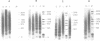Abstract
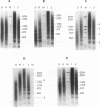
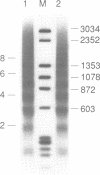
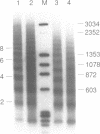
A defined in vitro chromatin assembly system was used to examine the nucleosome alignment induced by histone H5 throughout a 12 kilobase pair chicken genomic DNA fragment containing the ovalbumin gene. In contrast with total fragmented chicken DNA and several anonymous cloned fragments, much of the gene permitted histone H5 to space nucleosomes at physiological intervals in an extended array. Nucleosomes at the 3'-end of the gene and on approximately 4 kilobase pairs of 5'-flanking ovalbumin sequence did not become aligned to appreciable extents. Analysis of cloned 2-3 kilobase pair subfragments suggested that a strong nucleosome alignment signal, specifying a 196 +/- 5 base pair repeat exists in intron E. A second discrete region of the gene, which mapped approximately to intron A, exhibited nucleosome alignment with a spacing periodicity of about 200 base pairs. The ovalbumin cDNA did not permit nucleosome alignment. These findings suggest that some of the introns contain signals that direct nucleosome alignment over the ovalbumin gene in a way conducive to its regulation.
Full text
PDF

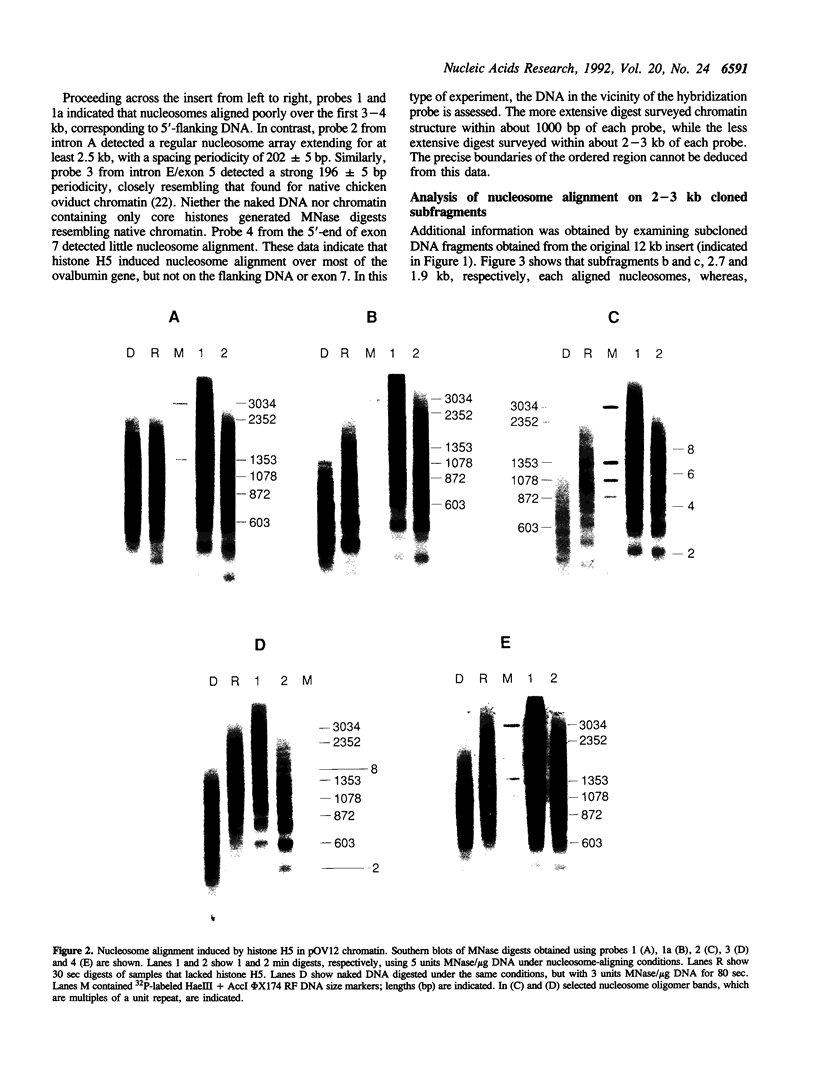
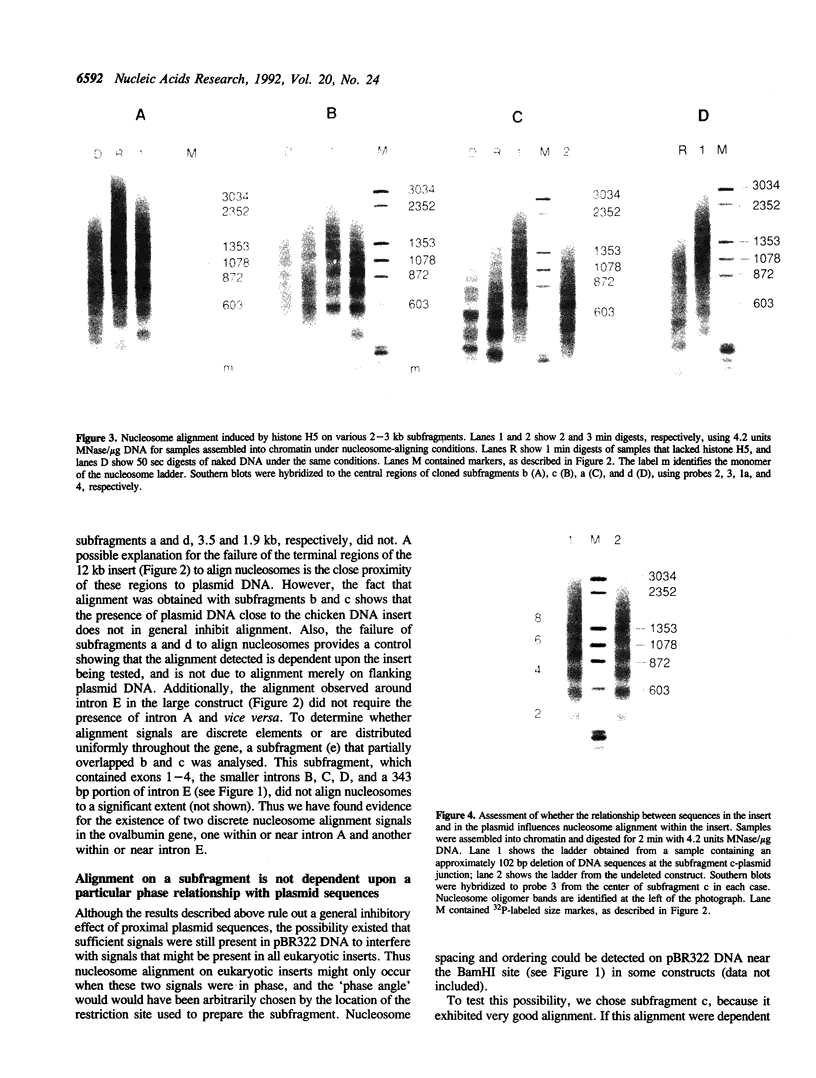
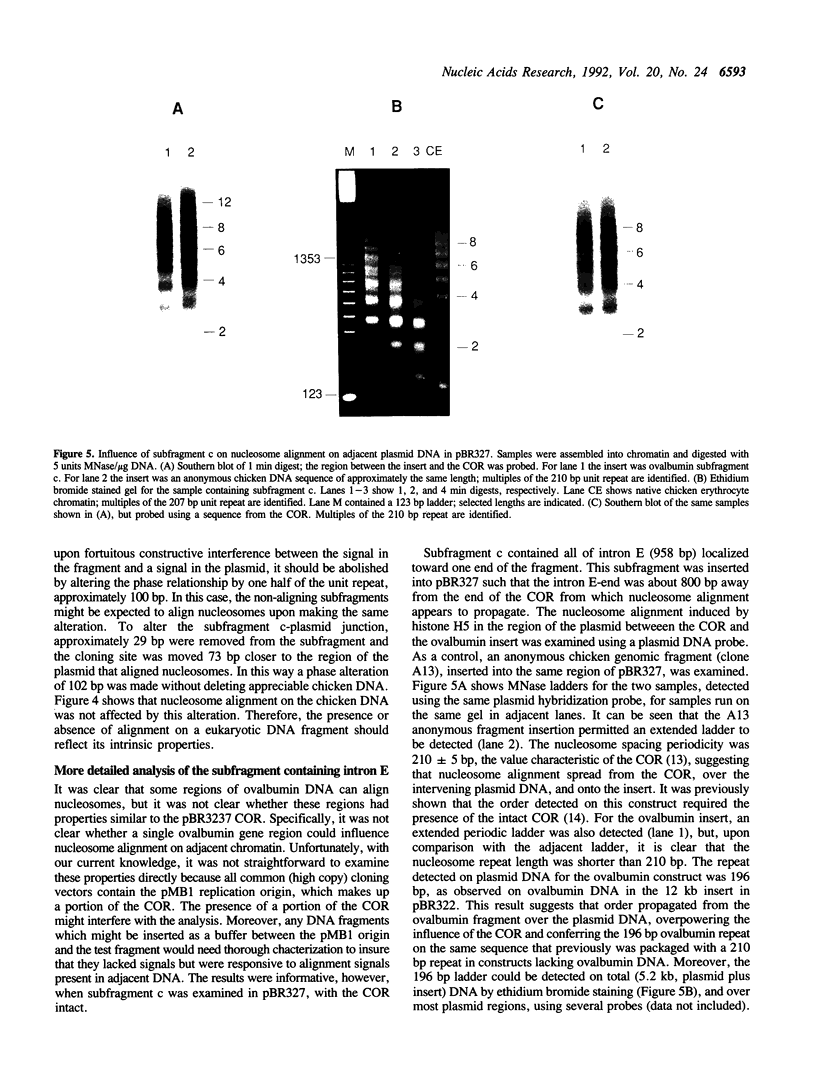



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., Davis T., Rinaldi A., Eason R. CpG deficiency, dinucleotide distributions and nucleosome positioning. Eur J Biochem. 1987 May 15;165(1):107–115. doi: 10.1111/j.1432-1033.1987.tb11200.x. [DOI] [PubMed] [Google Scholar]
- Beckmann J. S., Trifonov E. N. Splice junctions follow a 205-base ladder. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2380–2383. doi: 10.1073/pnas.88.6.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Dretzen G., Bellard F., Kaye J. S., Pratt-Kaye S., Chambon P. Hormonally induced alterations of chromatin structure in the polyadenylation and transcription termination regions of the chicken ovalbumin gene. EMBO J. 1986 Mar;5(3):567–574. doi: 10.1002/j.1460-2075.1986.tb04248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Dretzen G., Bellard F., Oudet P., Chambon P. Disruption of the typical chromatin structure in a 2500 base-pair region at the 5' end of the actively transcribed ovalbumin gene. EMBO J. 1982;1(2):223–230. doi: 10.1002/j.1460-2075.1982.tb01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K. S., Anderson J. N. Hormonal regulation of the conformation of the ovalbumin gene in chick oviduct chromatin. J Biol Chem. 1982 Nov 10;257(21):13018–13027. [PubMed] [Google Scholar]
- Brinster R. L., Allen J. M., Behringer R. R., Gelinas R. E., Palmiter R. D. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci U S A. 1988 Feb;85(3):836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown I. R., Sutcliffe J. G. Atypical nucleosome spacing of rat neuronal identifier elements in non-neuronal chromatin. Nucleic Acids Res. 1987 Apr 24;15(8):3563–3571. doi: 10.1093/nar/15.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert D. A., Knoll B. J., Woo S. L., Mace M. L., Tsai M. J., O'Malley B. W. Differential hormonal responsiveness of the ovalbumin gene and its pseudogenes in the chick oviduct. Biochemistry. 1980 Nov 25;19(24):5586–5592. doi: 10.1021/bi00565a020. [DOI] [PubMed] [Google Scholar]
- Compton J. L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugaiczyk A., Woo S. L., Colbert D. A., Lai E. C., Mace M. L., Jr, O'Malley B. W. The ovalbumin gene: cloning and molecular organization of the entire natural gene. Proc Natl Acad Sci U S A. 1979 May;76(5):2253–2257. doi: 10.1073/pnas.76.5.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. The formation and function of DNase I hypersensitive sites in the process of gene activation. J Biol Chem. 1988 Dec 25;263(36):19259–19262. [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme gene. Nature. 1984 Sep 13;311(5982):163–165. doi: 10.1038/311163a0. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. DNase I-hypersensitive sites in the chromatin structure of the lysozyme gene in steroid hormone target and non-target cells. Biol Chem Hoppe Seyler. 1987 Feb;368(2):111–119. doi: 10.1515/bchm3.1987.368.1.111. [DOI] [PubMed] [Google Scholar]
- Gannon F., O'Hare K., Perrin F., LePennec J. P., Benoist C., Cochet M., Breathnach R., Royal A., Garapin A., Cami B. Organisation and sequences at the 5' end of a cloned complete ovalbumin gene. Nature. 1979 Mar 29;278(5703):428–434. doi: 10.1038/278428a0. [DOI] [PubMed] [Google Scholar]
- Gottschling D. E., Palen T. E., Cech T. R. Different nucleosome spacing in transcribed and non-transcribed regions of the ribosomal RNA gene in Tetrahymena thermophila. Nucleic Acids Res. 1983 Apr 11;11(7):2093–2109. doi: 10.1093/nar/11.7.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig R., Perrin F., Gannon F., Mandel J. L., Chambon P. The ovalbumin gene family: structure of the X gene and evolution of duplicated split genes. Cell. 1980 Jul;20(3):625–637. doi: 10.1016/0092-8674(80)90309-8. [DOI] [PubMed] [Google Scholar]
- Jeong S. W., Lauderdale J. D., Stein A. Chromatin assembly on plasmid DNA in vitro. Apparent spreading of nucleosome alignment from one region of pBR327 by histone H5. J Mol Biol. 1991 Dec 20;222(4):1131–1147. doi: 10.1016/0022-2836(91)90597-y. [DOI] [PubMed] [Google Scholar]
- Keene M. A., Elgin S. C. Patterns of DNA structural polymorphism and their evolutionary implications. Cell. 1984 Jan;36(1):121–129. doi: 10.1016/0092-8674(84)90080-1. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Lorch Y. Irresistible force meets immovable object: transcription and the nucleosome. Cell. 1991 Nov 29;67(5):833–836. doi: 10.1016/0092-8674(91)90354-2. [DOI] [PubMed] [Google Scholar]
- Lowman H., Bina M. Correlation between dinucleotide periodicities and nucleosome positioning on mouse satellite DNA. Biopolymers. 1990;30(9-10):861–876. doi: 10.1002/bip.360300902. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D., Brinster R. L. Germ-line transformation of mice. Annu Rev Genet. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter R. D., Sandgren E. P., Avarbock M. R., Allen D. D., Brinster R. L. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):478–482. doi: 10.1073/pnas.88.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell S. C., Drew H. R., Travers A. A. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986 Oct 20;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- Smith R. D., Seale R. L., Yu J. Transcribed chromatin exhibits an altered nucleosomal spacing. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5505–5509. doi: 10.1073/pnas.80.18.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Thompson R. J. Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell. 1977 Apr;10(4):633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N., Sussman J. L. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeponteau B., Brawley J., Martinson H. G. Nucleosome spacing is compressed in active chromatin domains of chick erythroid cells. Biochemistry. 1992 Feb 11;31(5):1554–1563. doi: 10.1021/bi00120a037. [DOI] [PubMed] [Google Scholar]