Abstract
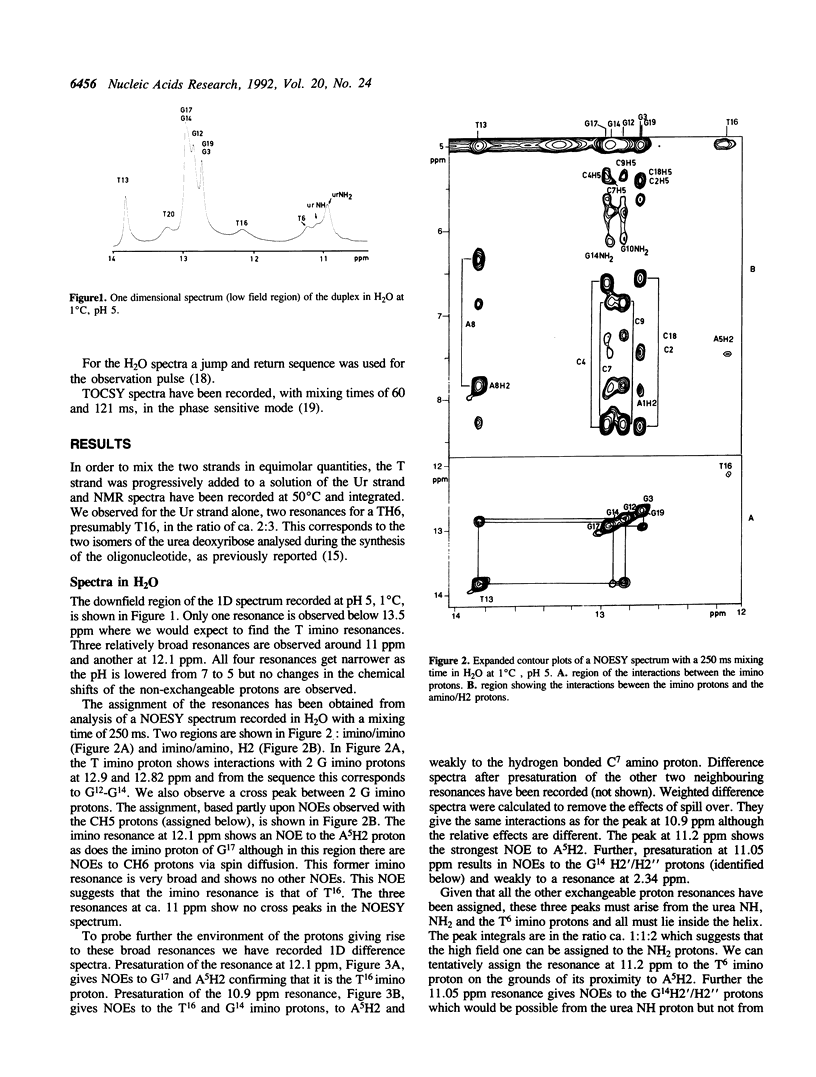
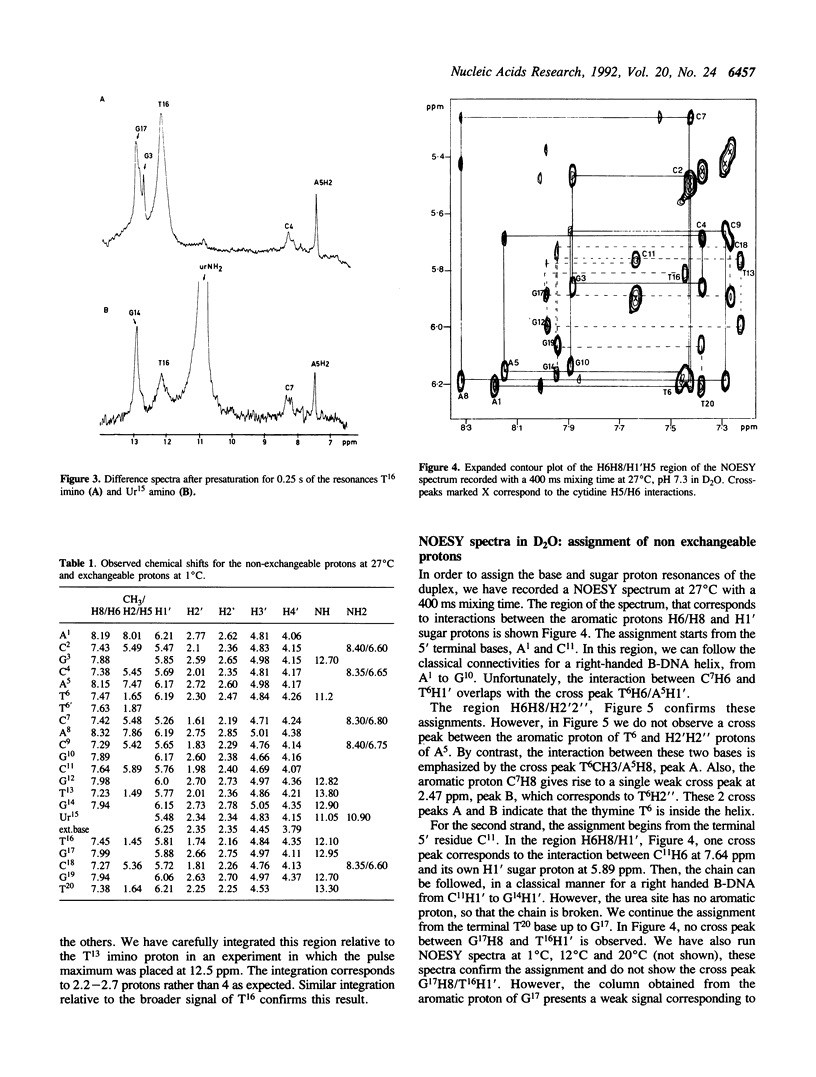
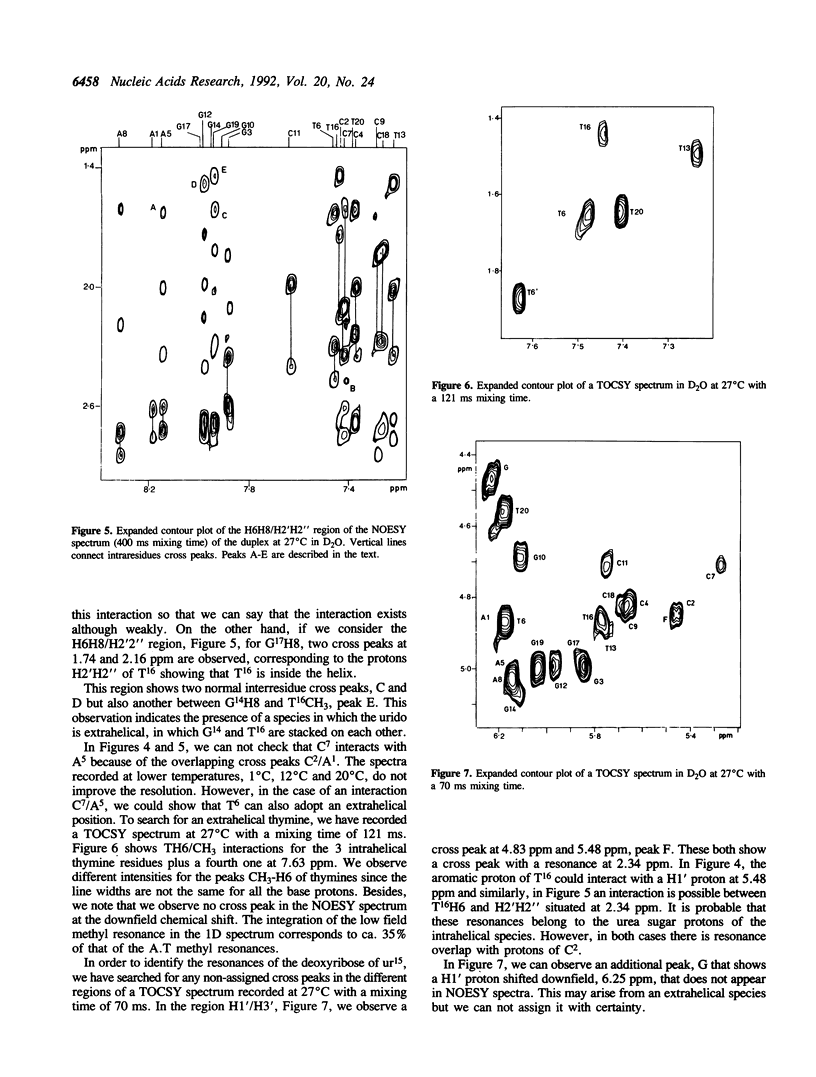
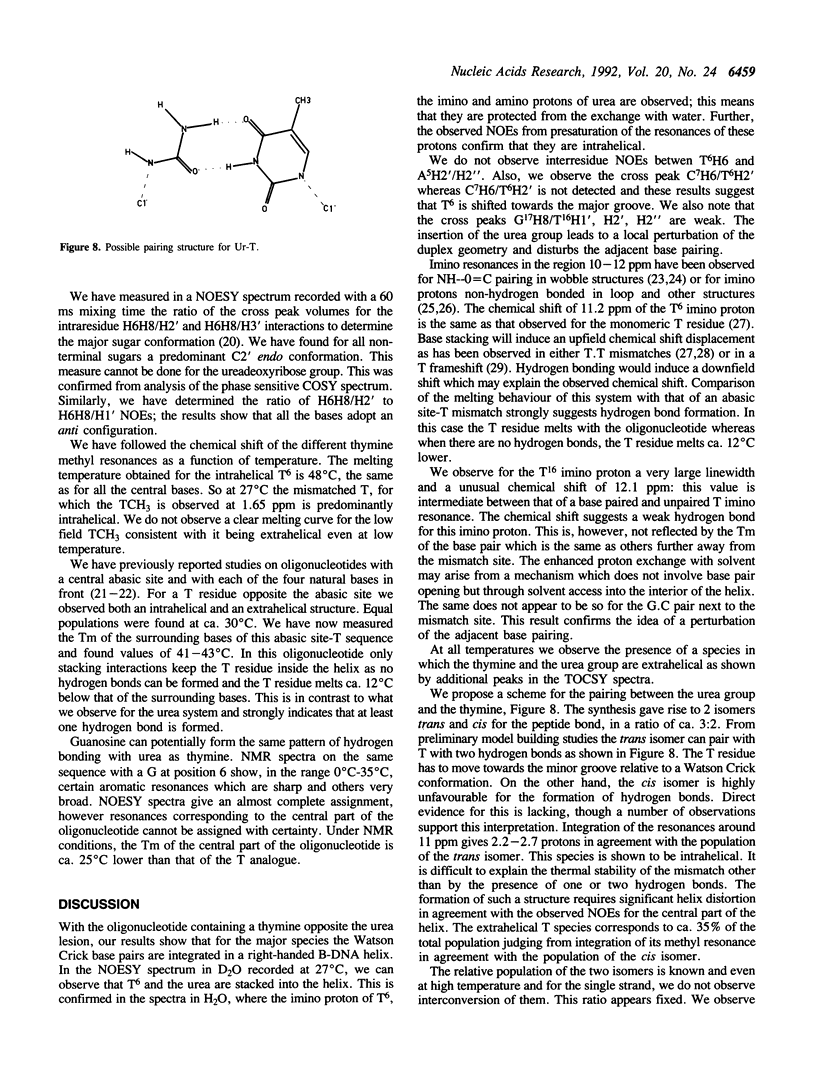
Urea residues are produced by ionizing radiation on thymine residues in DNA. We have studied an oligodeoxynucleotide containing a thymine opposite the urea residue, by one and two dimensional NMR spectroscopy. The urea deoxyribose exists as two isomers with respect to the orientation about the peptide bond. For the trans isomer we find that the thymine and urea site are positioned within the helix and are probably hydrogen bonded. The oligonucleotide adopts a globally B form structure although conformational changes are observed around the mismatch site. A minor species is observed, in which the urea deoxyribose and the opposite base adopt an extrahelical position and this corresponds to the isomer cis for the peptide bond.
Full text
PDF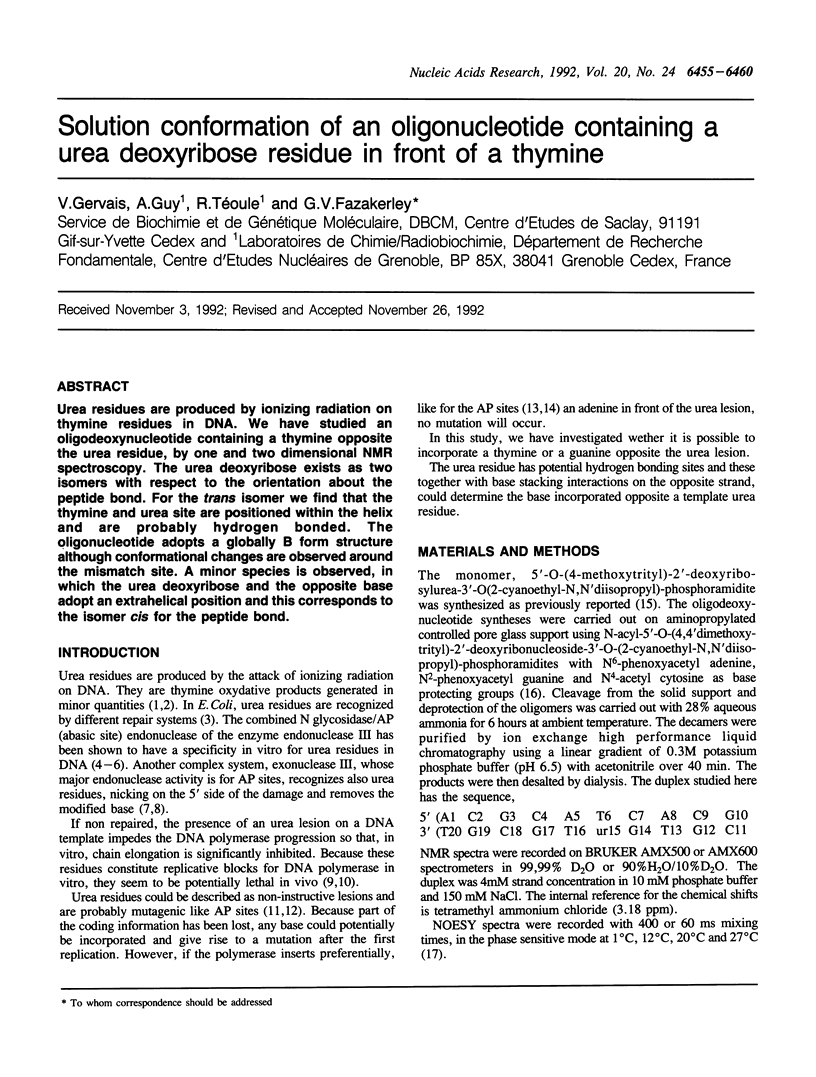

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boulard Y., Gabarro-Arpa J., Cognet J. A., Le Bret M., Guy A., Téoule R., Guschlbauer W., Fazakerley G. V. The solution structure of a DNA hairpin containing a loop of three thymidines determined by nuclear magnetic resonance and molecular mechanics. Nucleic Acids Res. 1991 Oct 11;19(19):5159–5167. doi: 10.1093/nar/19.19.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breimer L., Lindahl T. A DNA glycosylase from Escherichia coli that releases free urea from a polydeoxyribonucleotide containing fragments of base residues. Nucleic Acids Res. 1980 Dec 20;8(24):6199–6211. doi: 10.1093/nar/8.24.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnaux C., Fazakerley G. V., Sowers L. C. An NMR structural study of deaminated base pairs in DNA. Nucleic Acids Res. 1990 Jul 25;18(14):4075–4081. doi: 10.1093/nar/18.14.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis A. G., Haasnoot J. H., den Hartog J. F., de Rooij M., van Boom J. H., Cornelis A. Local destabilisation of a DNA double helix by a T--T wobble pair. Nature. 1979 Sep 20;281(5728):235–236. doi: 10.1038/281235a0. [DOI] [PubMed] [Google Scholar]
- Cuniasse P., Fazakerley G. V., Guschlbauer W., Kaplan B. E., Sowers L. C. The abasic site as a challenge to DNA polymerase. A nuclear magnetic resonance study of G, C and T opposite a model abasic site. J Mol Biol. 1990 May 20;213(2):303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- Cuniasse P., Sowers L. C., Eritja R., Kaplan B., Goodman M. F., Cognet J. A., LeBret M., Guschlbauer W., Fazakerley G. V. An abasic site in DNA. Solution conformation determined by proton NMR and molecular mechanics calculations. Nucleic Acids Res. 1987 Oct 12;15(19):8003–8022. doi: 10.1093/nar/15.19.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide H., Kow Y. W., Wallace S. S. Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res. 1985 Nov 25;13(22):8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katcher H. L., Wallace S. S. Characterization of the Escherichia coli X-ray endonuclease, endonuclease III. Biochemistry. 1983 Aug 16;22(17):4071–4081. doi: 10.1021/bi00286a013. [DOI] [PubMed] [Google Scholar]
- Kouchakdjian M., Li B. F., Swann P. F., Patel D. J. Pyrimidine.pyrimidine base-pair mismatches in DNA. A nuclear magnetic resonance study of T.T pairing at neutral pH and C.C pairing at acidic pH in dodecanucleotide duplexes. J Mol Biol. 1988 Jul 5;202(1):139–155. doi: 10.1016/0022-2836(88)90526-8. [DOI] [PubMed] [Google Scholar]
- Kow Y. W. Mechanism of action of Escherichia coli exonuclease III. Biochemistry. 1989 Apr 18;28(8):3280–3287. doi: 10.1021/bi00434a024. [DOI] [PubMed] [Google Scholar]
- Kow Y. W., Wallace S. S. Exonuclease III recognizes urea residues in oxidized DNA. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8354–8358. doi: 10.1073/pnas.82.24.8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kow Y. W., Wallace S. S. Mechanism of action of Escherichia coli endonuclease III. Biochemistry. 1987 Dec 15;26(25):8200–8206. doi: 10.1021/bi00399a027. [DOI] [PubMed] [Google Scholar]
- Laspia M. F., Wallace S. S. Excision repair of thymine glycols, urea residues, and apurinic sites in Escherichia coli. J Bacteriol. 1988 Aug;170(8):3359–3366. doi: 10.1128/jb.170.8.3359-3366.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A. Apurinic sites as mutagenic intermediates. Cell. 1985 Mar;40(3):483–484. doi: 10.1016/0092-8674(85)90191-6. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Dallas J., Itakura K., Breslauer K. J. Structure, dynamics, and energetics of deoxyguanosine . thymidine wobble base pair formation in the self-complementary d(CGTGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- Randall S. K., Eritja R., Kaplan B. E., Petruska J., Goodman M. F. Nucleotide insertion kinetics opposite abasic lesions in DNA. J Biol Chem. 1987 May 15;262(14):6864–6870. [PubMed] [Google Scholar]
- Sagher D., Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983 Sep 13;22(19):4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Kunkel T. A., Loeb L. A. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc Natl Acad Sci U S A. 1983 Jan;80(2):487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers L. C., Goodman M. F., Eritja R., Kaplan B., Fazakerley G. V. Ionized and wobble base-pairing for bromouracil-guanine in equilibrium under physiological conditions. A nuclear magnetic resonance study on an oligonucleotide containing a bromouracil-guanine base-pair as a function of pH. J Mol Biol. 1989 Jan 20;205(2):437–447. doi: 10.1016/0022-2836(89)90353-7. [DOI] [PubMed] [Google Scholar]
- Teoule R., Bert C., Bonicel A. Thymine fragment damage retained in the DNA polynucleotide chain after gamma irradiation in aerated solutions. II. Radiat Res. 1977 Nov;72(2):190–200. [PubMed] [Google Scholar]
- Wallace S. S. The biological consequences of oxidized DNA bases. Br J Cancer Suppl. 1987 Jun;8:118–128. [PMC free article] [PubMed] [Google Scholar]
- Woodson S. A., Crothers D. M. Preferential location of bulged guanosine internal to a G.C tract by 1H NMR. Biochemistry. 1988 Jan 12;27(1):436–445. doi: 10.1021/bi00401a065. [DOI] [PubMed] [Google Scholar]
- van den Hoogen Y. T., van Beuzekom A. A., van den Elst H., van der Marel G. A., van Boom J. H., Altona C. Extra thymidine stacks into the d(CTGGTGCGG).d(CCGCCCAG) duplex. An NMR and model-building study. Nucleic Acids Res. 1988 Apr 11;16(7):2971–2986. doi: 10.1093/nar/16.7.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]



