Abstract
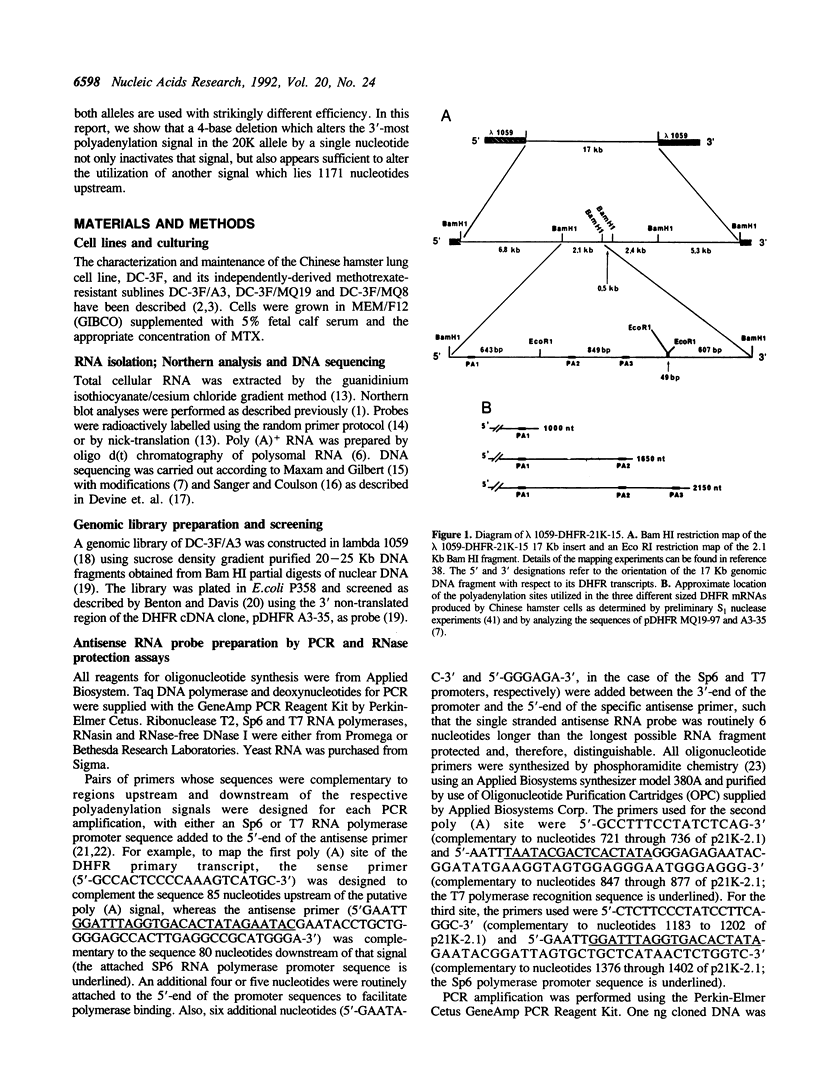
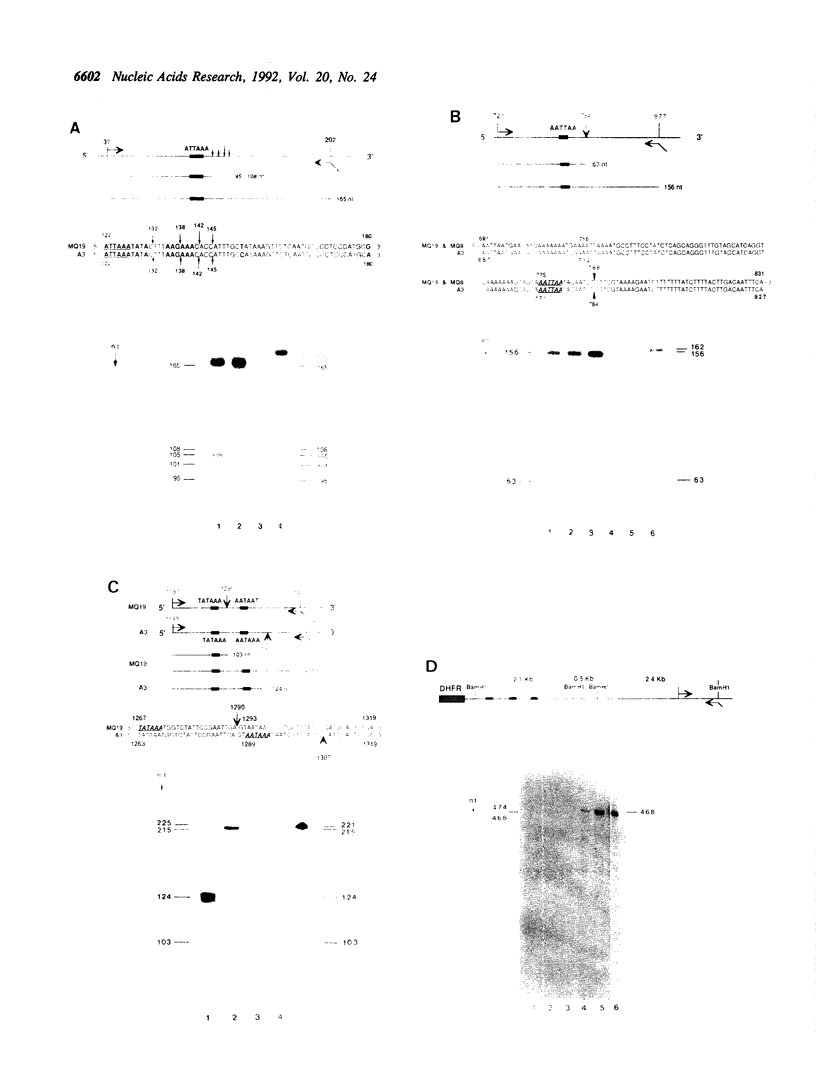
The Chinese hamster cell line, DC-3F, is heterozygous at the DHFR locus, and each allele can be distinguished on the basis of a unique DNA restriction pattern, protein isoelectric profile and in the abundancy of the DHFR mRNAs it expresses. Although each allele produces four transcripts, 1000, 1650 and 2150 nucleotides [corrected] in length, the relative distribution of these RNAs differs for each; the 2150 nt mRNA represents the major (60%) species generated from one allele, while the 1000 nt mRNA is the major species generated from the other. The allele that predominantly expresses the 2150 nt transcript is preferentially overexpressed when DC-3F cells are subjected to selection in methotrexate. We have analyzed the 3' ends of both DHFR alleles and have found that the three major mRNAs arise by readthrough of multiple polyadenylation signals. A four base deletion in one allele changes the consensus polyadenylation signal AAUAAA to AAUAAU, resulting in the utilization of a cryptic polyadenylation signal lying 21 bp upstream. Surprisingly, this mutation in the third polyadenylation signal appears to affect not only the utilization of this signal, but also the efficiency with which the first signal, located 1171 bp upstream from the third site, is utilized.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Biedler J. L., Albrecht A. M., Hutchison D. J., Spengler B. A. Drug response, dihydrofolate reductase, and cytogenetics of amethopterin-resistant Chinese hamster cells in vitro. Cancer Res. 1972 Jan;32(1):153–161. [PubMed] [Google Scholar]
- Biedler J. L., Albrecht A. M., Spengler B. A. Biochemical and karyological properties of cells resistant to the quinazoline antifolate, methasquin. Eur J Cancer. 1978 Jan;14(1):41–49. doi: 10.1016/0014-2964(78)90136-6. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Ellis N., Chasin L. A. Structure of the dihydrofolate reductase gene in Chinese hamster ovary cells. Nucleic Acids Res. 1983 Apr 11;11(7):1997–2012. doi: 10.1093/nar/11.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C. N., Stacy T. P. Identification of sequences in the herpes simplex virus thymidine kinase gene required for efficient processing and polyadenylation. Mol Cell Biol. 1985 Aug;5(8):2104–2113. doi: 10.1128/mcb.5.8.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway L., Wickens M. A sequence downstream of A-A-U-A-A-A is required for formation of simian virus 40 late mRNA 3' termini in frog oocytes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3949–3953. doi: 10.1073/pnas.82.12.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denome R. M., Cole C. N. Patterns of polyadenylation site selection in gene constructs containing multiple polyadenylation signals. Mol Cell Biol. 1988 Nov;8(11):4829–4839. doi: 10.1128/mcb.8.11.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine S. E., Hussain A., Davide J. P., Melera P. W. Full length and alternatively spliced pgp1 transcripts in multidrug-resistant Chinese hamster lung cells. J Biol Chem. 1991 Mar 5;266(7):4545–4555. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3'-end formation. 1984 Nov 29-Dec 5Nature. 312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- Hamlin J. L., Milbrandt J. D., Heintz N. H., Azizkhan J. C. DNA sequence amplification in mammalian cells. Int Rev Cytol. 1984;90:31–82. doi: 10.1016/s0074-7696(08)61487-4. [DOI] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Butler E. T., Roulland D., Chamberlin M. J. Bacteriophage SP6-specific RNA polymerase. II. Mapping of SP6 DNA and selective in vitro transcription. J Biol Chem. 1982 May 25;257(10):5779–5788. [PubMed] [Google Scholar]
- Lewis J. A., Davide J. P., Melera P. W. Selective amplification of polymorphic dihydrofolate reductase gene loci in Chinese hamster lung cells. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6961–6965. doi: 10.1073/pnas.79.22.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. A., Kurtz D. T., Melera P. W. Molecular cloning of Chinese hamster dihydrofolate reductase-specific cDNA and the identification of multiple dihydrofolate reductase mRNAs in antifolate-resistant Chinese hamster lung fibroblasts. Nucleic Acids Res. 1981 Mar 25;9(6):1311–1322. doi: 10.1093/nar/9.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L. Polyadenylation of mRNA precursors. Biochim Biophys Acta. 1988 May 6;950(1):1–12. doi: 10.1016/0167-4781(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melera P. W., Davide J. P., Hession C. A., Scotto K. W. Phenotypic expression in Escherichia coli and nucleotide sequence of two Chinese hamster lung cell cDNAs encoding different dihydrofolate reductases. Mol Cell Biol. 1984 Jan;4(1):38–48. doi: 10.1128/mcb.4.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melera P. W., Davide J. P., Oen H. Antifolate-resistant Chinese hamster cells. Molecular basis for the biochemical and structural heterogeneity among dihydrofolate reductases produced by drug-sensitive and drug-resistant cell lines. J Biol Chem. 1988 Feb 5;263(4):1978–1990. [PubMed] [Google Scholar]
- Melera P. W., Hession C. A., Davide J. P., Scotto K. W., Biedler J. L., Meyérs M. B., Shanske S. Antifolate-resistant Chinese Hamster Cells. mRNA directed overproduction of multiple dihydrofolate reductases from a series of independently derived sublines containing amplified dihydrofolate reductase genes. J Biol Chem. 1982 Nov 10;257(21):12939–12949. [PubMed] [Google Scholar]
- Melera P. W., Lewis J. A., Biedler J. L., Hession C. Antifolate-resistant Chinese hamster cells. Evidence for dihydrofolate reductase gene amplification among independently derived sublines overproducing different dihydrofolate reductases. J Biol Chem. 1980 Jul 25;255(14):7024–7028. [PubMed] [Google Scholar]
- Melera P. W., Wolgemuth D., Biedler J. L., Hession C. Antifolate-resistant chinese hamster cells. Evidence from independently derived sublines for the overproduction of two dihydrofolate reductases encoded by different mRNAs. J Biol Chem. 1980 Jan 25;255(2):319–322. [PubMed] [Google Scholar]
- Milbrandt J. D., Azizkhan J. C., Greisen K. S., Hamlin J. L. Organization of a Chinese hamster ovary dihydrofolate reductase gene identified by phenotypic rescue. Mol Cell Biol. 1983 Jul;3(7):1266–1273. doi: 10.1128/mcb.3.7.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Skolnik-David H., Sharp P. A. Analysis of RNA cleavage at the adenovirus-2 L3 polyadenylation site. EMBO J. 1986 Aug;5(8):1929–1938. doi: 10.1002/j.1460-2075.1986.tb04446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandi C., Masters J. N., Mottes M., Attardi G. Multiple forms of human dihydrofolate reductase messenger RNA. Cloning and expression in Escherichia coli of their DNA coding sequence. J Mol Biol. 1982 Apr 15;156(3):583–607. doi: 10.1016/0022-2836(82)90268-6. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978 Dec;15(4):1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci. 1989 Mar;14(3):105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- Qian Z. W., Wilusz J. An RNA-binding protein specifically interacts with a functionally important domain of the downstream element of the simian virus 40 late polyadenylation signal. Mol Cell Biol. 1991 Oct;11(10):5312–5320. doi: 10.1128/mcb.11.10.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky M., Connelly S., Manley J. L., Alwine J. C. Identification of a sequence element on the 3' side of AAUAAA which is necessary for simian virus 40 late mRNA 3'-end processing. Mol Cell Biol. 1985 Oct;5(10):2713–2719. doi: 10.1128/mcb.5.10.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Nunberg J. H., Schimke R. T. Size heterogeneity in the 3' end of dihydrofolate reductase messenger RNAs in mouse cells. Cell. 1980 Nov;22(2 Pt 2):361–370. doi: 10.1016/0092-8674(80)90346-3. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Schimke R. T. Nucleotide sequence surrounding multiple polyadenylation sites in the mouse dihydrofolate reductase gene. J Biol Chem. 1982 May 10;257(9):5143–5147. [PubMed] [Google Scholar]
- Sheets M. D., Ogg S. C., Wickens M. P. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990 Oct 11;18(19):5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets M. D., Stephenson P., Wickens M. P. Products of in vitro cleavage and polyadenylation of simian virus 40 late pre-mRNAs. Mol Cell Biol. 1987 Apr;7(4):1518–1529. doi: 10.1128/mcb.7.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990 Jul;15(7):277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Woychik R. P., Lyons R. H., Post L., Rottman F. M. Requirement for the 3' flanking region of the bovine growth hormone gene for accurate polyadenylylation. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3944–3948. doi: 10.1073/pnas.81.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen J. Y., Kellems R. E. Independent 5' and 3'-end determination of multiple dihydrofolate reductase transcripts. Mol Cell Biol. 1987 Oct;7(10):3732–3739. doi: 10.1128/mcb.7.10.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D., Stephenson P., Sheets M., Wickens M. The AAUAAA sequence is required both for cleavage and for polyadenylation of simian virus 40 pre-mRNA in vitro. Mol Cell Biol. 1986 Jul;6(7):2317–2323. doi: 10.1128/mcb.6.7.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]