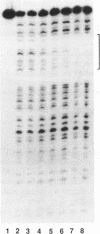
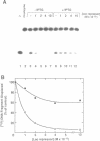
Abstract
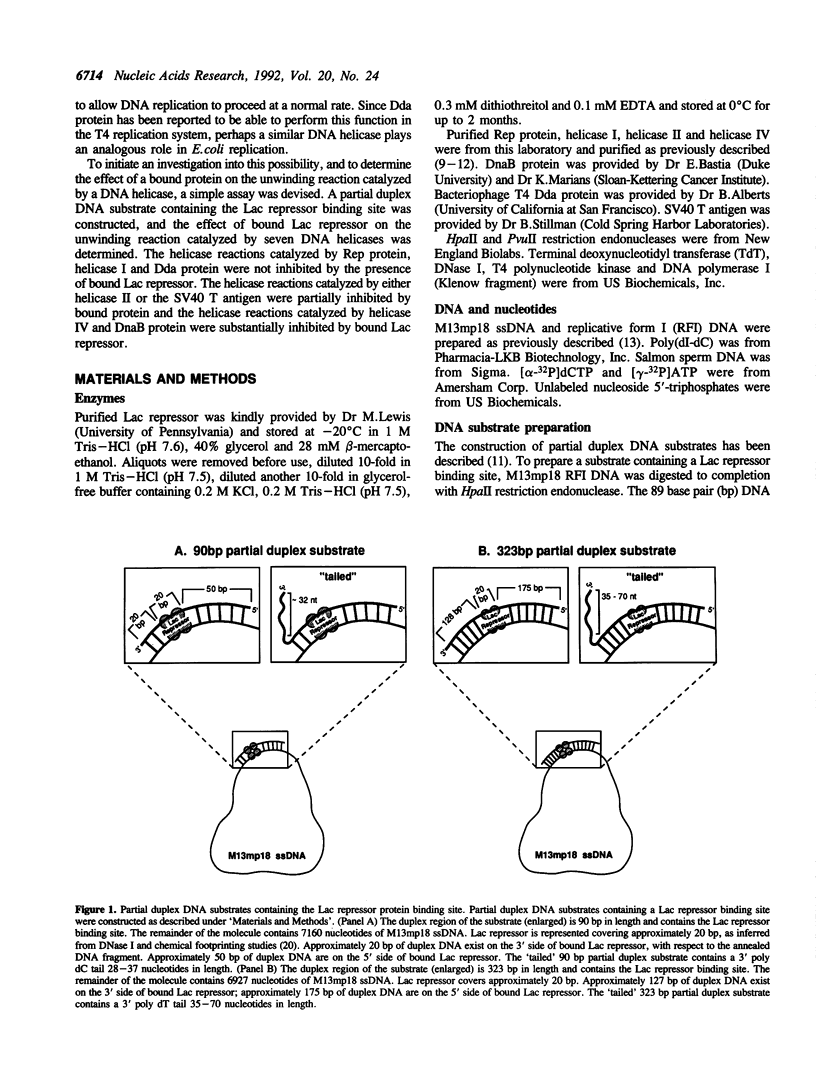
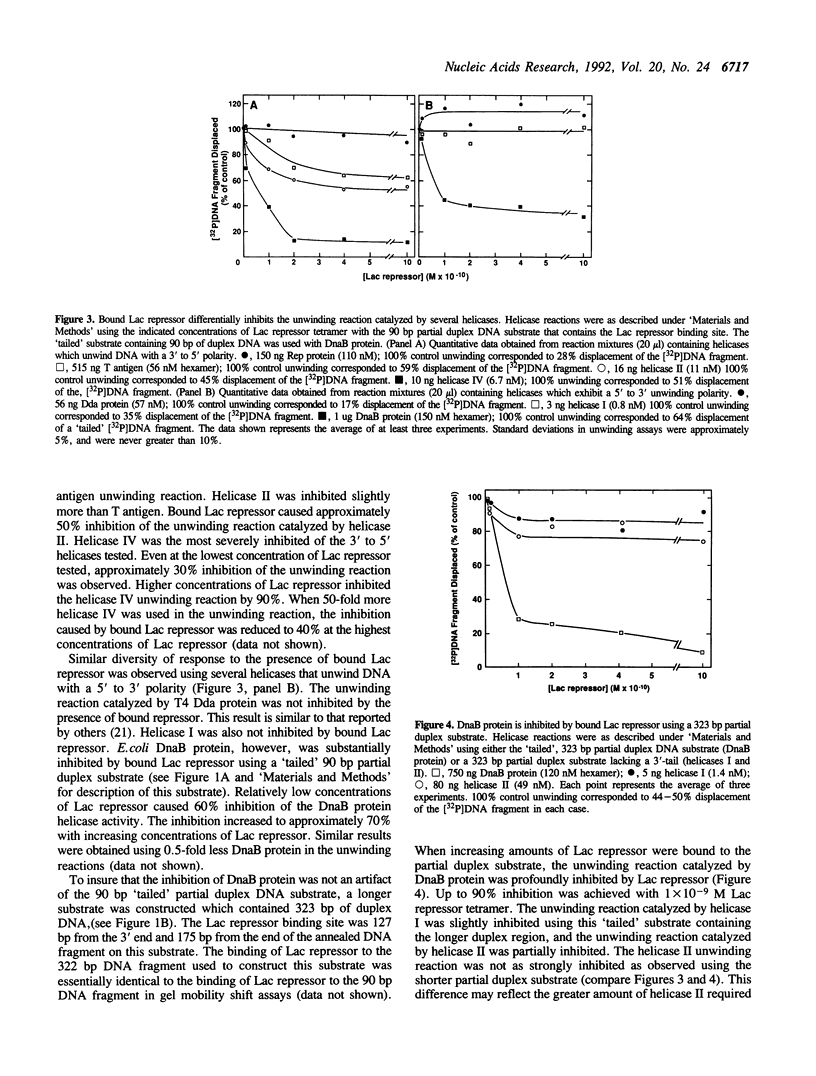
A partial duplex DNA substrate containing the Lac repressor binding site, within the duplex region, was constructed to examine the effect of bound Lac repressor on the unwinding reaction catalyzed by several DNA helicases. The substrate contained 90 base pairs of double-stranded DNA and, in the absence of Lac repressor, was effectively unwound by each of the seven helicases tested. The unwinding reactions catalyzed by Escherichia coli Rep protein, bacteriophage T4 Dda protein and E. coli DNA helicase I were not inhibited by the presence of bound Lac repressor. Both SV40 T antigen and E. coli helicase II were partially inhibited by bound repressor at the highest repressor concentrations tested. The helicase reactions catalyzed by E. coli DnaB protein and helicase IV were substantially inhibited by the presence of bound protein. When the length of the duplex region was increased to 323 base pairs the inhibition spectrum caused by bound Lac repressor on the unwinding reactions catalyzed by DnaB protein, helicase I and helicase II was essentially the same as that observed using the shorter partial duplex molecule. Inhibition of the unwinding reaction was due to the presence of bound Lac repressor as evidenced by the substantially weaker inhibition of helicase IV by Lac repressor in the presence of IPTG. In addition, we have shown that Rep protein displaces the bound repressor protein during the course of an unwinding reaction.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedinger P., Hochstrasser M., Jongeneel C. V., Alberts B. M. Properties of the T4 bacteriophage DNA replication apparatus: the T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell. 1983 Aug;34(1):115–123. doi: 10.1016/0092-8674(83)90141-1. [DOI] [PubMed] [Google Scholar]
- Bonne-Andrea C., Wong M. L., Alberts B. M. In vitro replication through nucleosomes without histone displacement. Nature. 1990 Feb 22;343(6260):719–726. doi: 10.1038/343719a0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brewer B. J. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988 Jun 3;53(5):679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- Calendar R., Lindqvist B., Sironi G., Clark A. J. Characterization of REP- mutants and their interaction with P2 phage. Virology. 1970 Jan;40(1):72–83. doi: 10.1016/0042-6822(70)90380-6. [DOI] [PubMed] [Google Scholar]
- Colasanti J., Denhardt D. T. The Escherichia coli rep mutation. X. Consequences of increased and decreased Rep protein levels. Mol Gen Genet. 1987 Sep;209(2):382–390. doi: 10.1007/BF00329669. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T., Dressler D. H., Hathaway A. THE ABORTIVE REPLICATION OF PhiX174 DNA IN A RECOMBINATION-DEFICIENT MUTANT OF Escherichia coli. Proc Natl Acad Sci U S A. 1967 Mar;57(3):813–820. doi: 10.1073/pnas.57.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T., Iwaya M., Larison L. L. The rep mutation. II. Its effect on Escherichia coli and on the replication of bacteriophage phi X174. Virology. 1972 Aug;49(2):486–496. doi: 10.1016/0042-6822(72)90500-4. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz G. S., Dean F. B., Hurwitz J., Matson S. W. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J Biol Chem. 1988 Jan 5;263(1):383–392. [PubMed] [Google Scholar]
- Hiasa H., Marians K. J. Differential inhibition of the DNA translocation and DNA unwinding activities of DNA helicases by the Escherichia coli Tus protein. J Biol Chem. 1992 Jun 5;267(16):11379–11385. [PubMed] [Google Scholar]
- Hill T. M., Marians K. J. Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2481–2485. doi: 10.1073/pnas.87.7.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongeneel C. V., Bedinger P., Alberts B. M. Effects of the bacteriophage T4 dda protein on DNA synthesis catalyzed by purified T4 replication proteins. J Biol Chem. 1984 Oct 25;259(20):12933–12938. [PubMed] [Google Scholar]
- Kodaira K. I., Taketo A. Conversion of bacteriophage G4 single-stranded viral DNA to double-stranded replicative form in dna mutants of Escherichia coli. Biochim Biophys Acta. 1977 May 17;476(2):149–155. doi: 10.1016/0005-2787(77)90091-0. [DOI] [PubMed] [Google Scholar]
- Lahue E. E., Matson S. W. Escherichia coli DNA helicase I catalyzes a unidirectional and highly processive unwinding reaction. J Biol Chem. 1988 Mar 5;263(7):3208–3215. [PubMed] [Google Scholar]
- Lane H. E., Denhardt D. T. The rep mutation. III. Altered structure of the replicating Escherichia coli chromosome. J Bacteriol. 1974 Nov;120(2):805–814. doi: 10.1128/jb.120.2.805-814.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane H. E., Denhardt D. T. The rep mutation. IV. Slower movement of replication forks in Escherichia coli rep strains. J Mol Biol. 1975 Sep 5;97(1):99–112. doi: 10.1016/s0022-2836(75)80025-8. [DOI] [PubMed] [Google Scholar]
- LeBowitz J. H., McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986 Apr 5;261(10):4738–4748. [PubMed] [Google Scholar]
- Lechner R. L., Richardson C. C. A preformed, topologically stable replication fork. Characterization of leading strand DNA synthesis catalyzed by T7 DNA polymerase and T7 gene 4 protein. J Biol Chem. 1983 Sep 25;258(18):11185–11196. [PubMed] [Google Scholar]
- Lohman T. M. Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol Microbiol. 1992 Jan;6(1):5–14. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- Matson S. W. DNA helicases of Escherichia coli. Prog Nucleic Acid Res Mol Biol. 1991;40:289–326. doi: 10.1016/s0079-6603(08)60845-4. [DOI] [PubMed] [Google Scholar]
- Matson S. W., George J. W. DNA helicase II of Escherichia coli. Characterization of the single-stranded DNA-dependent NTPase and helicase activities. J Biol Chem. 1987 Feb 15;262(5):2066–2076. [PubMed] [Google Scholar]
- Matson S. W., Kaiser-Rogers K. A. DNA helicases. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970 Nov 14;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Runyon G. T., Bear D. G., Lohman T. M. Escherichia coli helicase II (UvrD) protein initiates DNA unwinding at nicks and blunt ends. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6383–6387. doi: 10.1073/pnas.87.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. F., Kornberg A. Purification of the rep protein of Escherichia coli. An ATPase which separates duplex DNA strands in advance of replication. J Biol Chem. 1978 May 10;253(9):3292–3297. [PubMed] [Google Scholar]
- Thömmes P., Hübscher U. Eukaryotic DNA helicases. FEBS Lett. 1990 Aug 1;268(2):325–328. doi: 10.1016/0014-5793(90)81279-w. [DOI] [PubMed] [Google Scholar]
- Wessel R., Müller H., Hoffmann-Berling H. Electron microscopic analysis of DNA forks generated by Escherichia coli DNA helicase II. Eur J Biochem. 1990 Sep 24;192(3):695–701. doi: 10.1111/j.1432-1033.1990.tb19278.x. [DOI] [PubMed] [Google Scholar]
- Wiekowski M., Schwarz M. W., Stahl H. Simian virus 40 large T antigen DNA helicase. Characterization of the ATPase-dependent DNA unwinding activity and its substrate requirements. J Biol Chem. 1988 Jan 5;263(1):436–442. [PubMed] [Google Scholar]
- Wong I., Lohman T. M. Allosteric effects of nucleotide cofactors on Escherichia coli Rep helicase-DNA binding. Science. 1992 Apr 17;256(5055):350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- Wood E. R., Matson S. W. Purification and characterization of a new DNA-dependent ATPase with helicase activity from Escherichia coli. J Biol Chem. 1987 Nov 5;262(31):15269–15276. [PubMed] [Google Scholar]
- Yancey-Wrona J. E., Wood E. R., George J. W., Smith K. R., Matson S. W. Escherichia coli Rep protein and helicase IV. Distributive single-stranded DNA-dependent ATPases that catalyze a limited unwinding reaction in vitro. Eur J Biochem. 1992 Jul 15;207(2):479–485. doi: 10.1111/j.1432-1033.1992.tb17074.x. [DOI] [PubMed] [Google Scholar]
- Yarranton G. T., Gefter M. L. Enzyme-catalyzed DNA unwinding: studies on Escherichia coli rep protein. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1658–1662. doi: 10.1073/pnas.76.4.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]