ABSTRACT
Microorganisms growing near the boiling point have enormous biotechnological potential but only recently have molecular engineering tools become available for them. We have engineered the hyperthermophilic archaeon Pyrococcus furiosus, which grows optimally at 100°C, to switch its end products of fermentation in a temperature-controlled fashion without the need for chemical inducers. The recombinant strain (LAC) expresses a gene (ldh) encoding lactate dehydrogenase from the moderately thermophilic Caldicellulosiruptor bescii (optimal growth temperature [Topt] of 78°C) controlled by a “cold shock” promoter that is upregulated when cells are transferred from 98°C to 72°C. At 98°C, the LAC strain fermented sugar to produce acetate and hydrogen as end products, and lactate was not detected. When the LAC strain was grown at 72°C, up to 3 mM lactate was produced instead. Expression of a gene from a moderately thermophilic bacterium in a hyperthermophilic archaeon at temperatures at which the hyperthermophile has low metabolic activity provides a new perspective to engineering microorganisms for bioproduct and biofuel formation.
IMPORTANCE Extremely thermostable enzymes from microorganisms that grow near or above the boiling point of water are already used in biotechnology. However, the use of hyperthermophilic microorganisms themselves for biotechnological applications has been limited by the lack of their genetic accessibility. Recently, a genetic system for Pyrococcus furiosus, which grows optimally near 100°C, was developed in our laboratory. In this study, we present the first heterologous protein expression system for a microorganism that grows optimally at 100°C, a first step towards the potential expression of genes involved in biomass degradation or biofuel production in hyperthermophiles. Moreover, we developed the first system for specific gene induction in P. furiosus. As the cold shock promoter for protein expression used in this study is activated at suboptimal growth temperatures of P. furiosus, it is a powerful genetic tool for protein expression with minimal interference of the host’s metabolism and without the need for chemical inducers.
IMPORTANCE
Extremely thermostable enzymes from microorganisms that grow near or above the boiling point of water are already used in biotechnology. However, the use of hyperthermophilic microorganisms themselves for biotechnological applications has been limited by the lack of their genetic accessibility. Recently, a genetic system for Pyrococcus furiosus, which grows optimally near 100°C, was developed in our laboratory. In this study, we present the first heterologous protein expression system for a microorganism that grows optimally at 100°C, a first step towards the potential expression of genes involved in biomass degradation or biofuel production in hyperthermophiles. Moreover, we developed the first system for specific gene induction in P. furiosus. As the cold shock promoter for protein expression used in this study is activated at suboptimal growth temperatures of P. furiosus, it is a powerful genetic tool for protein expression with minimal interference of the host’s metabolism and without the need for chemical inducers.
Introduction
Since the discovery of hyperthermophiles in the 1980s (1), hyperthermophiles have attracted a great deal of attention due to their ability to grow optimally at temperatures above 80°C. Virtually all are classified within the archaeal domain rather than the bacterial domain. In addition to their evolutionary implications, hyperthermostable enzymes are of high biotechnological interest (2–4), since many industrial processes are facilitated by elevated temperatures and organisms that grow under such conditions can be used without risk of contamination (3). Although the ability to metabolically engineer microorganisms is a prerequisite for their utility as whole-cell biocatalysts, the genetic manipulation of hyperthermophiles is a very recent development. Targeted modifications of the chromosome have so far been reported only for those microorganisms growing optimally near 80°C or so, which include Sulfolobus acidocaldarius (optimal growth temperature [Topt] of 80°C) and the related species Sulfolobus solfataricus (Topt of 75°C) (5, 6) and Thermococcus kodakarensis (Topt of 85°C) (7). Very recently, however, we developed a genetic system for Pyrococcus furiosus, the first for an organism that grows optimally near 100°C (8). P. furiosus is one of the best-studied hyperthermophiles, belonging to the same family as T. kodakarensis but with a much higher optimal growth temperature (9). P. furiosus is a strict anaerobe and obtains carbon and energy for growth by the fermentation of carbohydrates and peptides with organic acids, CO2, and H2 as end products (10).
A variant of wild-type P. furiosus (DSM 3638) was recently isolated. This strain takes up DNA very efficiently to create a genetically modified P. furiosus strain (COM1) by the targeted and markerless deletion of the pyrF gene that encodes orotidine-5′-monophosphate (OMP) decarboxylase, which is essential for uracil biosynthesis (8). Moreover, linear DNA could be used to transform P. furiosus, and the DNA was integrated into the chromosome by homologous recombination. The COM1 strain was also used to overexpress a minimal form of the cytoplasmic hydrogenase (SHI) of P. furiosus (11). This study utilized a new selective marker for P. furiosus based on previous work with T. kodakarensis (12) in which the deletion of the gene (pdaD) encoding pyruvoyl-dependent arginine decarboxylase led to an auxotrophy for agmatine, an essential intermediate in polyamine biosynthesis (13) and in translation (14). Agmatine is not present in complex growth substrates, such as yeast extract and casein, thus allowing the use of rich media for P. furiosus mutant selection.
The availability of a genetic system for an organism growing optimally at 100°C opens up the possibility of engineering the temperature-dependent heterologous expression of genes encoding enzymes that are active at suboptimal growth temperatures for the host. Depending on the temperature, the host organism can have lower metabolic activity or be virtually inactive. For example, the generation time of P. furiosus increases from less than 1 h at 98°C to about 7 h at 72°C (15), with little growth below 65°C (9). Production of enzymes optimally active near 70°C or so could give P. furiosus new metabolic capabilities at this temperature that it does not have at the optimum near 100°C where the heterologously produced enzymes would likely be inactive. Similarly, at even lower temperatures (≤60°C), P. furiosus could be a nonmetabolizing host, and chemical conversions could be accomplished only by the heterologously produced enzymes. While heterologous gene expression has already been reported using the related hyperthermophile, T. kodakarensis (16, 17), this involved genes from archaeal species that grow at temperatures comparable to that of T. kodakarensis. In this study, our goal was to heterologously express in P. furiosus a gene from a bacterium that grows at significantly lower temperature and demonstrate temperature-dependent generation of a new end product in the absence of any chemical inducer for gene expression.
The use of temperature to change metabolism might be feasible if, at the higher temperature, the heterologously produced enzymes are unstable and degraded by the hyperthermophile. However, it would obviously be more efficient if the genes that encode the foreign proteins were transcribed only at suboptimal temperatures. In a previous study, we showed that when P. furiosus was grown at 72°C, rather than near the optimum at 98°C, or if cells growing at 98°C were “cold shocked” by transferring them to 72°C, the expression of numerous genes was upregulated significantly (15). One of the most highly expressed genes encoded a membrane-bound glycoprotein, the so-called cold-induced protein A, or CipA (PF0190), which was upregulated 26-fold (as determined by quantitative PCR [qPCR] analysis) at 72°C (15). The cipA promoter, PcipA, was therefore chosen for the attempt to induce expression of a bacterial gene at suboptimal growth temperature for the host.
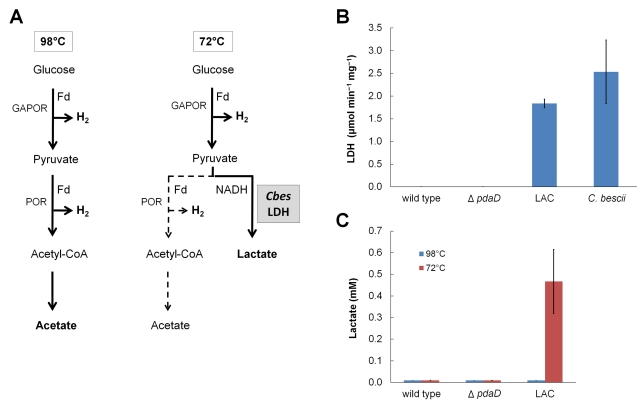
For proof of principle of a temperature-dependent metabolic switch in P. furiosus, we selected a bacterial gene that has no homolog in the P. furiosus genome and one that is involved in the metabolism of a compound that P. furiosus is not known to produce. The anaerobic bacterium Caldicellulosiruptor bescii grows optimally at 78°C by sugar fermentation and produces lactate at millimolar concentrations as the main end product (18). Lactate is generated by the reduction of pyruvate catalyzed by an NADH-dependent lactate dehydrogenase (LDH) encoded by Cbes_1918 (ldh). In contrast, while P. furiosus also ferments sugars to pyruvate, its genome does not contain a gene encoding an LDH homolog, and the organism oxidizes pyruvate by pyruvate ferredoxin oxidoreductase to produce acetate, CO2, and H2 as the primary products (Fig. 1A). The goal was therefore to express the LDH gene of C. bescii in P. furiosus under control of the PcipA promoter and determine whether any lactate is produced during growth at 72°C, but not at 98°C (Fig. 1A).
FIG 1 .
Recombinant expression of lactate dehydrogenase (LDH) in P. furiosus strain LAC changes its fermentation pattern. (A) Concept of temperature-dependent switch in end product formation by P. furiosus. Abbreviations: GAPOR, glyceraldehyde-3-phosphate ferredoxin oxidoreductase; POR, pyruvate ferredoxin oxidoreductase; Fd, ferredoxin; acetyl-CoA, acetyl coenzyme A; Cbes LDH, C. bescii LDH. (B) Specific activity of lactate dehydrogenase in the protein extract of C. bescii DSM 6725, P. furiosus DSM 3638 (wild type), P. furiosus ΔpdaD host strain, and P. furiosus LAC obtained from 400-ml batch cultures. (C) Lactate production in the same P. furiosus cultures. Values given are averages ± standard deviations (SD) (error bars) of three independent biological cultures.
RESULTS
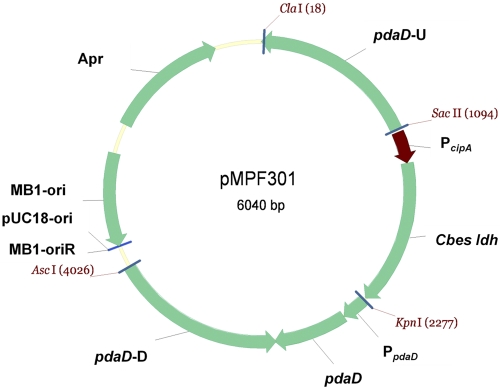
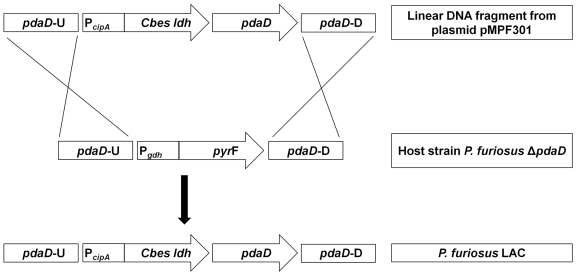
To construct a P. furiosus strain containing the C. bescii ldh gene under control of the PcipA promoter, the PcipACbes-ldh gene fusion was cloned in the plasmid vector pSPF300 in Escherichia coli (Fig. 2 and Table 1). The agmatine-requiring P. furiosus mutant strain, ΔpdaD strain, was used as the host (11). This strain is deficient in agmatine biosynthesis, as the pdaA gene is replaced by pyrF, an essential gene for uracil biosynthesis (Table 1). The linearized plasmid containing PcipACbes-ldh (Fig. 2) was used to complement the pdaD gene into the P. furiosus chromosome by replacing the pyrF gene by homologous recombination. The resulting transformants (ΔpyrF) required uracil but did not require agmatine for growth (Fig. 3). Plasmid integration was confirmed by PCR and for one colony by DNA sequencing. The recombinant P. furiosus strain was named LAC (Table 1). To investigate the expression of C. bescii ldh in P. furiosus and the production of lactate, the LAC strain was grown in batch culture under three different conditions: (i) in closed, static cultures (400-ml scale) at 72°C and at 98°C with no pH control; (ii) in Ar-sparged, stirred cultures (15-liter scale) at 72°C with pH control; and (iii) in Ar-sparged, stirred cultures at 94°C with a pH control (15-liter scale) followed by rapid cooling of the culture to 72°C within 10 min (cold shock).
FIG 2 .
Plasmid vector pMPF301 containing the pdaD PcipACbes-ldh cassette, 1-kb upstream and downstream flanking regions of the pdaD gene and the apr gene as a selective marker in Escherichia coli (apramycin resistance). Plasmid diagrams were constructed using Vector NTI software (Invitrogen).
TABLE 1 .
Pyrococcus furiosus strains used in this study
| Strain | Relevant genotype | Parent strain | Requirement | Source |
|---|---|---|---|---|
| DSM 3638 | Wild type | NAa | NA | 9 |
| COM1 | ΔpyrF | DSM 3638 | Uracil (20 µM) | 8 |
| ΔpdaD | ΔpyrF ΔpdaD::PgdhpyrF | COM1 | Agmatine (4 mM) | This study |
| LAC | ΔpyrF ΔpdaD::pdaD PcipACbes-ldh | ΔpdaD | Uracil (20 µM) | This study |
NA, not available.
FIG 3 .
Cloning strategy for the mutant strain P. furiosus LAC. The fusion product PcipACbes-ldh was obtained by overlapping PCR and integrated into vector pSPF300 (11). The new vector, pMPF301 (Fig. 2), additionally carried the pdaD gene essential for agmatine biosynthesis and 1-kb upstream and downstream flanking regions of the pdaD gene. Linearized DNA was used for transformation of the P. furiosus ΔpdaD host strain. The pdaD PcipACbes-ldh cassette integrated into the genome by homologous recombination, replacing the PgdhpyrF cassette. Therefore, the resulting new strain, P. furiosus LAC, exhibits a uracil auxotrophy, but does not, in contrast to the host, require agmatine for growth.
The recombinant strains of P. furiosus were grown at 98°C and at 72°C in closed, static cultures without a pH control. The ΔpdaD and LAC strains grew at 98°C to comparable cell densities after 12 h (>108 cells/ml, >50 µg protein/ml), but at 72°C growth was still very poor even after 45 h (<108 cells/ml, <20 µg protein/ml). Interestingly, cells changed their morphology at 72°C, becoming light refracting, and larger (up to 5 µm), in accordance with the upregulation of the expression of a wide range of genes, including those encoding numerous membrane-bound proteins (15). To determine whether recombinant protein production was successful, the cells were lysed by sonication, and the activity of NAD-dependent lactate dehydrogenase (LDH) was determined in cell-free extracts at 75°C. The activity of NAD-dependent glutamate dehydrogenase (GDH), a well-characterized cytoplasmic enzyme of P. furiosus (19), was used as a control. The wild-type, ΔpdaD, and LAC strains had comparable specific activities of GDH when grown at 72°C (0.09 to 0.11 U mg−1) and when grown at 98°C (0.14 to 0.27 U mg−1). LDH activity was not detected (<0.05 U mg−1) in cell-free extracts of any strain grown at 98°C or in extracts of cells of the wild-type and parent strains grown at 72°C. However, extracts of the LAC strain grown at 72°C had high LDH activity (1.8 ± 0.1 U mg−1). C. bescii ldh is the first bacterial gene to be expressed and to yield an active enzyme in P. furiosus (Fig. 1A). Remarkably, the specific activity of LDH in P. furiosus was comparable to that measured in cell-free extracts of cellobiose-grown C. bescii (2.5 ± 0.7 U mg−1; Fig. 1B), conditions under which C. bescii produces lactate as the major metabolic product. Moreover, while lactate was not detected (<20 µM) in the growth medium of any of the P. furiosus strains grown at 98°C or in the wild-type and parent strains grown at 72°C, the medium of the LAC strain contained 0.47 ± 0.14 mM lactate (Fig. 1C).
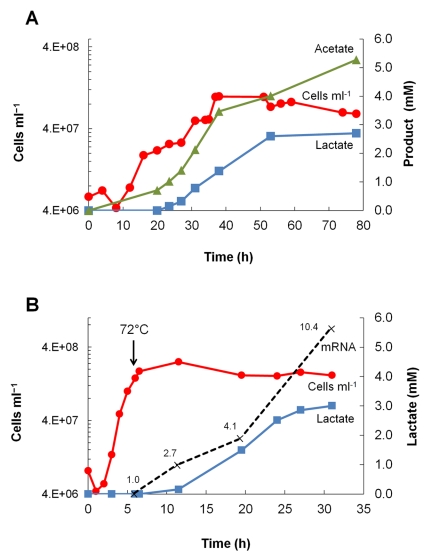
Growth of the P. furiosus LAC strain at 72°C was scaled up in a stirred, pH-controlled fermentor (15 liters), conditions under which good growth of P. furiosus is obtained even at this low temperature (15). The organism reached a maximum cell density after approximately 50 h (1 × 108 ml−1, 60 µg ml−1) (Fig. 4A) and remained stable in stationary phase for a further 28 h. The specific activity of LDH in the cell extract was the same as that measured in the small-scale cultures and remained unchanged (2.0 ± 0.4 U mg−1) in exponential (at 38 h [Fig. 4A]), early stationary (at 59 h), and late stationary (at 78 h) growth phase. Consequently, the amount of lactate produced paralleled the cell density. This reached a concentration near 3 mM in stationary phase, which was approximately half of the concentration of acetate that was produced (Fig. 4A). Therefore, we conclude that the P. furiosus LAC strain is robust and can be cultivated in large volumes with a specific LDH activity comparable to that measured in uncontrolled batch cultures but with higher yields of both total protein and lactate.
FIG 4 .
(A and B) Lactate production (blue squares), acetate production (green triangles), cell density (red circles), and relative mRNA fold expression levels (broken lines) in 15-liter fermentor cultures of P. furiosus LAC. One culture was grown at 72°C (A), while another culture was grown at 94°C and rapidly cooled to 72°C after a cell density of 1.5 × 108 was reached (indicated by the black arrow) (B). After the temperature switch, higher mRNA levels for the heterologous gene Cbes-ldh, high specific activity of lactate dehydrogenase, and a high rate of lactate formation were observed.
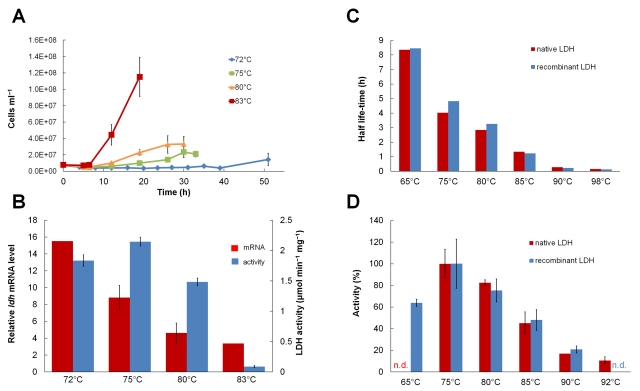
LDH activity and transcription of the C. bescii LDH gene were measured in the P. furiosus LAC strain over the growth temperature range from 72 to 83°C. Transcription of the ldh gene from C. bescii (Cbes-ldh) is controlled by the PcipA promoter, and the corresponding CipA protein was reported previously to be produced at 72°C (15), although no data are available on its expression at other temperatures. While the growth rate of P. furiosus LAC drastically increases with increasing temperature, the highest relative Cbes-ldh mRNA level was found in cultures grown at 72°C (Fig. 5A). In addition, the highest specific LDH activities were detected in cultures grown in the 72 to 75°C range (Fig. 5B). Obviously, the promoter induces transcription, and this leads to protein production at those low temperatures and to reasonable amounts and activities of the recombinant protein, even though the culture itself exhibited relatively poor growth (Fig. 5A).
FIG 5 .
Recombinant expression and activity of C. bescii lactate dehydrogenase in P. furiosus at different temperatures. (A and B) Cell density (A) and relative Cbes-ldh mRNA level and specific activity of lactate dehydrogenase (LDH) (B) in cell extracts of P. furiosus LAC grown at different temperatures (for 72°C, n = 2; for 98°C, n = 1). Although growth was negligible at 72°C and 75°C, the highest ldh mRNA level and lactate dehydrogenase activities were observed at these growth temperatures. (C and D) Thermostability (C) and temperature dependence of lactate dehydrogenase activity (D) in protein extracts of C. bescii DSM 6725 (native LDH) and P. furiosus strain LAC (recombinant LDH) grown at 75°C and harvested in the stationary phase. Values given are averages ± SD of three independent biological cultures (B) or three independent enzymatic measurements (D), unless denoted otherwise. n.d., not determined.
To confirm that producing C. bescii LDH in P. furiosus at 72°C was comparable to producing the enzyme in C. bescii, we determined the properties of the recombinant LDH produced in P. furiosus with those of the native LDH produced in C. bescii (Fig. 5C and D). Both forms of the enzyme had temperature optima near 75°C, close to the optimal growth temperature of C. bescii (18), and both forms had only barely detectable activity above 90°C, which is above the maximal growth temperature of C. bescii (90°C). Moreover, both forms of the enzyme had a relatively long half-life of about 5 h at the temperature optimum (75°C). Such stability is comparable to that of the most thermostable LDH previously reported, the enzyme from Thermotoga maritima, an organism that has growth properties similar to that of C. bescii (Topt of 80°C) (20).
In terms of temperature-dependent bioprocessing, a useful approach would be to grow P. furiosus to a high cell density under conditions that are nearly optimal for growth in the absence of heterologous gene expression and then cold shock the culture for bioproduction generation as a result of heterologous gene expression. The LAC strain was grown at 94°C, conditions known not to lead to detectable C. bescii LDH activity or detectable amounts of ldh mRNA, to a cell density of 2 × 108 ml−1, and the culture was rapidly cooled to 72°C (over 10 min). At this point, lactate could not be detected in the culture medium. However, 5 h after the switch, mRNA corresponding to C. bescii ldh was detected and lactate was measured in the growth medium (Fig. 4B). Moreover, the concentration of both ldh mRNA (relative to the level of the gamma subunit of the constitutively expressed pyruvate-ferredoxin oxidoreductase) and lactate increased over the following 25 h (Fig. 4B), leading to the production of approximately 3 mM lactate. Cells contained C. bescii LDH with a specific activity of 1.9 ± 0.6 U mg−1. The latter value is comparable to those determined with cells grown in batch culture at 72°C (Fig. 1B), showing that cold shock bioproduct generation is a valid experimental approach.
DISCUSSION
We have demonstrated that a microorganism (in this case, from the domain Archaea) that grows optimally near 100°C transcribes mRNA and produces the corresponding enzyme, LDH, from another microorganism (in this case, from the domain Bacteria) that grows optimally at 78°C but does so only under the conditions where the foreign protein shows significant catalytic activity, namely, below 80°C. The activity of the heterologously expressed LDH in P. furiosus might be the result of processes at both the RNA and protein level. First, the relative ldh mRNA level increased due to the cold-induced promoter, with an upregulation about 10-fold at 72°C. Although cold-responsive promoters have been previously reported in mesophilic bacteria, they were utilized to facilitate protein folding at low temperature (reference 21 and references therein) rather than to exploit temperature induction for biotechnological purposes such as biofuel production. Second, the stability of the protein and its activity decreased with increasing temperature above 80°C. Interestingly, only two LDHs have been previously characterized from thermophiles, and they are homooligomeric enzymes (20, 22). The finding that C. bescii LDH produced in P. furiosus and C. bescii were similarly thermostable suggests that the P. furiosus version was correctly assembled into its multimeric form.
Recombinant production of the C. bescii LDH represents the first bacterial protein to be expressed in a hyperthermophilic microorganism from the domain Archaea and one of the first heterologously expressed proteins in archaea in general (16, 17, 23). It provides interesting options for the future production of other bacterial proteins, particularly ones involved with lignocellulosic biomass degradation, since an archaeon that can degrade crystalline cellulose has yet to be reported (2, 3). Indeed, the lactate-producing strain described here offers a potential platform to enhance the temperature limit for lactate production from lignocellulosic substrates, a process of industrial interest (24).
P. furiosus has therefore been metabolically engineered to change its end products of fermentation without the need for the addition of any chemical inducer, and thus any indirect impact on its metabolism or the accumulation of inducer products. Moreover, we demonstrate that temperature is an effective means of regulation even using cells grown rapidly to high cell density, particularly since the corresponding mRNA, enzyme activity, or product (lactate) could not be detected until the temperature was lowered. The unusual cold shock response of P. furiosus could be a powerful tool for biotechnological applications.
MATERIALS AND METHODS
Strains and media.
Pyrococcus furiosus strains used in this study are listed in Table 1. In the transformation experiments, P. furiosus (DSM 3638) was cultured with 5 g liter−1 maltose as the primary electron donor on liquid and solid complex medium as previously described (8). In all other experiments, the same medium was used, except that it contained no casein, but a yeast extract concentration of 2 g liter−1 (15). For the cultivation of the ΔpdaD mutant strain, 4 mM agmatine (Sigma Chemical, St. Louis, MO) was added, while the medium for the COM1 and LAC strains was supplemented with 20 µM uracil (Table 1). Adapted growth at 72°C and the temperature shock experiment were performed in a 20-liter custom fermentor as described previously (15). In the temperature shock experiment, the whole culture (15 liters) was rapidly cooled from 94°C to 72°C within 10 min. Caldicellulosiruptor bescii was grown on complex medium with 5 g liter−1 cellobiose as an electron donor as described previously (25). Culture growth was in general followed by cell counting and by determination of protein concentration in subsamples.
Genetic manipulations.
Extraction of DNA from C. bescii was performed by the method of Zhou et al. (26). Extraction of DNA from P. furiosus, transformation of P. furiosus, and selection of genetically modified strains were performed as previously described (8). P. furiosus COM1 served as the parent strain for genetic manipulations. A deletion of the pyruvoyl-dependent arginine decarboxylase (pdaD) gene (PF1623) was achieved by homologous recombination with the PgdhpyrF cassette (11). The resulting strain, P. furiosus ΔpdaD strain, was used as the parent strain for the heterologous expression of the putative l-lactate dehydrogenase of C. bescii (Cbes1918; Cbes-ldh). Cbes-ldh was amplified by PCR using the primer set Cbes1918-F (F stands for forward) and Cbes1918-KpnI-R (R stands for reverse). The cold-induced promoter PcipA was amplified from genomic DNA from P. furiosus DSM3638 with the primer set PcipA-SacII-F and PcipA-Cbes1918-R. Finally, the fusion product PcipACbes1918 was obtained by overlapping PCR using both products from the PCRs above and the primers PcipA-SacII-F and Cbes1918-KpnI-R. The fusion product was introduced between the SacII site and the KpnI site of the plasmid vector pSPF300 (11), which additionally contained the pdaD gene and 1 kb upstream and downstream regions of pdaD. The resulting plasmid pMPF301 (Fig. 2) was amplified in Escherichia coli XL1 Blue-MRF′ (Stratagene, now Agilent Technologies, Santa Clara, CA) applying general genetic techniques (27). The plasmid was digested with the ClaI and AscI restriction endonucleases, the larger fragment including the pdaD PcipACbes-ldh cassette was purified with the Strataprep DNA gel extraction kit (Agilent Technologies) and subsequently used for transformation of the P. furiosus ΔpdaD strain (Fig. 3). Verification of the insertion of the pdaD PcipACbes-ldh cassette into the chromosome was achieved by PCR with the primer set PF1623L-F and PF1623R-R located upstream and downstream of the cassette and subsequent sequencing. All primers used for PCR are listed in Table 2.
TABLE 2 .
Primers used in this study for PCR amplification and qPCR
| Primer | Sequence (5′–3′) | Source |
|---|---|---|
| PcipA-SacII-F | GAATCCCCGCGGTGACCTTTTATCCATTACTAACTTGC | This study |
| PcipA-Cbes1918-R | CAATAATTACAATTTTACCCGGTTTTCTCATTGCATATCACCTGCCAGGTATCTC | This study |
| Cbes1918-F | CCTGGCAGGTGATATGCAATGAGAAAACCGGGTAAAATTGTAATTATTGGAAC | This study |
| Cbes1918-KpnI-R | TCGGTTGGTACCAGCCTCCTATTATAGTTTTAAAGACTCTATCACAC | This study |
| PF1623L-F | GGAGCTCTGTTGCTTCTGCTAGAG | This study |
| PF1623R-R | CTTTTCACCTACTATCTGCTCAAATGC | This study |
| PF0971-qF | CGTTGTTGTTGTGCTAGATCC | 8 |
| PF0971-qR | GATGGCTTCCTCTATGCTCTC | 8 |
| Cbes1918-qF | GGGCGAACATGGAGACAGTGAAATTG | This study |
| Cbes1918-qR | GCCAATGCAATGGCGTAATATGTTGC | This study |
Preparation of cell extracts and enzyme assays.
P. furiosus and C. bescii cells were harvested by centrifugation for 10 min at 6,000 × g. C. bescii cells were resuspended in 50 mM Tris (pH 8) and disrupted by sonication (five times, 2 min each time, maximum of 36 W and discontinuous operation at 50% of time). The P. furiosus cells were lysed by osmotic shock in 50 mM Tris HCl (pH 8.0) and 2 mM sodium dithionite. The lysis buffer contained 50 mg/ml DNase I (Sigma) to decrease the viscosity of the protein extract. Fractionation of the resulting protein extract into the soluble (cytoplasmic) fraction and the membrane fraction was achieved by ultracentrifugation at 100,000 × g for 1 h. The membrane fraction was washed once with 50 mM Tris (pH 8.0) in order to minimize contamination with soluble proteins. Lactate dehydrogenase (LDH) (EC 1.1.1.27) activity was determined photometrically by the oxidation of NADH (340 nm) concomitant with lactate formation according to the following chemical equation: NADH + pyruvate + H+ → NAD+ + lactate. The assays were performed aerobically in closed glass cuvettes at 75°C, which contained 2.5 mM NADH in 50 mM sodium phosphate buffer (pH 7.0). The rate of nonspecific oxidation of NADH was determined before the reaction was started by the addition of 5 mM pyruvate. As internal controls for the quality of the P. furiosus protein extracts, glutamate dehydrogenase (GDH) (EC 1.4.1.2) activity was routinely measured by the formation of NADPH (340 nm) according to the following chemical equation: NADP+ + glutamate + H2O → 2-oxoglutarate + NH4+ + NADPH. The GDH assay was the same as for LDH except that NADH was exchanged for NADP+ (0.25 mM), and pyruvate was exchanged for glutamate (5 mM). The protein content of the cell-free extracts were determined by the method of Bradford (28).
RNA extraction and quantitative PCR.
Cells were harvested for RNA extraction in the late logarithmic to early stationary phase of the growth curve unless noted otherwise. Cells were centrifuged for 10 min at 6,000 × g and frozen until further processing. RNA was extracted using the Absolute RNA miniprep kit (Agilent Technologies), including a DNA digestion step with Turbo DNase (Ambion, Austin, TX) for 30 min at 37°C. cDNA was prepared using the Affinity Script cDNA synthesis kit (Agilent Technologies). All quantitative reverse transcription-PCRs (qRT-PCRs) were performed with an Mx3000P instrument (Stratagene), using the Brilliant Sybr green QPCR master mix (Agilent Technologies). The gamma subunit of the constitutively transcribed gene encoding the pyruvate-ferredoxin oxidoreductase (29) (PF0971) was used as an internal control to calculate the relative mRNA level of Cbes-ldh. Primers for qRT-PCR were designed using the VectorNTI software (Invitrogen). The amplicon sizes were 194 bp and 267 bp for Cbes-ldh and PF0971, respectively. Primers were tested for nonspecific products, and all experiments included controls without the addition of reverse transcriptase in the cDNA synthesis step to test for DNA contamination. The comparative cycle threshold method was used to analyze the resulting data, which are expressed as a ratio of gene expression change (n-fold). All primers used in qRT-PCR experiments are listed in Table 2.
Chemical analyses.
l-Lactic acid was determined by using the Megazyme l-lactic assay kit (Megazyme, Wicklow, Ireland). Acetate was determined by high-performance liquid chromatography (HPLC) on a model 2690 separations module (Waters, Milford, MA) equipped with an Aminex HPX-87H column (300 mm by 7.8 mm; Bio-Rad, Hercules, CA) and a photodiode array detector (model 996; Waters). The system was operated with 5 mM H2SO4 as the eluent at a flow rate of 0.6 ml min−1. Samples for HPLC were acidified with 0.1 M H2SO4 and centrifuged before analysis to remove particles. Hydrogen was determined on a GC-8A gas chromatograph (Shimadzu, Kyoto, Japan) equipped with a thermal conductivity detector and a molecular sieve column (model 5A 80/100; Alltech, Deerfield, IL) with argon as the carrier gas.
ACKNOWLEDGMENTS
This work was supported by a grant (DE-PS02-06ER64304) from the Bioenergy Science Center (BESC), Oak Ridge National Laboratory, a U.S. Department of Energy (DOE) Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science, and by a grant (DE-AR0000081) from the ARPA-E Electrofuels Program of the U.S. Department of Energy.
We thank Christopher Hopkins, Sanjeev K. Chandrayan, Gerrit J. Schut, Irina Kataeva, Angeli L. Menon, Andrew Lancaster, and Farris Poole for helpful discussions.
M.B. and J.S. performed the genetic engineering. M.B. designed and performed experiments, analyzed the data, and wrote the manuscript. M.W.W.A. initiated the project, coordinated the research, and wrote the manuscript.
Footnotes
Citation Basen M, Sun J, Adams MWW. 2012. Engineering a hyperthermophilic archaeon for temperature-dependent product formation. mBio 3(2):e00053-12. doi:10.1128/mBio.00053-12.
REFERENCES
- 1. Stetter KO. 2006. History of discovery of the first hyperthermophiles. Extremophiles 10:357–362 [DOI] [PubMed] [Google Scholar]
- 2. Barnard D, Casanueva A, Tuffin M, Cowan D. 2010. Extremophiles in biofuel synthesis. Environ. Technol. 31:871–888 [DOI] [PubMed] [Google Scholar]
- 3. Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. 2008. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr. Opin. Biotechnol. 19:210–217 [DOI] [PubMed] [Google Scholar]
- 4. Atomi H, Sato T, Kanai T. 2011. Application of hyperthermophiles and their enzymes. Curr. Opin. Biotechnol. 22:618–626 [DOI] [PubMed] [Google Scholar]
- 5. Schelert J, et al. 2004. Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J. Bacteriol. 186:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wagner M, et al. 2009. Expanding and understanding the genetic toolbox of the hyperthermophilic genus Sulfolobus. Biochem. Soc. Trans. 37:97–101 [DOI] [PubMed] [Google Scholar]
- 7. Sato T, Fukui T, Atomi H, Imanaka T. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lipscomb GL, et al. 2011. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 77:2232–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fiala G, Stetter KO. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145:56–61 [Google Scholar]
- 10. Chou CJ, Jenney FE, Adams MW, Kelly RM. 2008. Hydrogenesis in hyperthermophilic microorganisms: implications for biofuels. Metab. Eng. 10:394–404 [DOI] [PubMed] [Google Scholar]
- 11. Hopkins RC, et al. 2011. Homologous expression of a subcomplex of Pyrococcus furiosus hydrogenase that interacts with pyruvate ferredoxin oxidoreductase. PLoS One 6:e26569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Santangelo TJ, Cubonová L, Reeve JN. 2010. Thermococcus kodakarensis genetics: TK1827-encoded beta-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl. Environ. Microbiol. 76:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fukuda W, Morimoto N, Imanaka T, Fujiwara S. 2008. Agmatine is essential for the cell growth of Thermococcus kodakaraensis. FEMS Microbiol. Lett. 287:113–120 [DOI] [PubMed] [Google Scholar]
- 14. Ikeuchi Y, et al. 2010. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat. Chem. Biol. 6:277–282 [DOI] [PubMed] [Google Scholar]
- 15. Weinberg MV, Schut GJ, Brehm S, Datta S, Adams MW. 2005. Cold shock of a hyperthermophilic archaeon: Pyrococcus furiosus exhibits multiple responses to a suboptimal growth temperature with a key role for membrane-bound glycoproteins. J. Bacteriol. 187:336–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matsumi R, Manabe K, Fukui T, Atomi H, Imanaka T. 2007. Disruption of a sugar transporter gene cluster in a hyperthermophilic archaeon using a host-marker system based on antibiotic resistance. J. Bacteriol. 189:2683–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takemasa R, Yokooji Y, Yamatsu A, Atomi H, Imanaka T. 2011. Thermococcus kodakarensis as a host for gene expression and protein secretion. Appl. Environ. Microbiol. 77:2392–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang SJ, et al. 2009. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl. Environ. Microbiol. 75:4762–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adams MW. 1993. Enzymes and proteins from organisms that grow near and above 100°C. Annu. Rev. Microbiol. 47:627–658 [DOI] [PubMed] [Google Scholar]
- 20. Ostendorp R, Auerbach G, Jaenicke R. 1996. Extremely thermostable L(+)-lactate dehydrogenase from Thermotoga maritima: cloning, characterization, and crystallization of the recombinant enzyme in its tetrameric and octameric state. Protein. Sci. 5:862–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jana S, Deb JK. 2005. Strategies for efficient production of heterologous proteins in Escherichia coli. Appl. Microbiol. Biotechnol. 67:289–298 [DOI] [PubMed] [Google Scholar]
- 22. Zhou Q, Shao WL. 2010. Molecular genetic characterization of the thermostable L-lactate dehydrogenase gene (ldhL) of Thermoanaerobacter ethanolicus JW200 and biochemical characterization of the enzyme. Biochemistry (Mosc) 75:526–530 [DOI] [PubMed] [Google Scholar]
- 23. Lessner DJ, Lhu L, Wahal CS, Ferry JG. 2010. An engineered methanogenic pathway derived from the domains Bacteria and Archaea. mBio 1:e00243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang QZ, Ingram LO, Shanmugam KT. 2011. Evolution of D-lactate dehydrogenase activity from glycerol dehydrogenase and its utility for D-lactate production from lignocellulose. Proc. Natl. Acad. Sci. U. S. A. 108:18920–18925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang SJ, et al. 2010. Classification of “Anaerocellum thermophilum” strain DSM 6725 as Caldicellulosiruptor bescii sp. nov. Int. J. Syst. Evol. Microbiol. 60:2011–2015 [DOI] [PubMed] [Google Scholar]
- 26. Zhou JZ, Fries MR, Cheesanford JC, Tiedje JM. 1995. Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth of toluene and description of Azoarcus tolulyticus sp. nov. Int. J. Syst. Bacteriol. 45:500–506 [DOI] [PubMed] [Google Scholar]
- 27. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor: Press, Cold Spring Harbor, NY. [Google Scholar]
- 28. Bradford MM. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 29. Schut GJ, Brehm SD, Datta S, Adams MW. 2003. Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J. Bacteriol. 185:3935–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]