Abstract
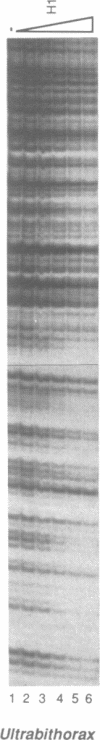
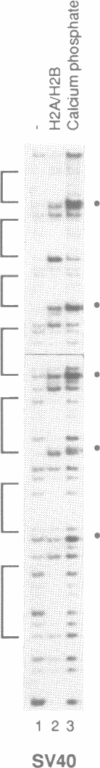
By using a DNase I footprinting assay, we have purified a factor by DNA affinity chromatography that binds to the minimal enhancer region of the Drosophila knirps gene and subsequently identified the protein as the core histone H2B. This inadvertent purification of a core histone as a putative sequence-specific DNA binding protein was due to a previously unknown property of H2B to interact with DNA in a periodic manner. Moreover, we found that each of the individual core histones, but not histone H1 or high mobility group protein 1, bound to the knirps enhancer to give a repetitive DNase I footprint pattern with a periodicity of about 10 base pairs, which is approximately one turn of the DNA helix. In addition, preparations containing the core histones H2A-H2B or H3-H4 yielded identical periodic DNase I footprint patterns on several different promoter and enhancer regions. These findings suggest that there are periodic, homotypic interactions between DNA-bound core histones that result from an alteration of the overall DNA structure such as the curvature rather than a specific sequence. We have also shown that histones H2A-H2B can repress initiation of transcription by RNA polymerase II. The phenomena described here may reflect histone-DNA interactions in non-nucleosomal stretches of chromatin and could be involved in some aspects of either rotational or translational positioning of nucleosomes. Furthermore, these findings indicate that a repeated 10 bp DNase I ladder, which has previously been considered to be a property of an intact nucleosome, can also be generated with subnucleosomal components. It will thus be necessary to reevaluate the criteria applied to the analysis of nucleosomes both in vivo and in vitro.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behe M., Zimmerman S., Felsenfeld G. Changes in the helical repeat of poly(dG-m5dC) . poly(dG-m5dC) and poly(dG-dC) . poly(dG-dC) associated with the B-Z transition. Nature. 1981 Sep 17;293(5829):233–235. doi: 10.1038/293233a0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988 Jun 3;53(5):699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Bresnick E. H., Bustin M., Marsaud V., Richard-Foy H., Hager G. L. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992 Jan 25;20(2):273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler P. J., Thomas J. O. Changes in chromatin folding in solution. J Mol Biol. 1980 Jul 15;140(4):505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Felsenfeld G. Supercoiling energy and nucleosome formation: the role of the arginine-rich histone kernel. Nucleic Acids Res. 1977;4(5):1159–1181. doi: 10.1093/nar/4.5.1159-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D. I., Leatherwood J., Carey M., Ptashne M., Kornberg R. D. Activation of yeast polymerase II transcription by herpesvirus VP16 and GAL4 derivatives in vitro. Mol Cell Biol. 1989 Nov;9(11):4746–4749. doi: 10.1128/mcb.9.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croston G. E., Kerrigan L. A., Lira L. M., Marshak D. R., Kadonaga J. T. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991 Feb 8;251(4994):643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. Chromatin structure and gene activity. Curr Opin Cell Biol. 1990 Jun;2(3):437–445. doi: 10.1016/0955-0674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- Hayes J. J., Bashkin J., Tullius T. D., Wolffe A. P. The histone core exerts a dominant constraint on the structure of DNA in a nucleosome. Biochemistry. 1991 Aug 27;30(34):8434–8440. doi: 10.1021/bi00098a022. [DOI] [PubMed] [Google Scholar]
- Hill C. S., Thomas J. O. Core histone-DNA interactions in sea urchin sperm chromatin. The N-terminal tail of H2B interacts with linker DNA. Eur J Biochem. 1990 Jan 12;187(1):145–153. doi: 10.1111/j.1432-1033.1990.tb15288.x. [DOI] [PubMed] [Google Scholar]
- Hoch M., Schröder C., Seifert E., Jäckle H. cis-acting control elements for Krüppel expression in the Drosophila embryo. EMBO J. 1990 Aug;9(8):2587–2595. doi: 10.1002/j.1460-2075.1990.tb07440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T. Assembly and disassembly of the Drosophila RNA polymerase II complex during transcription. J Biol Chem. 1990 Feb 15;265(5):2624–2631. [PubMed] [Google Scholar]
- Kadonaga J. T. Purification of sequence-specific binding proteins by DNA affinity chromatography. Methods Enzymol. 1991;208:10–23. doi: 10.1016/0076-6879(91)08004-2. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka R. T., Tyree C. M., Kadonaga J. T. Accurate and efficient RNA polymerase II transcription with a soluble nuclear fraction derived from Drosophila embryos. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):1024–1028. doi: 10.1073/pnas.88.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrigan L. A., Croston G. E., Lira L. M., Kadonaga J. T. Sequence-specific transcriptional antirepression of the Drosophila Krüppel gene by the GAGA factor. J Biol Chem. 1991 Jan 5;266(1):574–582. [PubMed] [Google Scholar]
- Kleinschmidt J. A., Franke W. W. Soluble acidic complexes containing histones H3 and H4 in nuclei of Xenopus laevis oocytes. Cell. 1982 Jul;29(3):799–809. doi: 10.1016/0092-8674(82)90442-1. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Lorch Y. Irresistible force meets immovable object: transcription and the nucleosome. Cell. 1991 Nov 29;67(5):833–836. doi: 10.1016/0092-8674(91)90354-2. [DOI] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991 Oct 11;254(5029):238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Carey M. F., Ptashne M., Green M. R. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988 Aug 26;54(5):659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Digestion of nucleosomes with deoxyribonucleases I and II. Methods Enzymol. 1989;170:264–269. doi: 10.1016/0076-6879(89)70051-3. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Nacheva G. A., Guschin D. Y., Preobrazhenskaya O. V., Karpov V. L., Ebralidse K. K., Mirzabekov A. D. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989 Jul 14;58(1):27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- Nelson P. P., Albright S. C., Wiseman J. M., Garrard W. T. Reassociation of histone H1 with nucleosomes. J Biol Chem. 1979 Nov 25;254(22):11751–11760. [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Pankratz M. J., Busch M., Hoch M., Seifert E., Jäckle H. Spatial control of the gap gene knirps in the Drosophila embryo by posterior morphogen system. Science. 1992 Feb 21;255(5047):986–989. doi: 10.1126/science.1546296. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Helical periodicity of DNA determined by enzyme digestion. Nature. 1980 Aug 7;286(5773):573–578. doi: 10.1038/286573a0. [DOI] [PubMed] [Google Scholar]
- Saltzman A. G., Weinmann R. Promoter specificity and modulation of RNA polymerase II transcription. FASEB J. 1989 Apr;3(6):1723–1733. doi: 10.1096/fasebj.3.6.2649403. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Sentenac A. RNA polymerase B (II) and general transcription factors. Annu Rev Biochem. 1990;59:711–754. doi: 10.1146/annurev.bi.59.070190.003431. [DOI] [PubMed] [Google Scholar]
- Simon R. H., Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979 Feb;6(2):689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Thoma F. Electron microscopy of chromatin. Methods Enzymol. 1989;170:142–165. doi: 10.1016/0076-6879(89)70045-8. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Nüsslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992 Jan 24;68(2):201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wampler S. L., Tyree C. M., Kadonaga J. T. Fractionation of the general RNA polymerase II transcription factors from Drosophila embryos. J Biol Chem. 1990 Dec 5;265(34):21223–21231. [PubMed] [Google Scholar]
- Wolffe A. P. New approaches to chromatin function. New Biol. 1990 Mar;2(3):211–218. [PubMed] [Google Scholar]
- Workman J. L., Taylor I. C., Kingston R. E. Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell. 1991 Feb 8;64(3):533–544. doi: 10.1016/0092-8674(91)90237-s. [DOI] [PubMed] [Google Scholar]