Abstract
The residence times of individual hydration water molecules in the major and minor grooves of DNA were measured by nuclear magnetic resonance (NMR) spectroscopy in aqueous solutions of d-(CGCGAATTCGCG)2 and d-(AAAAATTTTT)2. The experimental observations were nuclear Overhauser effects (NOE) between water protons and the protons of the DNA. The positive sign of NOEs with the thymine methyl groups shows that the residence times of the hydration water molecules near these protons in the major groove of the DNA must be shorter than about 500 ps, which coincides with the behavior of surface hydration water in peptides and proteins. Negative NOEs were observed with the hydrogen atoms in position 2 of adenine in both duplexes studied. This indicates that a 'spine of hydration' in the minor groove, as observed by X-ray diffraction in DNA crystals, is present also in solution, with residence times significantly longer than 1 ns. Such residence times are reminiscent of 'interior' hydration water molecules in globular proteins, which are an integral part of the molecular architecture both in solution and in crystals.
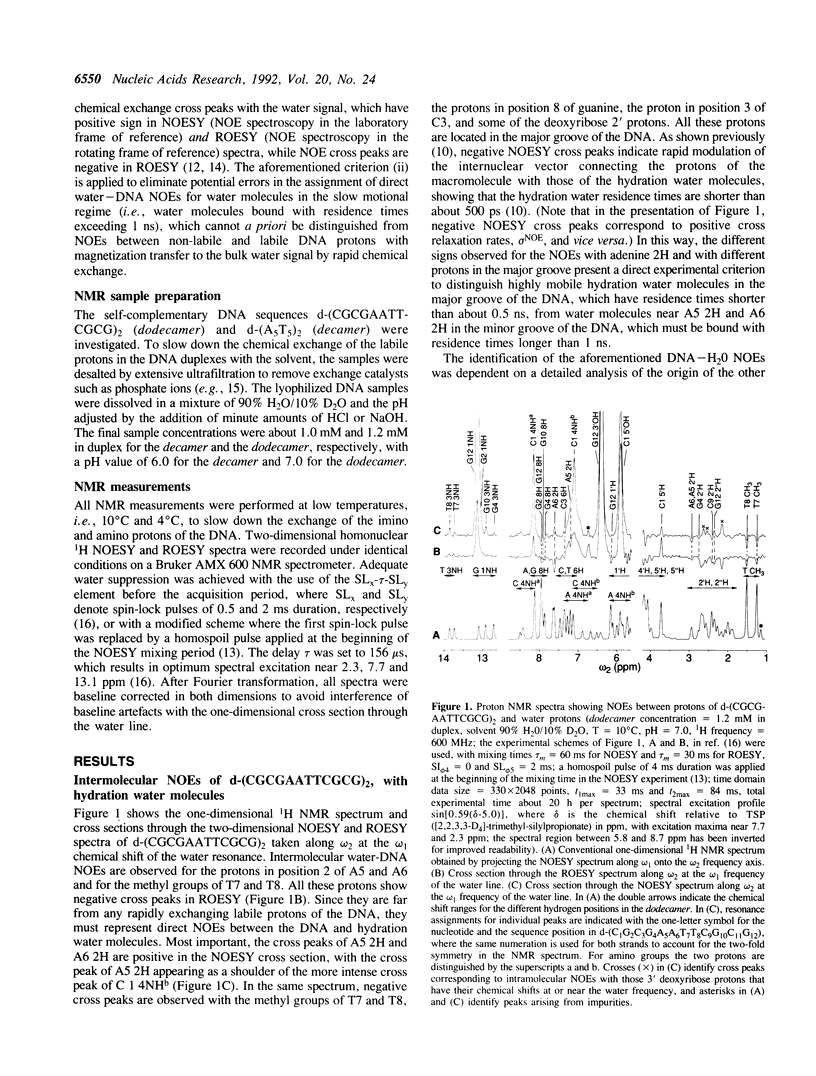
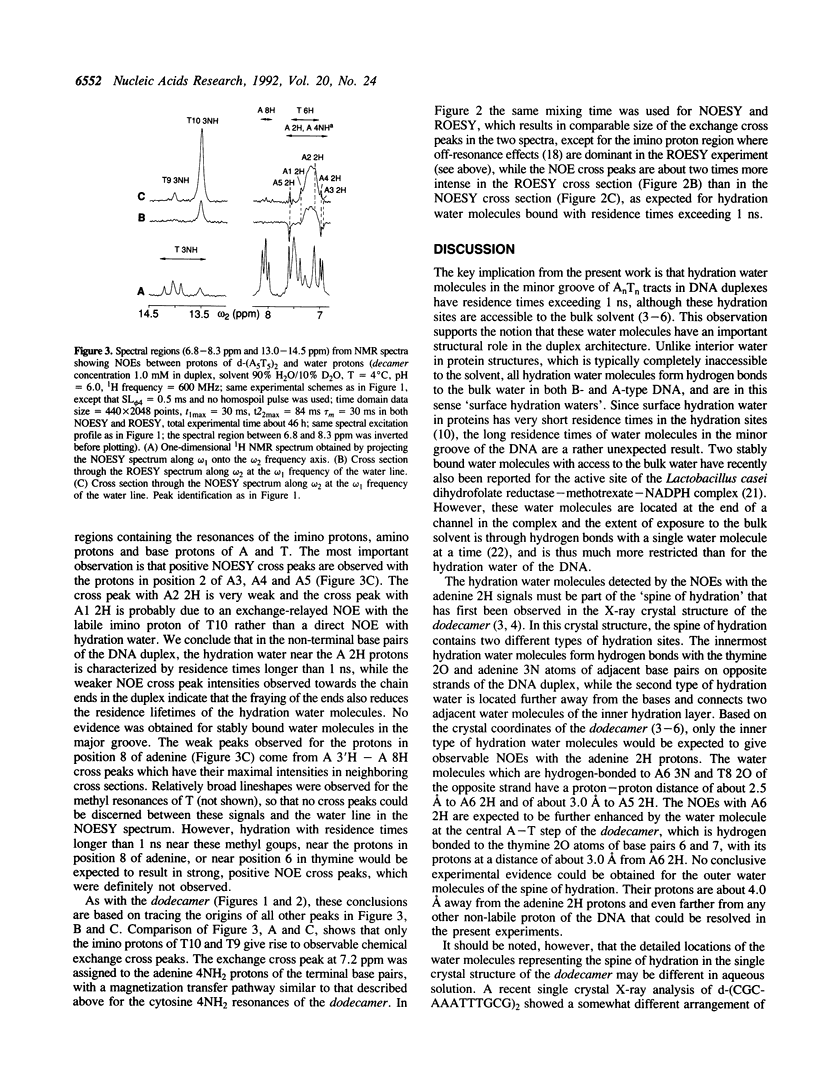
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Bolin J. T., Filman D. J., Matthews D. A., Hamlin R. C., Kraut J. Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. I. General features and binding of methotrexate. J Biol Chem. 1982 Nov 25;257(22):13650–13662. [PubMed] [Google Scholar]
- Celda B., Widmer H., Leupin W., Chazin W. J., Denny W. A., Wüthrich K. Conformational studies of d-(AAAAATTTTT)2 using constraints from nuclear overhauser effects and from quantitative analysis of the cross-peak fine structures in two-dimensional 1H nuclear magnetic resonance spectra. Biochemistry. 1989 Feb 21;28(4):1462–1471. doi: 10.1021/bi00430a006. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P. Anomalous structure and properties of poly (dA).poly(dT). Computer simulation of the polynucleotide structure with the spine of hydration in the minor groove. Nucleic Acids Res. 1987 Jan 12;15(1):293–311. doi: 10.1093/nar/15.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Edwards K. J., Brown D. G., Spink N., Skelly J. V., Neidle S. Molecular structure of the B-DNA dodecamer d(CGCAAATTTGCG)2. An examination of propeller twist and minor-groove water structure at 2.2 A resolution. J Mol Biol. 1992 Aug 20;226(4):1161–1173. doi: 10.1016/0022-2836(92)91059-x. [DOI] [PubMed] [Google Scholar]
- Gerothanassis I. P., Birdsall B., Bauer C. J., Frenkiel T. A., Feeney J. Nuclear magnetic resonance detection of bound water molecules in the active site of Lactobacillus casei dihydrofolate reductase in aqueous solution. J Mol Biol. 1992 Jul 20;226(2):549–554. doi: 10.1016/0022-2836(92)90967-o. [DOI] [PubMed] [Google Scholar]
- Guéron M., Kochoyan M., Leroy J. L. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 1987 Jul 2;328(6125):89–92. doi: 10.1038/328089a0. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Fratini A. V., Drew H. R., Dickerson R. E. Ordered water structure around a B-DNA dodecamer. A quantitative study. J Mol Biol. 1983 Jan 5;163(1):129–146. doi: 10.1016/0022-2836(83)90033-5. [DOI] [PubMed] [Google Scholar]
- Otting G., Liepinsh E., Farmer B. T., 2nd, Wüthrich K. Protein hydration studied with homonuclear 3D 1H NMR experiments. J Biomol NMR. 1991 Jul;1(2):209–215. doi: 10.1007/BF01877232. [DOI] [PubMed] [Google Scholar]
- Otting G., Liepinsh E., Wüthrich K. Protein hydration in aqueous solution. Science. 1991 Nov 15;254(5034):974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- Subramanian P. S., Beveridge D. L. A theoretical study of the aqueous hydration of canonical B d(CGCGAATTCGCG): Monte Carlo simulation and comparison with crystallographic ordered water sites. J Biomol Struct Dyn. 1989 Jun;6(6):1093–1122. doi: 10.1080/07391102.1989.10506539. [DOI] [PubMed] [Google Scholar]
- Subramanian P. S., Swaminathan S., Beveridge D. L. Theoretical account of the 'spine of hydration' in the minor groove of duplex d(CGCGAATTCGCG). J Biomol Struct Dyn. 1990 Apr;7(5):1161–1165. doi: 10.1080/07391102.1990.10508553. [DOI] [PubMed] [Google Scholar]
- Westhof E., Prangé T., Chevrier B., Moras D. Solvent distribution in crystals of B- and Z-oligomers. Biochimie. 1985 Jul-Aug;67(7-8):811–817. doi: 10.1016/s0300-9084(85)80172-3. [DOI] [PubMed] [Google Scholar]
- Westhof E. Re-refinement of the B-dodecamer d(CGCGAATTCGCG) with a comparative analysis of the solvent in it and in the Z-hexamer d(5BrCG5BrCG5BrCG). J Biomol Struct Dyn. 1987 Dec;5(3):581–600. doi: 10.1080/07391102.1987.10506414. [DOI] [PubMed] [Google Scholar]
- Zhang R. G., Joachimiak A., Lawson C. L., Schevitz R. W., Otwinowski Z., Sigler P. B. The crystal structure of trp aporepressor at 1.8 A shows how binding tryptophan enhances DNA affinity. Nature. 1987 Jun 18;327(6123):591–597. doi: 10.1038/327591a0. [DOI] [PubMed] [Google Scholar]



