not it would improve risk prediction and stratification compared with CRFs alone. There was no statistical evidence of interaction between these SNPs (see Supplementary material online, Table 3), and the AROC for these two SNPs alone was 0.577 (95% CI 0.540–0.614). The AROC for the model that included both CRFs and genotypes was 0.64 (95% CI 0.598–0.675; Figure 2), which was a small but statistically significant improvement when compared with the value achieved by CRFs alone (P= 0.03). The numbers of patients reclassified using the model that included CRFs and the two SNPs are shown in Table 2. The NRI was 11.1% which was statistically significant (P= 0.007). A Kaplan–Meier plot of freedom from CHD with increasing numbers of risk alleles carried is shown in Figure 3, demonstrating the added effect on risk of carriage of more than one risk allele.
Figure 2.
Receiver-operating characteristic curve for different prediction models in NPHS-II men. The area under the receiver-operating characteristic curve increases from 0.61 to 0.64 when the gene score is added to the Framingham variables (P= 0.03).
Table 2.
Reclassification based on the Framingham risk score + gene score
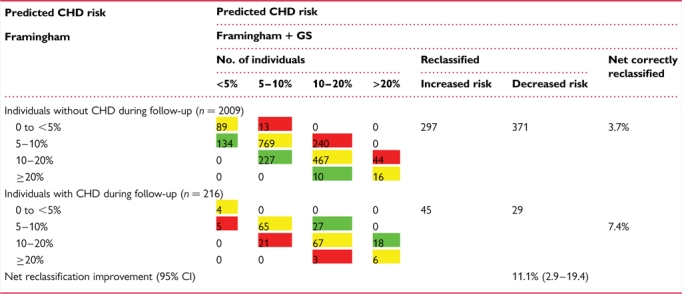 |
The percentage correctly reclassified by adding gene score to the Framingham variables is 11.1% (P= 0.007). Green signifies correctly reclassified, yellow no reclassification and red incorrectly reclassified.
Figure 3.
The Kaplan–Meier plot for coronary heart disease and the number of risk alleles carried. Compared with the reference group of those with no risk alleles, the age- and practice-adjusted hazard ratio for those with only one allele = 1.19 (0.78–1.84), P = 0.42, for any two alleles = 1.79 (1.16–2.75), P = 0.008, and for three and four alleles = 1.75 (1.07–2.85) P = 0.025. Overall P = 0.005.
Discussion
This study is the first to replicate the association between rs7025486 and the risk of CHD following the initial publication.1 Figure 1A and B demonstrate that the effect size in different groups of subjects is extremely consistent, even with differing underlying aetiologies such as T2DM or FH, and in case–control and prospective studies, including previous published GWAS. Although only 1 of the 15 studies included in the meta-analysis passed a conventional P-value threshold of 0.05 for single SNP-disease association, this is not unexpected given the minor-allele frequency (MAF) and modest effect size. The fact that in all 15 studies the minor allele increased the risk of CHD with a comparable effect suggests that this association is unlikely to be a false-positive, reflected by the combined P-value. Indeed, it is becoming clear that the initial wave of GWAS has detected only SNPs with the largest effect and favourable MAFs (the so-called ‘low-hanging fruit’). Even meta-analysis of GWAS, involving many thousands of subjects, may be underpowered to detect all SNPs with modest effects sizes. Therefore, single SNP studies guided by the findings of GWAS are likely to be an important method to decipher which of the many associations that fail to reach genome-wide levels of significance are actually true positives.
Furthermore, although the fact that the effect sizes of newly discovered variants such as this are small, this does not preclude important biological insights being made. The fact that this variant has a similar effect in differing CHD aetiologies, and that there is no association with established biomarkers, implies that this variant is acting through pathways independent of those classically associated with CHD such as lipid metabolism. DAB2IP is involved in regulating cell survival and senescence, and the association seen here adds to emerging evidence (seen with the strong association between the 9p21 locus and CHD) that genetic variants in genes that regulate the cell cycle may be an important mediators of CVD. The additive effect on risk of CHD seen with these two SNPs implies that the accumulation of variants of modest effect in this pathway is an important independent mediator of CHD development (as suggested by the common variant hypothesis) and highlights potential targets for the development of novel therapeutic options.
The original report found an association with VTE and pulmonary embolism,1 so we hypothesized that this variant may act through pathways that promote thrombosis but found no significant associations with a panel of haemostatic markers including fibrinogen, PAI-1, and numerous clotting factors. Shorter telomeres are associated with premature CHD; however, no consistent association between this SNP and mean LTL was found in 2012 subjects examined.
It is unlikely that a single SNP of modest effect will improve the prediction of CVD when compared with a risk score based upon multiple CRFs.25 It is much more likely, however, that variants found to be associated with CHD, which do not act through intermediate phenotypes such as lipid traits, which are already included in such risk scores, will be of use when combined with CRFs. It has been reported that addition of the single chromosome 9 SNP (rs10757278) to the FRS did not improve prediction (as measured by the AROC).25 In this study, we show that addition of two SNPs to the FRS improved the AROC by 0.03 which was statistically significant (P= 0.001). Although the two SNPs show no statistical evidence of interaction, the study has limited power to detect such interactive effects, and possibly because of small numbers, risk was not highest in men carrying all four risk alleles. Such effects will need to be confirmed in larger studies. Because of the high frequency of both of the risk alleles, 38% of these UK men carry one risk allele, 34% carry two, and 14% carry three or four risk alleles, with those carrying two or more having an HR of over 1.7.
The AROC is a commonly used statistic to describe the discriminative ability of a diagnostic test, but the number itself provides little clinically useful information. In the UK, the FRS is used to classify the population into risk groups that guide clinicians when planning treatment. In the UK, treatment with a statin is recommended in individuals with a 10-year risk >20%.26 In the NPHS-II at recruitment, adopting this policy would result in just 35 (1.6%) of men being offered treatment, in part because men with prevalent CHD or being treated with preventative medications were excluded.17 We found that addition of the two-SNP gene score to the FRS improved risk stratification as measured by the NRI of 11.1% (P= 0.007, 95% CI 2.9–19.4). If a policy was adapted whereby people with an FRS > 20% or an FRS + GS > 20% in the NPHS-II cohort were offered treatment, then the number of men who would qualify increases three-fold in the group who went on to develop CHD. It has been observed that the FRS tends to over-predict CVD in high-risk groups but under-predict CVD in low-risk groups.27,28 Compared with the general population, the participants in NPHS-II are a healthy cohort which is one reason why the FRS performs poorly as measured by the AROC. We have shown that in this group of patients, the addition of genotypes to the prediction model improved performance as measured by the AROC and the more clinically relevant NRI. Further research is required to assess whether or not these results are applicable on a population level.
Conclusion
This study has confirmed the effects of a common variant in DAB2IP (rs7025486) on the development of CHD. Prediction models which include CRFs and genotypes, which act through pathways independent of the CRFs, are likely to improve their performance. Even the small increment in AROC seen in this study has potential clinical utility when thresholds used for treatment in the UK are considered. Understanding the mechanism by which this variant exerts its effect should be the focus of ongoing research.
Supplementary material
Supplementary material is available at European Heart Journal online.
Funding
S.C.H. is supported by the BHF as a Chair Scholar, and S.E.H., J.A.C., and K.L. are funded by the British Heart Foundation RG2008/08. Reecha Sofat is supported by a British Heart Foundation (Schillingford) Clinical Training Fellowship (FS/07/011). The NPHS-II study was supported by the Medical Research Council, the US National Institutes of Health (NIH 33014), and Du Pont Pharma. Diabetes UK supported J.W.S. (BDA: RD01/0001357) and the creation of UDACS. The HIFMECH study was supported by the European Commission (BMH4-CT96–0272), the Swedish Medical Research Council, the Swedish Heart–Lung Foundation, INSERM, Université de la Méditerranée (INSERM U626), Foundation pour la Recherche Médicale (FRM), and Programme Hospitalier de Recherche Clinique (PHRC 1996). HIFMECH co-investigators are A.H., S.E.H., Irène Juhan-Vague, Maurizio Margaglione, Giovanni di Minno, John Yudkin, and Elena Tremoli. We would like to thank the members of the Simon Broome BHF study for access to patients’ samples: Dr Rossi Naoumova, Prof. Gil Thompson, Dr Mary Seed, Prof. Paul Durrington, Dr Paul Miller, and Prof. John Betteridge. The Wellcome Trust Case Control Consortium was funded by the Wellcome Trust (076113/B/04/Z). Funding to pay the Open Access publication charges for this article was provided by the British Heart Foundation.
Conflict of interest: none declared.
Supplementary Material
Acknowledgements
We acknowledge the contribution of the late Professor George Miller (1939–2006) who was the PI on the NPHS-II study and Phil Howard for his technical support. We also thank all the medical staff and patients who contributed to the NPHS-II study and the Office for National Statistics (NHS) Central Registry for provision of mortality data.
References
- 1.Gretarsdottir S, Baas AF, Thorleifsson G, Holm H, den Heijer M, de Vries JP, Kranendonk SE, Zeebregts CJ, van Sterkenburg SM, Geelkerken RH, van Rij AM, Williams MJ, Boll AP, Kostic JP, Jonasdottir A, Walters GB, Masson G, Sulem P, Saemundsdottir J, Mouy M, Magnusson KP, Tromp G, Elmore JR, Sakalihasan N, Limet R, Defraigne JO, Ferrell RE, Ronkainen A, Ruigrok YM, Wijmenga C, Grobbee DE, Shah SH, Granger CB, Quyyumi AA, Vaccarino V, Patel RS, Zafari AM, Levey AI, Austin H, Girelli D, Pignatti PF, Olivieri O, Martinelli N, Malerba G, Trabetti E, Becker LC, Becker DM, Reilly MP, Rader DJ, Mueller T, Dieplinger B, Haltmayer M, Urbonavicius S, Lindblad B, Gottsater A, Gaetani E, Pola R, Wells P, Rodger M, Forgie M, Langlois N, Corral J, Vicente V, Fontcuberta J, Espana F, Grarup N, Jorgensen T, Witte DR, Hansen T, Pedersen O, Aben KK, de Graaf J, Holewijn S, Folkersen L, Franco-Cereceda A, Eriksson P, Collier DA, Stefansson H, Steinthorsdottir V, Rafnar T, Valdimarsson EM, Magnadottir HB, Sveinbjornsdottir S, Olafsson I, Magnusson MK, Palmason R, Haraldsdottir V, Andersen K, Onundarson PT, Thorgeirsson G, Kiemeney LA, Powell JT, Carey DJ, Kuivaniemi H, Lindholt JS, Jones GT, Kong A, Blankensteijn JD, Matthiasson SE, Thorsteinsdottir U, Stefansson K. Genome-wide association study identifies a sequence variant within the DAB2IP gene conferring susceptibility to abdominal aortic aneurysm. Nat Genet. 2010;42:692–697. doi: 10.1038/ng.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie D, Gore C, Liu J, Pong RC, Mason R, Hao G, Long M, Kabbani W, Yu L, Zhang H, Chen H, Sun X, Boothman DA, Min W, Hsieh JT. Role of DAB2IP in modulating epithelial-to-mesenchymal transition and prostate cancer metastasis. Proc Natl Acad Sci USA. 2010;107:2485–2490. doi: 10.1073/pnas.0908133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, Robbins C, Isaacs SD, Cheng Y, Li G, Sun J, Chang BL, Marovich L, Wiley KE, Balter K, Stattin P, Adami HO, Gielzak M, Yan G, Sauvageot J, Liu W, Kim JW, Bleecker ER, Meyers DA, Trock BJ, Partin AW, Walsh PC, Isaacs WB, Gronberg H, Xu J, Carpten JD. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 4.Xie D, Gore C, Zhou J, Pong RC, Zhang H, Yu L, Vessella RL, Min W, Hsieh JT. DAB2IP coordinates both PI3K-Akt and ASK1 pathways for cell survival and apoptosis. Proc Natl Acad Sci USA. 2009;106:19878–19883. doi: 10.1073/pnas.0908458106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bown MJ, Braund PS, Thompson J, London NJ, Samani NJ, Sayers RD. Association between the coronary artery disease risk locus on chromosome 9p21.3 and abdominal aortic aneurysm. Circ Cardiovasc Genet. 2008;1:39–42. doi: 10.1161/CIRCGENETICS.108.789727. [DOI] [PubMed] [Google Scholar]
- 6.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, Jaaskelainen JE, Kyo Y, Lenk GM, Sakalihasan N, Kostulas K, Gottsater A, Flex A, Stefansson H, Hansen T, Andersen G, Weinsheimer S, Borch-Johnsen K, Jorgensen T, Shah SH, Quyyumi AA, Granger CB, Reilly MP, Austin H, Levey AI, Vaccarino V, Palsdottir E, Walters GB, Jonsdottir T, Snorradottir S, Magnusdottir D, Gudmundsson G, Ferrell RE, Sveinbjornsdottir S, Hernesniemi J, Niemela M, Limet R, Andersen K, Sigurdsson G, Benediktsson R, Verhoeven EL, Teijink JA, Grobbee DE, Rader DJ, Collier DA, Pedersen O, Pola R, Hillert J, Lindblad B, Valdimarsson EM, Magnadottir HB, Wijmenga C, Tromp G, Baas AF, Ruigrok YM, van Rij AM, Kuivaniemi H, Powell JT, Matthiasson SE, Gulcher JR, Thorgeirsson G, Kong A, Thorsteinsdottir U, Stefansson K. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 7.Thompson AR, Golledge J, Cooper JA, Hafez H, Norman PE, Humphries SE. Sequence variant on 9p21 is associated with the presence of abdominal aortic aneurysm disease but does not have an impact on aneurysmal expansion. Eur J Hum Genet. 2009;17:391–394. doi: 10.1038/ejhg.2008.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samani NJ, Raitakari OT, Sipila K, Tobin MD, Schunkert H, Juonala M, Braund PS, Erdmann J, Viikari J, Moilanen L, Taittonen L, Jula A, Jokinen E, Laitinen T, Hutri-Kahonen N, Nieminen MS, Kesaniemi YA, Hall AS, Hulkkonen J, Kahonen M, Lehtimaki T. Coronary artery disease-associated locus on chromosome 9p21 and early markers of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:1679–1683. doi: 10.1161/ATVBAHA.108.170332. [DOI] [PubMed] [Google Scholar]
- 10.Cunnington MS, Mayosi BM, Hall DH, Avery PJ, Farrall M, Vickers MA, Watkins H, Keavney B. Novel genetic variants linked to coronary artery disease by genome-wide association are not associated with carotid artery intima-media thickness or intermediate risk phenotypes. Atherosclerosis. 2009;203:41–44. doi: 10.1016/j.atherosclerosis.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cluett C, McDermott MM, Guralnik J, Ferrucci L, Bandinelli S, Miljkovic I, Zmuda JM, Li R, Tranah G, Harris T, Rice N, Henley W, Frayling TM, Murray A, Melzer D. The 9p21 myocardial infarction risk allele increases risk of peripheral artery disease in older people. Circ Cardiovasc Genet. 2009;2:347–353. doi: 10.1161/CIRCGENETICS.108.825935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarinova O, Stewart AF, Roberts R, Wells G, Lau P, Naing T, Buerki C, McLean BW, Cook RC, Parker JS, McPherson R. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671–1677. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Sanoff HK, Cho H, Burd CE, Torrice C, Mohlke KL, Ibrahim JG, Thomas NE, Sharpless NE. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4:e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salpea KD, Nicaud V, Tiret L, Talmud PJ, Humphries SE. The association of telomere length with paternal history of premature myocardial infarction in the European Atherosclerosis Research Study II. J Mol Med. 2008;86:815–824. doi: 10.1007/s00109-008-0347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brouilette S, Singh RK, Thompson JR, Goodall AH, Samani NJ. White cell telomere length and risk of premature myocardial infarction. Arterioscler Thromb Vasc Biol. 2003;23:842–846. doi: 10.1161/01.ATV.0000067426.96344.32. [DOI] [PubMed] [Google Scholar]
- 17.Cooper JA, Miller GJ, Humphries SE. A comparison of the PROCAM and Framingham point-scoring systems for estimation of individual risk of coronary heart disease in the Second Northwick Park Heart Study. Atherosclerosis. 2005;181:93–100. doi: 10.1016/j.atherosclerosis.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 18.Juhan-Vague I, Morange PE, Aubert H, Henry M, Aillaud MF, Alessi MC, Samnegard A, Hawe E, Yudkin J, Margaglione M, Di Minno G, Hamsten A, Humphries SE. Plasma thrombin-activatable fibrinolysis inhibitor antigen concentration and genotype in relation to myocardial infarction in the north and south of Europe. Arterioscler Thromb Vasc Biol. 2002;22:867–873. doi: 10.1161/01.atv.0000015445.22243.f4. [DOI] [PubMed] [Google Scholar]
- 19.Neil HA, Seagroatt V, Betteridge DJ, Cooper MP, Durrington PN, Miller JP, Seed M, Naoumova RP, Thompson GR, Huxley R, Humphries SE. Established and emerging coronary risk factors in patients with heterozygous familial hypercholesterolaemia. Heart. 2004;90:1431–1437. doi: 10.1136/hrt.2003.022764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wootton PT, Arora NL, Drenos F, Thompson SR, Cooper JA, Stephens JW, Hurel SJ, Hurt-Camejo E, Wiklund O, Humphries SE, Talmud PJ. Tagging SNP haplotype analysis of the secretory PLA2-V gene, PLA2G5, shows strong association with LDL and oxLDL levels, suggesting functional distinction from sPLA2-IIA: results from the UDACS study. Hum Mol Genet. 2007;16:1437–1444. doi: 10.1093/hmg/ddm094. [DOI] [PubMed] [Google Scholar]
- 21.Brull DJ, Montgomery HE, Sanders J, Dhamrait S, Luong L, Rumley A, Lowe GD, Humphries SE. Interleukin-6 gene −174g > c and −572g > c promoter polymorphisms are strong predictors of plasma interleukin-6 levels after coronary artery bypass surgery. Arterioscler Thromb Vasc Biol. 2001;21:1458–1463. doi: 10.1161/hq0901.094280. [DOI] [PubMed] [Google Scholar]
- 22.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pencina MJ, D'Agostino RB, Sr., D'Agostino RB, Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929. discussion 207–112. [DOI] [PubMed] [Google Scholar]
- 25.Talmud PJ, Cooper JA, Palmen J, Lovering R, Drenos F, Hingorani AD, Humphries SE. Chromosome 9p21.3 coronary heart disease locus genotype and prospective risk of CHD in healthy middle-aged men. Clin Chem. 2008;54:467–474. doi: 10.1373/clinchem.2007.095489. [DOI] [PubMed] [Google Scholar]
- 26.JBS 2: Joint British Societies’ guidelines on prevention of cardiovascular disease in clinical practice. Heart. 2005;91(Suppl. 5):v1–v52. doi: 10.1136/hrt.2005.079988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. http://www.screening.nhs.uk/cms.php?folder=2718 .
- 28.Brindle P, Beswick A, Fahey T, Ebrahim S. Accuracy and impact of risk assessment in the primary prevention of cardiovascular disease: a systematic review. Heart. 2006;92:1752–1759. doi: 10.1136/hrt.2006.087932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.