Abstract
Plant-derived polyphenols such as curcumin hold promise as a therapeutic agent in the treatment of chronic liver diseases. However, its development is plagued by poor aqueous solubility resulting in poor bioavailability. To circumvent the suboptimal bioavailability of free curcumin, we have developed a polymeric nanoparticle formulation of curcumin (NanoCurc™) that overcomes this major pitfall of the free compound. In this study, we show that NanoCurc™ results in sustained intrahepatic curcumin levels that can be found in both hepatocytes and non-parenchymal cells. NanoCurc™ markedly inhibits carbon tetrachloride-induced liver injury, production of pro-inflammatory cytokines and fibrosis. It also enhances antioxidant levels in the liver and inhibits pro-fibrogenic transcripts associated with activated myofibroblasts. Finally, we show that NanoCurc™ directly induces stellate cell apoptosis in vitro. Our results suggest that NanoCurc™ might be an effective therapy for patients with chronic liver disease.
Keywords: carbon tetrachloride, cirrhosis, curcumin, cytokines, liver fibrosis, myofibroblasts, NanoCurc™, polymeric nanoparticle
Cirrhosis is a wound-healing response to liver injury from a variety of viral, toxin or metabolic insults. Cirrhosis is responsible for as many as 35 000 annual deaths in the United States, unless the progressive degeneration in liver function can be rescued with orthotopic liver transplantation. Histologically, cirrhosis is characterized by excessive extracellular matrix deposition (ie, fibrosis), which surrounds regenerative hepatocellular nodules. In addition to deterioration in liver function, these nodules are also a seedbed for the formation of hepatocellular carcinoma. Hepatocyte injury and chronic inflammation appear to be essential in initiating events for liver fibrosis, leading to activated myofibroblasts. Myofibroblasts can be derived from hepatic stellate or immune cells, or the epithelial-to-mesenchymal transition of hepatocytes or cholangiocytes.1 In response to the transforming growth factor-β (TGF-β) ligand, the myofibroblasts produce collagen matrix proteins, eventually culminating in fibrosis.1,2 Agents that ameliorate hepatic injury or inflammation can serve as a potential antifibrotic therapeutic.
Curcumin or diferuloylmethane is a yellow polyphenol extracted from the rhizome of turmeric (Curcuma longa), a plant grown in tropical Southeast Asia. For centuries, turmeric has been used as a spice and coloring agent in Indian food, as well as a therapeutic agent in traditional Indian medicine.3 Curcumin has been explored as a promising antineoplastic,4 antioxidant,5 anti-inflammatory6,7 and antidepressant8,9 in animal models. In the liver, Chen and others have shown that oral administration of free curcumin (FC) inhibits liver fibrosis in vivo.10,11 Although curcumin has a direct effect on preventing stellate cell activation in vitro,12–15 it is not clear if curcumin’s antifibrotic activity in vivo is a direct effect in the liver or an indirect systemic effect through the tubular gastrointestinal system.
The potential for using curcumin as a systemic therapeutic has been plagued by poor bioavailability.16 In rodents, oral gavage with as much as 340 mg/kg of FC results in no detectable curcumin in the liver, although it is present in the gastrointestinal mucosa.17 Further, oral doses reaching 1000 mg/kg in rodents result in only submicromolar serum concentrations, and even at these levels, there is still no detectable curcumin in the liver.18 Comparable data exist in limited clinical trials performed in human beings, where ‘mega’ doses of daily oral FC (up to 8 g per day) mostly results in uptake that is limited to the colonic mucosa, and little, if any, of the active compound reaches the bloodstream or visceral sites.19–21
To circumvent the suboptimal systemic bioavailability of FC, we have developed a polymeric nanoparticle formulation of curcumin (NanoCurc™) that overcomes this major pitfall of the free compound.22 In contrast to curcumin, NanoCurc™ is fully soluble in aqueous media, and shows essentially no toxicity upon daily systemic administration through the intraperitoneal route in mice. Further, at comparable dosages, parenteral NanoCurc™ shows significant intrahepatic bioavailability, which can be found in both hepatocytes and non-hepatocytes, also referred to as nonparenchymal cells (NPCs). Finally, in this study, we confirm that parenteral NanoCurc™ can successfully ameliorate the effects of carbon tetrachloride (CCl4)-induced liver injury and subsequent fibrosis in rodents. These effects are associated with inhibition of pro-inflammatory cytokines production, enhancing intrahepatic antioxidant levels and reducing pro-fibrogenic transcripts. Finally, we show that the nanoparticle formulation retains the potency of the free compound in inducing apoptosis of pro-fibrotic stellate cells.
MATERIALS AND METHODS
Materials
Ultra-pure curcumin (>99% diferuloylmethane) was purchased from Sabinsa Corporation (Piscataway, NJ, USA); this source of curcumin has been used for both preclinical and clinical studies in the past.23,24 Monomers for polymer nanoparticle synthesis—specifically N-isopropylacrylamide (NIPAAM), vinylpyrrolidone (VP) and acrylic acid (AA)—were obtained from Sigma Aldrich (St Louis, MO, USA). Reagents for the polymerization step, including NN′ methylene-bis-acrylamide, ammonium persulfate (APS) and ferrous sulfate (FeSO4), were also procured from Sigma.
Synthesis of NanoCurc™
Polymer nanoparticles comprising of NIPAAM, VP and AA were synthesized via free radical mechanism, according to the detailed synthesis method we have described previously.25,26 The pre-distilled monomers of NIPAAM, VP and AA are mixed together in a molar ratio of 60:20:20, respectively; hence, the acronym ‘NVA622’ for the resulting polymer. Polymerization was performed for 24 h at 30°C under an inert (nitrogen) atmosphere, using APS and FeSO4 as initiator and activator, respectively. After complete polymerization, the total aqueous solution of polymer was purified using dialysis, and then lyophilized for post-loading of curcumin, as described.26 Typically, a 10 ml stock solution of polymeric nanoparticles (100 mg) was slowly mixed with 150 µl of curcumin solution in chloroform (10 mg/ml), and gently stirred for 15–20 min on low heating, to load curcumin and evaporate chloroform simultaneously. The resulting solution, corresponding to 1.5% (w/w) loading of curcumin in nanoparticles, was then snap frozen on a dry ice/acetone bath, and lyophilized. The lyophilized NanoCurc™ powder is stored at 4°C until further use, whereupon simple reconstitution in an aqueous phase is required before parenteral administration.
Nanocurc™ Bioavailability Protocol
Male C57BL/6 mice 6–10 weeks of age weighing approximately 25 g were purchased from Charles River (Wilmington, MA, USA). All animals were housed at The Johns Hopkins University animal facility, and handled according to NIH guidelines and protocols approved by the Institutional Animal Care and Use Committee. Mice had free access to food and water in 50–60% relative humidity and 12 h light cycle. Mice received one or three doses of either 1667 mg/kg body weight dose of void nanoparticles and NanoCurc™ (equivalent to 25 mg/kg body weight of FC) or control phosphate-buffered saline (PBS) via intraperitoneal injection over a 24 h period. Curcumin bioavailability was determined by high-performance liquid chromatography in liver tissue obtained 6 or 24 h after treatment as detailed in Results and Figure 1 legend. Oral gavage was performed by dissolving 25 mg of curcumin in 200 µl of PBS and instilling the resulting fluid into the stomach using a curved gavage needle and syringe. Parenchymal (hepatocytes) and NPCs were isolated by clearing the liver of blood with 50 ml PBS with Ca2+ through a portal vein rinse and digesting the liver with 50 ml of collagenase (1.6 mg/ml) in situ. The digested liver was removed, weighed and pressed through a wire mesh into 40 ml of PBS. The resultant homogenate was centrifuged at 50 g for 5 min at 4°C The top half of the supernate containing NPC was collected in 50 ml tube and kept on ice for subsequent processing of NPC. The hepatocyte pellet was resuspended in cold PBS and re-pelleted and washed three times with 50 g centrifugations for 5 min at 4°C. The supernate NPC fraction was centrifuged at 800 g for 5 min. The resultant pellet was incubated with 5ml ACK lysis buffer at room temperature to remove the RBCs. It was washed with cold PBS three times. The isolated NPC and parenchymal fractions were counted using a hemocytometer and smeared onto slides, formalin-fixed and stained with hematoxylin–eosin (H&E) histochemical stain for counting to estimate cell purity.
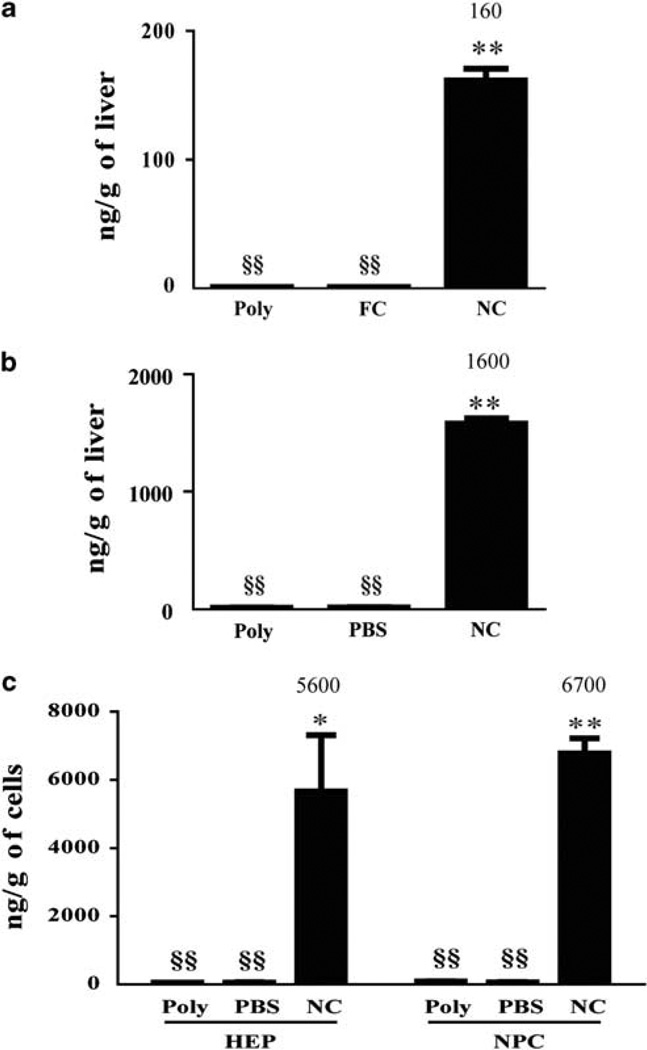
Figure 1.
Bioavailability of NanoCurc™ in liver tissue. (a) Mice (n = 3 in each group) were treated with single intraperitoneal (i.p.) 25 mg/kg dose of NanoCurc™ (NC), void nanoparticles (Poly) or an equivalent oral gavage of 25 mg/kg of free curcumin (FC). Curcumin bioavailability was determined by high-performance liquid chromatography (HPLC) in liver tissue obtained 12 h after treatment. (b) Mice (n = 3 in each group) were treated with three i.p. 25 mg/kg doses equally spaced over 24 h of NanoCurc™, Poly and phosphate-buffered saline (PBS). Curcumin bioavailability in whole liver tissue was determined by HPLC 6 h after the last dose. (c) Mice (n = 3 in each group) were treated with three i.p. 25 mg/kg doses equally spaced over 24 h of NC, poly and PBS. Hepatocytes (HEPs) and non-parenchymal cells (NPCs) were isolated from the liver. Curcumin bioavailability in HEPs and NPCs was determined by HPLC 6 h after the last dose. Data were expressed as mean ± s.e.m.,*P < 0.01, **P < 0.001, t-test, from two experiments. §§indicates below limit of detection.
Estimation of Liver Curcumin Levels
Liver and intracellular curcumin levels were measured at the Johns Hopkins Pharmacology Analytical core facility in tissue lysates, using a recently described liquid chromatography/tandem mass spectrometric (LC/MS/MS) approach.22
CCl4 Treatment Protocols
CCl4 was administered dissolved in olive oil at a 1:9 ratio through intraperitoneal injection at a dose of 1 µg/g of body weight on days 5, 7, 10 and 12. NanoCurc™ dissolved in pyrogen free sterile water (Life Technologies, Carlsbad, CA, USA) was given once a day equivalent to 25 mg/kg body weight of FC via intraperitoneal injection on days 1–4, 6, 8, 9, 11 and 13. Mice were harvested on day 14, 24 h after the last NanoCurc™ injection.
Serum Alanine Aminotransferase
Serum alanine aminotransaminase were determined with a colorimetric kit (Biotron Diagnostics, Hemet, CA, USA), according to the manufacturer’s protocol. Briefly, 100 µl of pre-warmed ALT reagent substrate and 20 µl of diluted serum samples were incubated for 30 min at 37°C in a water bath. Sodium pyruvate (1.5 mM) was used in graduated concentrations for generating a standard curve. Then, 100 µl of colorimetric reagent (dinitrophenyl hydrazine) was added to all samples and standards and incubated at room temperature. A measure of 1 ml sodium hydroxide was added to each tube, and the absorbance measured at 520 nm.
Enzyme-Linked Immunosorbent Assays
At the time of harvest, liver tissue was weighed and homogenized with 500 µl of RIPA buffer containing complete mini protease inhibitors (Roche Diagnostics, Indianapolis, IN, USA). The lysate was centrifuged at 21 000 g at 4°C for 10 min and the clarified supernate was collected and subjected to total protein BCA assay (Pierce, Rockford, IL, USA). Equal total protein was used in an enzyme-linked immunosorbent assay according to the manufacturer’s instructions(R & D Systems Minneapolis, MN, USA).
Liver mRNA Isolation and Real-Time RT-PCR
Liver tissue was harvested and collected in RNAlater (Sigma Life Science, St Louis, MO, USA). Total RNA was extracted by using RNeasy Min Kit according to the manufacturer’s protocol (Qiagen, Valencia, CA, USA). DNase I column digestion was carried out to eliminate the contamination of genomic DNA. cDNA synthesis was carried out using AffinityScript Multi-Temperature cDNA Synthesis Kit (Stratagene, Santa Clara, CA, USA) using random primers and following the manufacturer’s protocol.
Quantitative real-time RT-PCR amplification was performed with TaqMan Fast Universal PCR Master Mix with (Applied Biosystem, Carlsbad, CA, USA) without AmpErase UNG using 250 ng of total RNA in a 96-well plate. The real-time RT-PCR was performed with an Applied Biosystem Step One Plus system. The assay ID, the reference sequence, and the exon boundaries used to design the probes and primers are shown in Table 1. Genes hprt and H2AX were used as references for TNF-α and IL-6, whereas gene hprt was used as reference for COL A, FN-1, TGF-β and PPARG.
Table 1.
Real-time RT-PCR primer sets
| Target gene | Assay ID | Reference sequence | Exon boundary |
|---|---|---|---|
| TGF-β | Mm01178820_m1 | NM_011577.1 | 5–6 |
| COL A | Mm00801666_g1 | NM_007742.3 | 49–50 |
| FN-1 | Mm01256744_m1 | NM_010233.1 | 44–45 |
| α-SMA | Mm01546133_m1 | NM_007392.2 | 1–2 |
| Hprt1 | Mm00446968_m1 | NM_013556.2 | 6–7 |
| IL-6 | Mm00446190_m1 | NM_031168.1 | 2–3 |
| H2AZ | Mm02018760_g1 | NM_016750.2 | 1–1 |
| H2AV | Mm01181326_m1 | AI854262.1 | 3–4 |
Liver Leukocyte Isolation and Flow Cytometry
NanoCurc™ dissolved in pyrogen free sterile water was given once a day equivalent to 25 mg/kg body weight of FC via intraperitoneal injection on days 1, 4 and 6, whereas CCl4 (1 µg/g of body weight) was administered on days 5 and 7. Mice were harvested on day 8, 24 h after the last CCl4 injection. Liver leukocytes were isolated as described previously.27 Livers were weighed before lymphocyte isolation and total number of isolated leukocytes was determined with a hemocytometer. Equal number of lymphocytes were stained with anti-CD3, Gr1.1 and NK1.1 (BD Bioscience) and subjected to flow cytometry (FACSCalibur™, BD Bioscience). Analysis was performed with the FlowJo™ software (Tree Star, Ashland, OR, USA).
Histology and Sirius Red Stain for Fibrosis
The medial liver lobe was collected, fixed in methacarn (60% methanol, 30% chloroform, 10% glacial acetic acid), embedded in paraffin and 5-µm sections stained for H&E. For Sirius Red staining (Sigma Aldrich), unstained sections were de-paraffinized in warm xylene and hydrated in graded alcohol and water. Thereafter, nuclei were counterstained with Weigert’s hematoxylin for 30 min, and then the slides were rinsed in tap water for 3 min, followed by Sirius Red staining for an hour. This was followed by washing in two changes of acetic acid water, dehydrated in three changes of 100% ethanol, cleared in xylene and mounted in a resinous medium.
Fibrosis Quantification
Ten fields containing no significant staining artifacts (folds or scratches) were randomly selected from each slide. Each field was digitally acquired with 10× objective as a 1280 pixel × 960 pixel JPEG image using a Nikon Eclipse 50i microscope equipped with a Nikon Digital Sight DS-5M camera. To correct for uneven illumination (vignetting), five additional fields were acquired in empty areas of the slide and averaged to produce a single background illumination image. A median filter (radius = 8) was applied to the background illumination image to further reduce potential noise. Custom macros in ImageJ 1.42q (http://www.rsbweb.nih.gov/ij/) were used to measure percent fibrosis. Each image was first converted to 8-bit gray-scale and then corrected for uneven illumination (‘shading correction’). Shading correction consisted of dividing an image by its associated background illumination image and then multiplying the result by the median gray value of the background illumination image. After shading correction, an observer manually selected a gray-scale threshold value that best segments the fibrotic areas from the background image. Percent fibrosis was calculated as the area of fibrosis divided by the total image area multiplied by 100.
Estimation of Free and Total Glutathione Levels in Liver Tissue Lysates
Corresponding void polymeric nanoparticles and NanoCurc™-treated liver tissue slices were homogenized, extracts were prepared and equal volumes of the extract were subjected to estimation of free and total glutathione (GSH) levels. The results were normalized with protein and amounts of free GSH were measured using the fluorescent-based NADPH substrate and other appropriate reagents (Arbor assay, MI, USA). For estimation of total GSH levels (free plus oxidized), NADPH, GSH reductase and ThioStar reagent were added to the extract and fluorescent signal was read after 15 min, and the activity was calculated. The level of oxidized form of GSH disulfide (GSSG) was estimated from the difference between total vs free GSH. In all instances, a standard curve was prepared before assay performance. All assays were performed in triplicate, and mean and standard deviations were determined.
Hepatic Stellate Cell Line
The immortalized rat liver hepatic stellate cell line, HSC-T6, was a kind gift from Scott L Friedman (Mount Sinai School of Medicine, New York, NY, USA).28 This line is maintained in DMEM (Life Technologies) supplemented with 5% FBS and the antibiotics penicillin–streptomycin (Life Technologies). Incubation is carried out at 37°C in a humidified incubator with 5% CO2.
Apoptosis and Cell Survival Studies
Apoptosis in the hepatic stellate cells (HSC-T6) was detected by 4′,6-diamidino-2-phenylindole (DAPI) fluorescent dye (Invitrogen Corporation, Carlsbad, CA), caspase-3 apoptotic detection (Cell Signaling Technology, Danvers, MA, USA) and by Trypan blue dye exclusion methods. HSC-T6 cells were plated at 35 000 cells per cm2 overnight and then treated 20 h with varying concentrations (5,10, 15 and 20 µM) of FC dissolved in 0.1% DMSO or the equivalent amount of NanoCurc™ dissolved in sterile pyrogen free water. For DAPI staining: the adherent cells were washed in PBS (pH 7.2) and fixed in 3.7% paraformaldehyde in PBS (pH 7.2) for 15 min at room temperature. The cells were washed with PBS for 5 min at room temperature, rinsed in methanol for 5 min and after a final PBS wash the cells were covered with DAPI staining solution (2 µg/ml) and incubated 5 min and observed under a fluorescence microscope. A small densely stained (pyknotic) nucleus was regarded as apoptotic. Mitotic nuclear forms (metaphase, anaphase, etc) were regarded as a single non-apoptotic cell. For Trypan blue cell viability counts: cells were plated at 35 000 cells per cm2 in complete DMEM media overnight, treating 20 h with 20, 40 and 80 µM FC dissolved in 0.1% DMSO or equivalent amount of NanoCurc™ dissolved in sterile pyrogen free water. After 0.05% trypsinization, cells were collected and diluted 50:50 into a solution of 0.4% (w/v) Trypan blue. Live cells (exclude dye) and dead cells were counted using a hemocytometer. For caspase-3 detection: cells were seeded in a 24-well plate in complete medium overnight and treated for 20 h with varying concentrations (5, 10, 15 and 20 µM) of FC dissolved in 0.1% DMSO or the equivalent amount of NanoCurc™ dissolved in sterile pyrogen free water. Cells were washed with PBS twice and fixed with 3.7% paraformaldehyde at room temperature for 10 min, washed twice with PBS and denatured in 0.1% Triton X-100 in PBS at room temperature for 10 min. After a two PBS washes, the cells were blocked with 5% fetal bovine albumin at room temperature for 30 min, and then incubated with 1:100 anti-cleaved caspase-3 (Cell Signaling Technology) solution at room temperature for 1 h. Cells were washed twice with PBS and incubated with 1:300 secondary antibody, Alexa Fluor 568 goat anti-rabbit IgG labeled with bright, orange-red—fluorescent Alexa Fluor 568 dye (Invitrogen Corporation) at room temperature for 30 min. Cells were then washed twice with PBS, counterstained with DAPI and observed under a fluorescent microscope.
RESULTS
Enhanced Bioavailability of Systemic NanoCurc™ in Liver Tissue
It has been previously shown that oral intake of up to 400 mg/kg of curcumin may be required for amelioration of liver injury in CCl4-treated rodents.10,11 In terms of human consumption, these translate into ‘mega’ doses of several grams (between 8 and 12 g) of curcumin, which becomes untenable in a clinical setting.29,30 Furthermore, even intraperitoneal doses of FC result in only ephemeral (min) intrahepatic curcumin delivery.16–18
A recently published study using NanoCurc™ in cancer models shows that a single intraperitoneal dose of this formulation (containing 25 mg/kg of curcumin equivalent) results in peak curcumin plasma levels 4 h after treatment;22 however, the tissue biodistribution, especially in the liver, was not studied. Using LC-MS/MS, we found significantly greater intrahepatic curcumin concentrations (160 ± 9 ng/g of liver tissue, P < 0.001) 12 h after a single 25 mg/kg intraperitoneal injection of NanoCurc™ compared with control void nanoparticles (Figure 1a). As expected, an equivalent dose of FC delivered via oral gavage did not result in any detectable intrahepatic curcumin. As we planned to use multiple doses, we wanted to see if multi-dosing resulted in higher intrahepatic curcumin concentration than a single dose of NanoCurc™. Compared with control injection and void nanoparticles, we found significantly higher curcumin concentration (1600 ± 44 ng/g of liver tissue, P < 0.001) in the liver 6 h after three equally spaced 25 mg/kg intraperitoneal doses (Figure 1b). As the liver is a complex tissue made of many cell types, we next wanted to determine if NanoCurc™ delivered curcumin to parenchymal (hepatocytes) and/or non-parenchymal (all other liver cell types) cells. Mice received three 25 mg/kg NanoCurc™ doses 8 h apart and were harvested 6 h after the last dose of NanoCurc™. The liver was fractionated into parenchymal cells and NPCs. The purity of the NPC fraction (92%) and hepatocyte fraction (96%) was determined by examination of H&E-stained smear preparation of each fraction (Supplementary Figure 1). We detected significantly more curcumin in hepatocytes (5600 ± 1000 ng/g of hepatocytes, P < 0.01) and NPCs (6700 ± 440 ng/g NPCs, P < 0.001) compared with void polymer- and vehicle-treated controls (Figure 1c). We attributed the increase in concentration of curcumin in the hepatocytes and NPC-purified fractions compared with the intact liver (Figure 1c vs b) to regions, perhaps structural, non-cellular components, that contribute to liver weight, but do not absorb curcumin.
NanoCurc™ Attenuates Hepatocellular Injury and Levels of Pro-Inflammatory Cytokines
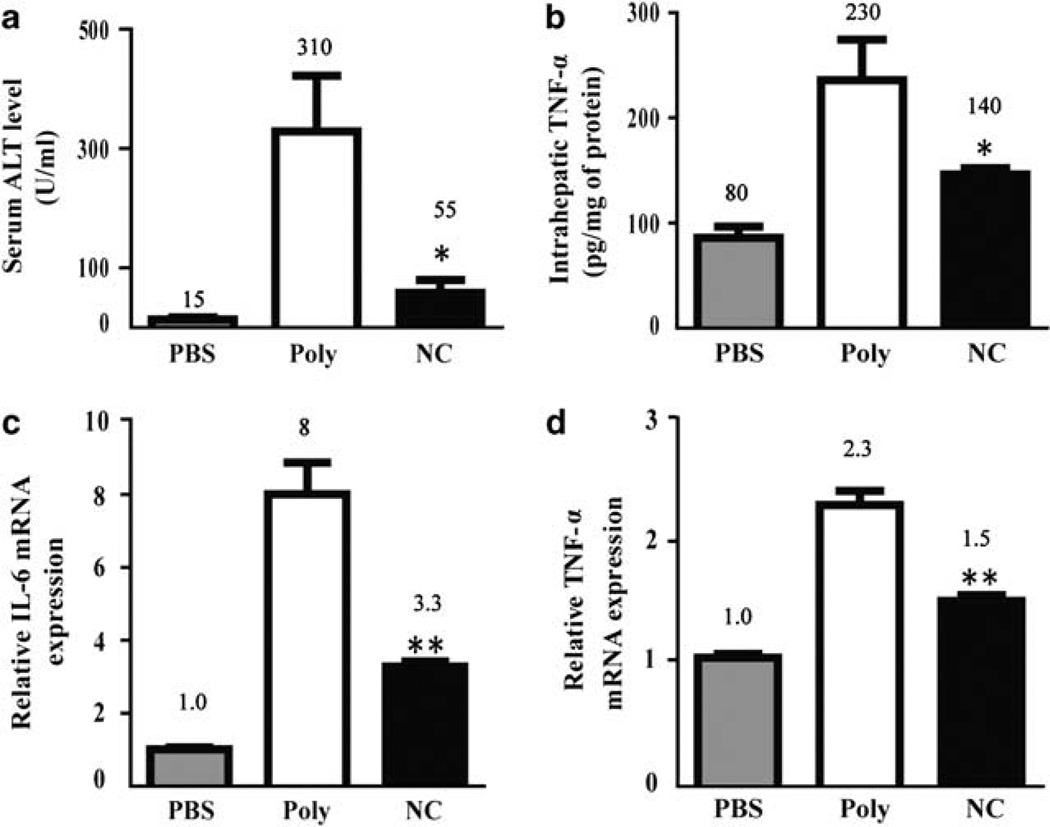
First, we wanted to determine if accumulation of intrahepatic curcumin administered as a nanoformulation was capable of suppressing CCl4-induced liver injury. Mice received 25 mg/kg intraperitoneal NanoCurc™ or control injection according to the treatment protocol detailed in Materials and Methods. To provide biochemical evidence of NanoCurc™ hepatoprotective effects, we collected serum 48 h after the last CCl4 treatment. Compared with control mice, those that received intraperitoneal NanoCurc™ showed biochemical evidence of reduced liver injury. Control-treated (310 ± 76U/ml) mice serum ALT activity was significantly higher compared with NanoCurc™-treated mice (55 ± 19 U/ml, P < 0.05) (Figure 2a). Next, we wanted to see if NanoCurc™ could suppress TNF-α, a pro-inflammatory cytokine associated with CCl4-induced liver injury.31 CCl4-induced TNF-α production is a powerful initiator of hepatocyte apoptosis and thought to be one of the important propagators of liver injury. Liver tissues collected 48 h after CCl4 treatment showed 1.6-fold significantly reduced TNF-α protein concentration in mice treated with NanoCurc™ compared with controls (P < 0.05) (Figure 2b). We also investigated IL-6, another inflammatory cytokine associated with hepatocellular injury, and TNF-α mRNA induction, and found a significant 2.4- and 1.5-fold reduction in IL-6 and TNF-α, respectively, in NanoCurc™-treated mice compared with controls (P < 0.001) (Figure 2c and d). The inflammatory cell milieu after liver injury is complex.32 The innate immune system is rapidly activated following injury and has been implicated as an important regulator of liver injury and fibrosis.33–35 We examined how NanoCurc™ treatment might alter the innate inflammatory response to CCl4. Livers leukocytes were isolated 24 h after the last NanoCurc™ treatment. There was a slight increase in the total number of inflammatory cells in void nanoparticle (1.9 ± 0.3 lymphocytes per g liver tissue) and NanoCurc™ (2.2 × 106 ± 0.4 lymphocytes per g liver tissue) compared with untreated control mice (1.0 × 106 ± 0.5 lymphocytes per g liver tissue); however, this was not statistically significant. Next, we examined CD3+ NK1.1+ natural killer T (NKT) cells whose absence makes mice resistant to liver fibrosis, whereas an excess accelerates fibrosis.36–39 Consistent to what others have reported, we found NKT cells decreased following CCl4 treatment. We found 24 ± 1% NKT cells in the liver of untreated wild-type mice, which fell to 12 ± 5% in void nanoparticle and 9 ± 1% in NanoCurc™-treated mice that also received CCl438 (Supplementary Figure 2). There was no significant change in the percentage of NK cells (Supplementary Figure 2), another innate immune cell implicated in antifibrosis.39–41 Lastly, we looked at neutrophil inflammation using Gr-1high staining.42 Consistent with what others have reported,38,43 CCl4 enhanced influx of Gr-1high cells into the liver. Untreated mice had 5% ± 3 Gr-1high neutrophils in the liver compared with 13 ± 1% in void nanoparticle-treated mice that also received CCl4 (Supplementary Figure 2). Interestingly, mice treated with CCl4 and NanoCurc™ also had a significant increase in Gr-1high cells compared with untreated mice (15 ± 8 vs 5 ± 3%) and was not different compared with mice treated with CCl4 and void nanoparticles (15 ± 8 vs 13 ± 2%). Taken together, these results support the notion that NanoCurc™ potently suppresses both hepatocellular injury and pro-inflammatory markers with relatively little effect on innate inflammatory cell infiltrate after CCl4 challenge.
Figure 2.
NanoCurc™ prevents carbon tetrachloride (CCl4) associated liver injury and inflammation. The mice were injected with either phosphate-buffered saline (PBS), void nanoparticles (Poly) or NanoCurc™ (NC) injections along with CCl4 according to the treatment protocol in Materials and Methods. (a) Liver damage was determined by measuring serum alanine aminotransferase (ALT) activity (expressed in Sigma Frankel units) and (b) intrahepatic tumor necrosis factor (TNF)-α was determined by enzyme-linked immunosorbent assay (ELISA). (c and d) Relative interleukin (IL)-6 and TNF-α mRNA expression was determined with real-time reverse transcription-polymerase chain reaction (RT-PCR) using the ΔΔCt method with histone 2AZ and hprt serving as a reference genes. PBS-negative control liver tissue was set to relative expression level of 1.0. Data were expressed as mean ± s.e.m., *P < 0.05, **P < 0.001, t-test, from two experiments.
Systemic NanoCurc™ Prevents Hepatic Fibrosis in the CCl4 Model of Liver Injury
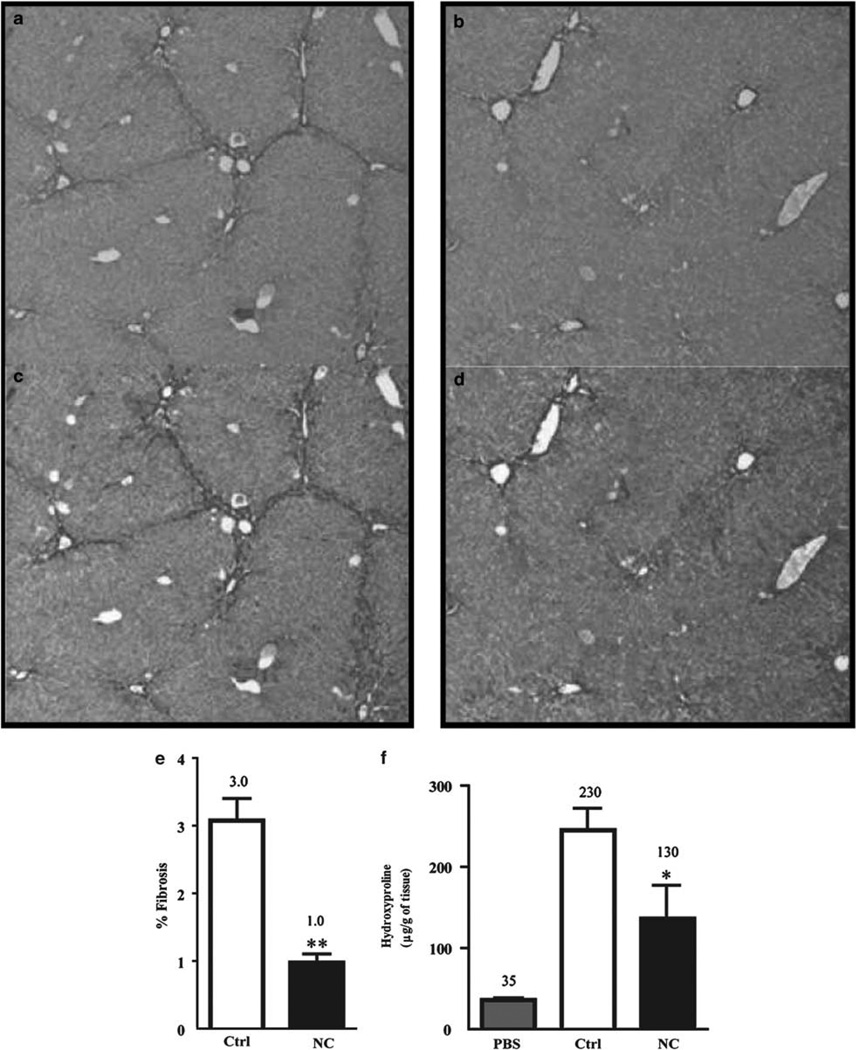
If NanoCurc™ protected against liver injury, it should also prevent the subsequent development of fibrosis that develops following low-dose, repeated CCl4 administration.44 Previous studies have established the inhibitory effects of FC on pro-fibrotic hepatic stellate cells in vitro.12–15 Nonetheless, given the minimal bioavailability of curcumin in the liver, the relevance of these studies to in vivo observations is not clear.18 NanoCurc™ treatment significantly suppressed liver fibrosis three-fold as determined by quantitative image analysis of Sirius Red tissue staining (Figure 3a–d). NanoCurc™ administered mice showed only 1.0 ± 0.1% quantifiable fibrosis, whereas control treatment showed 3.0 ± 0.3% (P < 0.001) (Figure 3e). We also found significantly less hydroxyproline in the liver of NanoCurc™- (130 ± 40 µg/g of liver) treated mice compared with void nanoparticle- (230 ± 20 µg/g of liver, P < 0.05) treated mice (Figure 3f).
Figure 3.
NanoCurc™ inhibits carbon tetrachloride (CCl4)-induced liver injury and fibrosis. Mice were treated with intraperitoneal control (a, c) or NanoCurc™ (b, d) injection and CCl4 according to the treatment protocol in Materials and Methods. (a, b) Methacarn-fixed liver tissues were stained with Sirius Red, which highlights collagen deposition in dark red. Representative photomicrographs (× 10 objective). (c, d) The extent of fibrosis was quantitatively determined using Threshold Image Analysis to highlight fibrosis (orange color) in 10 randomly selected photomicrographs (× 10 objective) after conversion to 8-bit gray scale. (e) The percentage of fibrosis calculated as the area of fibrosis divided by the total image area multiplied by 100. (f) Hydroxyproline assay was carried out to quantify the collagen content. Data were expressed as mean ± s.e.m.,*P < 0.05, **P < 0.001, t-test, from two experiments.
NanoCurc™ Enhances Levels of GSH in the Liver
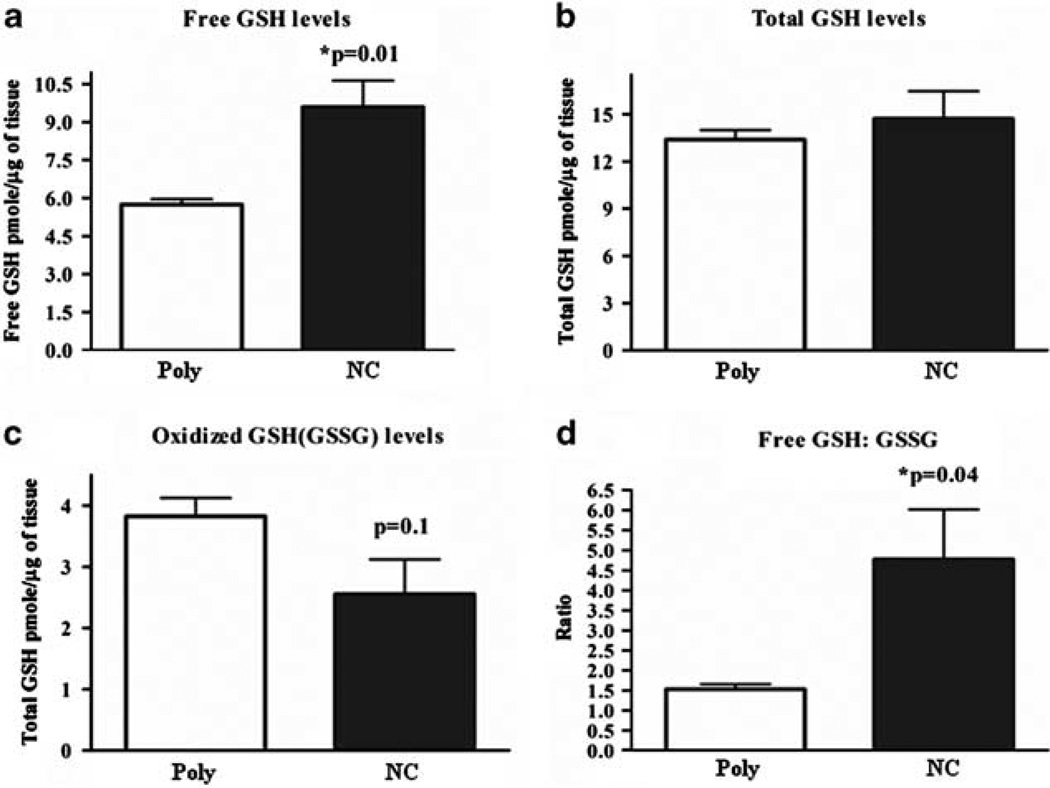
CCl4 is activated by centrilobular cytochrome P450 in the liver to form a trichloromethyl radical , which in the absence of oxygen exclusively causes hepatocyte injury through lipid peroxidation. If oxygen is present, the can rapidly change to a highly reactive peroxytrichloromethyl radical (CCl3OO•). This CCl3OO• can also cause hepatocyte injury through lipid peroxidation.45 Although the radical’s action can predominate in the liver’s oxygen poor centrilobular zone, GSH can significantly protect against CCl3OO• injury even in extremely low (1%) oxygen tensions.45 GSH is a major free radical scavenging species in hepatocytes that can protect hepatocytes from oxidative stress46 and is essential in causing stellate cell apoptosis.47 For these reasons, we sought to quantify the GSH levels in NanoCurc™-treated livers. We quantitatively assessed for levels of free and oxidized GSH in hepatic lysates post-CCl4 exposure in vehicle- and NanoCurc™-treated cohorts. NanoCurc™ treatment significantly increased levels of free GSH 1.75-fold vs control livers (P < 0.01) (Figure 4a). Interestingly, unlike free GSH, NanoCurc™ did not significantly alter levels of total GSH (Figure 4b). NanoCurc™ treatment did lower oxidized GSSG compared with void nanoparticle treatment, but this was not statistically significant (Figure 4c), suggesting that the is a major contributor to lipid peroxidation in our system. Notably, the ratio of free GSH to oxidized GSSG was significantly increased in the NanoCurc™-treated cohort vs the vehicle cohort (P < 0.04) (Figure 4d). Although the exact mechanism for the significantly increased free GSH levels in the liver of NanoCurc™-treated mice is unclear, the composite results indicate that NanoCurc™ favorably increases redox capacity of the cells, and thereby can protect liver cells from free radical insults.
Figure 4.
NanoCurc™ increases redox capacity of the hepatocytes. (a) Free glutathione (GSH), (b) oxidized GSH (c), total GSH and (d) the ratio of free GSH to oxidized glutathione disulfide (GSSG) were determined as described in Materials and Methods in liver lysates harvested 48 h after CCl4 treatment in void polymer- (Poly) and NanoCurc™- (NC) treated mice. Data were expressed as mean ± s.e.m., *P values as indicated, t-test, Poly n = 4, NC n = 5.
NanoCurc™ Attenuates Transcripts of Pro-Fibrogenic Cytokines Associated with Stellate Cell Activation
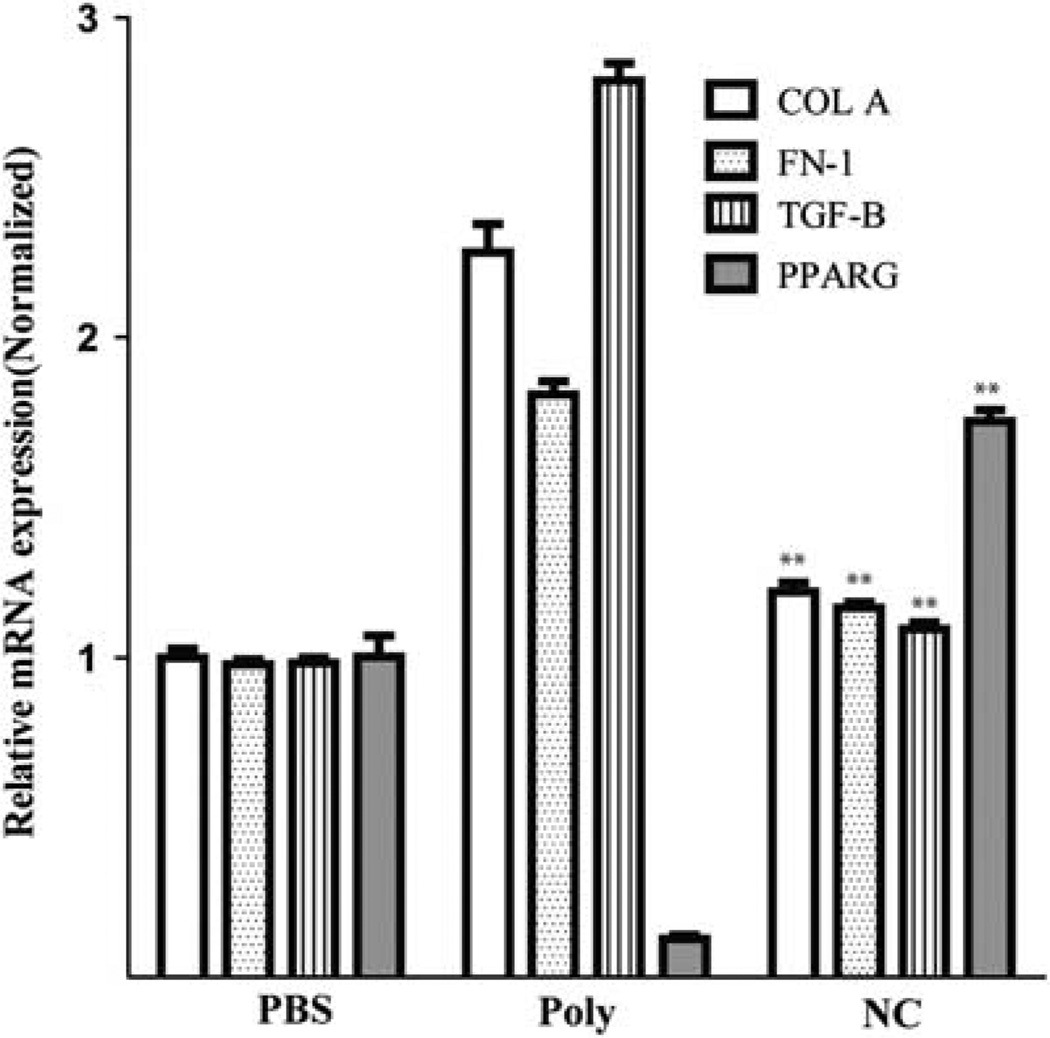
In addition to hepatocellular injury and inflammation, a critical initiating component of the fibrogenic responses in the liver is activation of collagen-producing myofibroblasts.1,48 To further understand the mechanisms by which NanoCurc™ might alter myofibroblast responses in the setting of liver injury, we investigated if NanoCurc™ disrupted production of pro-fibrotic cytokines transcripts. Type I α collagen49,50 and fibronectin 151,52 are pro-fibrotic proteins produced by activated myofibroblasts. We found that NanoCurc™ treatment significantly suppressed mRNA expression of collagen A and fibronectin 1.8- and 1.6–fold, respectively, compared with control void nanoparticle treatment (P < 0.001) (Figure 5). Notably, NanoCurc™ treatment reduced these pro-fibrotic transcripts to nearly the basal levels present in untreated mice (Figure 5). We then examined TGF-β, a molecule that has been strongly implicated in the activation of myofibroblast and stimulates collagen production.53,54 As expected, void nanoparticle-treated mice showed a 2.6-fold increase in TGF-β expression while NanoCurc™ treatment significantly and completely abrogated expression (P < 0.001) (Figure 5). In vitro studies have shown the importance of the nuclear transcription factor peroxisome proliferator-activated receptor γ (PPAR-γ) in controlling myofibroblast activation and its requirement in promoting curcumin-induced apoptosis.15,55–57 There was significant inhibition of PPAR-γ expression in void nanoparticle-treated mice, whereas NanoCurc™ significantly induced a 14 -fold expression of PPAR-γ compared with void nanoparticle treatment (P < 0.001) (Figure 5). Collectively, these data support NanoCurc™ ability to inhibit fibrosis through suppression of pro-fibrogenic transcripts and suggest that NanoCurc™ might induce myofibroblast apoptosis through activation of PPAR-γ.
Figure 5.
NanoCurc™ attenuates genes associated with stellate cell activation and fibrosis while stimulating pro-apoptotic gene expression. Liver tissue harvested from untreated mice (PBS), void polymer (Poly) or NanoCurc™ (NC) injections along with carbon tetrachloride (CCl4) according to the treatment protocol in Materials and Methods. Gene expression levels for transcripts encoding for Collagen A (COL A), fibronectin 1 (FN-1), transforming growth factor-β (TGFB) and peroxisome proliferator-activated receptor gamma (PPARG) were determined after isolation of RNA with real-time reverse transcription-polymerase chain reaction (RT-PCR). Gene expression was normalized with hprt and fold expression was determined using the ΔΔCt method in which untreated liver tissue was set to relative expression level of 1.0. Data were expressed as mean ± s.e.m., **P < 0.001, in comparing Ctrl vs NC, t-test; control n = 4, NC n = 5, representative of two experiments.
NanoCurc™ Treatment Induces Hepatic Stellate Cell Apoptosis
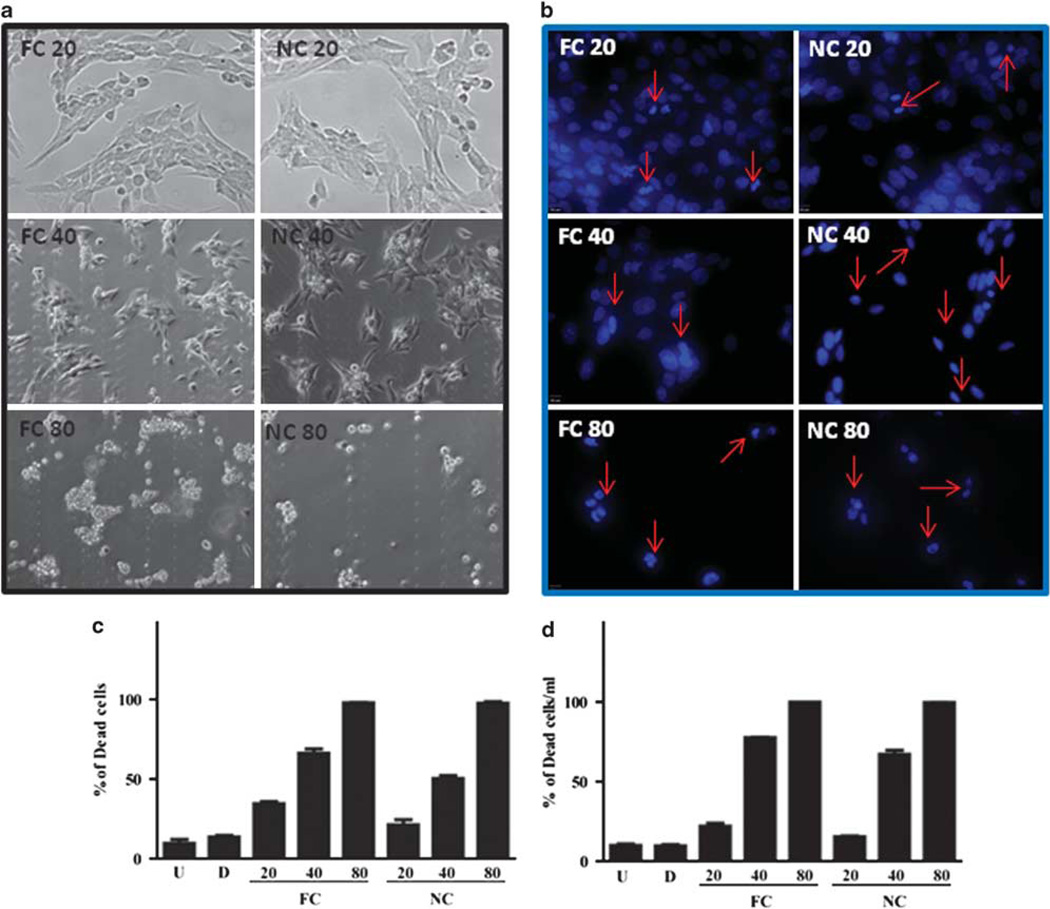
An important, but not the sole source of activated myofibroblasts is hepatic stellate cells.1 We wanted to determine if NanoCurc™ was capable of inducing stellate cell apoptosis as has been reported for FC.15,58 In vitro hepatic stellate cells were treated with equivalent concentrations of curcumin, NanoCurc™ or vehicle control for 24 h. We found a significant and dose-dependent decrease in the number of viable stellate cells in curcumin- and NanoCurc™-treated cultures compared with controls (Figure 6a–c). The same trend was found whether we assayed with dye exclusion (Figure 6d) or the apoptosis marker cleaved caspase-3 (Supplementary Figure 3). As expected, the nanoencapsulation of curcumin did not alter its biological activity in vitro.26
Figure 6.
In vitro NanoCurc™ treatment induces stellate cell apoptosis. Immortalized rat stellate cells, hepatic stellate cell line (HSC-T6), were plated and treated with the indicated doses of free curcumin (FC) dissolved in 0.1% dimethylsulfoxide (DMSO) or an equivalent dose of NanoCurc™ (NC) in plating media. Stellate cells were treated with 20, 40 and 80 µM of FC or NC. After treatment, cells were fixed with 3.7% paraformaldehyde, stained with 4′,6-diamidino-2-phenylindole (DAPI) and examined under ultraviolet (UV) light for morphological nuclear changes of apoptosis. Representative fluorescent photomicrographs show a dose-dependent increase in condensed apoptotic nuclei (red arrows) with nearly all apoptotic nuclei at the highest dose (80 µM). (a) Representative phase-contrast micrographs show shrinkage and condensation of the cells. (b, c) HSC-T6 were plated and treated with the indicated doses of FC, an equivalent dose of NC or 0.1% DMSO (d) and untreated (U). Stellate cells were treated with 20, 40 and 80 µM of FC or NC. After treatment, cells were fixed with 3.7% paraformaldehyde, and stained with DAPI. In all, 200 DAPI-positive cells were counted for live and dead cells. The percentage of the dead cells to the total cells was calculated as the total number of cells divided by the number of dead cell multiplied by 100. (d) HSC-T6 were plated and treated with the indicated doses of FC, an equivalent dose of NanoCurc™ (NC), 0.1% DMSO (d) and untreated (U). Stellate cells were treated with 20, 40 and 80 µM of FC or NanoCurc™. After treatment, the cells were trypsinized without ethylenediaminetetraacetic acid (EDTA) combined with an equal volume of Trypan blue and counted on hemocytometer for live and dead cells per ml. Percentage of dead cells were calculated by dividing Trypan-positive cells by the total number of cells multiplied by 100. All experiments are representative from two experiments.
DISCUSSION
Hepatic injury and fibrosis develops in response to a wide array of etiologies such as viral infection, toxin exposure, alcohol abuse and metabolic diseases.59 Persistent injury leads to inflammation, fibrosis and compensatory hepatocyte hyperplasia usually cumulating in cirrhosis. Any treatment that can prevent hepatocellular injury, inflammation or fibrosis is a potential therapeutic for preventing cirrhosis in the setting of chronic liver disease. Chronic liver disease is the third leading cause for hospital visits in the United States, resulting in 5.9 million visits in 2004 and nearly 30 000 annual deaths.60 Given this considerable disease burden, there is intense interest in developing therapies aimed at chronic liver disease.
Plant-derived polyphenols are increasingly being recognized for their medicinal potential.16 Curcumin is a yellow polyphenol extracted from the rhizome of turmeric (C. longa), which has been used as a spice, coloring agent and as a therapeutic agent in traditional Indian medicine.3 Curcumin’s promising antineoplastic,4 antioxidant,5 anti-inflammatory6,7 and antidepressant8,9 activities have been explored in animals. In vitro and in vivo studies have explored curcumin’s antifibrotic potential in the liver.10–15 However, its promise is plagued by the lack of bioavailable curcumin in liver even following massive oral doses.18 Although curcumin formulated with phosphatidylcholine (Meriva®) enhances oral absorption and results in detectable intrahepatic curcumin, it is only detectable up to 2 h after dosing.17 The lack of water solubility is the primary problem limiting curcumin’s therapeutic development.
To address the lack of solubility, we developed a biocompatible nanomaterial to provide an opportunity for harnessing the full potential of a potent, yet poorly soluble compounds like curcumin.61–63 We have developed polymeric nanoparticles comprised of NIPAAM, MMA and AA that are capable of solubilizing a broad range of poorly water-soluble drugs. Curcumin encapsulated in these polymeric nanoparticles, NanoCurc™, is fully soluble in aqueous media, and shows essentially no toxicity upon daily systemic administration through the intraperitoneal route in mice.
In this study, we found that NanoCurc™ delivers a substantial concentration of curcumin to the liver. As early as 6 h after multiple doses and as long as 12 h after a signal dose, a significant amount of curcumin is detected in the liver. The curcumin is detectable in hepatocytes isolated from the liver. This might explain why we see a significant reduction in CCl4-induced hepatocellular injury and production of proinflammatory cytokines (Figure 2). The exact mechanism by which curcumin induces a protective hepatocellular environment is not clear. The ability of GSH to prevent CCl4 liver injury is limited as the low oxygen tensions in the centrilobular zones limit formation of the CCl3OO• that GSH could reduce. Another possibility is the inhibition of the CCl3 free radical formation, which is required for CCl4-mediated liver injury. The formation of the free radical requires cytochrome P450, which is a known target of curcumin.64–66 Although we see the delivery of curcumin to hepatocytes as overcoming a significant limitation in the therapeutic development of curcumin, the need to deliver curcumin to hepatocytes is not clear. Others studies have shown large daily oral doses of FC can inhibit CCl4-induced fibrosis in rats.11 However, this delivery route results in detectable curcumin in the gastrointestinal tract, but not in the liver.10 This suggests that curcumin might work through multiple mechanisms.
We also found that NanoCurc™ treatment results in a significant amount of curcumin in the NPC compartment. Although the cellular make-up of this compartment is complex, it does contain pro-fibrotic stellate cells and myofibroblasts. Lipid-laden cells, likely stellate cells, were evident in the NPC fraction (Supplementary Figure 1, green arrows). This is important, as there is considerable in vitro evidence that curcumin directly induces stellate cell apoptosis and blocks stellate cell activation.10,67 These previous studies bolster our conclusions as we know that the NanoCurc™ formulation does not alter its biological activity. NanoCurc™ is equally effective as the unencapsulated form at inducing stellate cell apoptosis (Figure 6). However, this is not entirely surprising as NanoCurc™ bio-effectiveness is comparable to the free parent compound in a cancer system.26 We also provide in vivo evidence of NanoCurc™ effectiveness in preventing stellate cell activation as it induces PPAR-γ transcription, an essential mediator of curcumin’s effects on eliminating stellate cells.15
As our data show that NanoCurc™ protects the liver from injury, it could be argued that NanoCurc™ is more hepatoprotective than antifibrotic. However, it is likely that NanoCurc™ is working at multiple steps in the development of fibrosis. First, as already mentioned, NanoCurc™ prevents hepatocellular injury. However, this is not a complete suppression of injury. There is slight but measurable hepatocellular injury in NanoCurc™-treated mice as evidence by the increase in serum ALT and intrahepatic TNF-α compared with untreated mice (Figure 2). The same is true for the production of the potent pro-fibrotic cytokine TGF-β (Figure 5). We also present evidence of effects of curcumins on pro-fibrotic stellate cells through the induction of PPAR-γ (Figure 5). Collectively, our findings suggest that NanoCurc™ works at multiple steps in the injury and fibrosis pathway. This agent could be particularly useful in preventing the progression of chronic liver disease in patients with active on-going liver injury such as in viral or autoimmune hepatitis as it targets both liver injury and fibrosis mechanisms.
Hepatic inflammation is central to many liver diseases.32 Interestingly, our data show that NanoCurc™ has little effect on NKT cells and neutrophils in the liver. There is a comparable decrease in NKT and increase in Gr-1high cells in void nanoparticle and NanoCurc™-treated mice (Supplementary Figure 2). This is consistent with what has been reported in wild-type CCl4-treated mice.38 This could indicate that a low level of injury is sufficient to affect the immune system or that another important component of the innate immune system, particularly Kupffer cells,68 might be targeted by NanoCurc™. More immediate 24 h studies and analysis of chemokines might be more revealing in dissecting NanoCurc™’s potential role in altering the livers inflammatory milieu.
The results of this study show NanoCurc™ feasibility in treating chronic liver diseases. Being fully water soluble, NanoCurc™ overcomes the most important road block that prevented curcumin’s clinical development, further showing that NanoCurc™ prevents liver injury and fibrosis is a significant developmental step in bringing this natural compound to clinical trials.
Supplementary Material
ACKNOWLEDGEMENTS
This work was financially supported by R01DK080736 (RAA); R01DK081417 (RAA); R01CA113669 (AM); R01CA13767 (AM); P01CA134292 (AM); P30CA069773 (MAR); S10 RR026824 (MAR); Flight Attendants Medical Research Institute (FAMRI) (AM and MAR); Johns Hopkins CTSA Institute for Clinical and Translational Research (UL1RR025005) (MAR); The Sol Goldman Pancreatic Cancer Research Center (AM); Michael Rolfe Foundation for Pancreatic Cancer Research (AM and RAA); and SignPath Pharmaceuticals (AM). This publication was made possible by Grant Number UL1RR025005 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
Footnotes
Supplementary Information accompanies the paper on the Laboratory Investigation website (http://www.laboratoryinvestigation.org)
DISCLOSURE/CONFLICT OF INTEREST
NanoCurc is a registered trademark of SignPath Pharmaceuticals, Quakertown, PA, USA. AM is a member of the scientific advisory board of SignPath Pharma, and any conflicts of interest under this arrangement are handled in accordance with the Johns Hopkins University Office of Policy Coordination guidelines. SignPath Pharma has provided partial support for these studies through offsetting the costs of polymer synthesis. SB, GF and AM have filed a patent application (US 2008/0107749) that is relevant to the formulation described in this article. A report of invention to this effect has been filed with Johns Hopkins Technology Transfer and licensed by SignPath Pharma.
References
- 1.Brenner DA. Molecular pathogenesis of liver fibrosis. Trans Am Clin Climatol Assoc. 2009;120:361–368. [PMC free article] [PubMed] [Google Scholar]
- 2.Hellerbrand C, Stefanovic B, Giordano F, et al. The role of TGFbeta1 in initiating hepatic stellate cell activation in vivo. J Hepatol. 1999;30:77–87. doi: 10.1016/s0168-8278(99)80010-5. [DOI] [PubMed] [Google Scholar]
- 3.Sinha R, Anderson DE, McDonald SS, et al. Cancer risk and diet in India. J Postgrad Med. 2003;49:222–228. [PubMed] [Google Scholar]
- 4.Huang MT, Smart RC, Wong CQ, et al. Inhibitory effect of curcumin, chlorogenic acid, caffeic acid, and ferulic acid on tumor promotion in mouse skin by 12-O-tetradecanoylphorbol-13-acetate. Cancer Res. 1988;48:5941–5946. [PubMed] [Google Scholar]
- 5.Sharma OP. Antioxidant activity of curcumin and related compounds. Biochem Pharmacol. 1976;25:1811–1812. doi: 10.1016/0006-2952(76)90421-4. [DOI] [PubMed] [Google Scholar]
- 6.Rao TS, Basu N, Siddiqui HH. Anti-inflammatory activity of curcumin analogues. Indian J Med Res. 1982;75:574–578. [PubMed] [Google Scholar]
- 7.Brouet I, Ohshima H. Curcumin, an anti-tumour promoter and anti-inflammatory agent, inhibits induction of nitric oxide synthase in activated macrophages. Biochem Biophys Res Commun. 1995;206:533–540. doi: 10.1006/bbrc.1995.1076. [DOI] [PubMed] [Google Scholar]
- 8.Kulkarni SK, Bhutani MK, Bishnoi M. Antidepressant activity of curcumin: involvement of serotonin and dopamine system. Psychopharmacology (Berl) 2008;201:435–442. doi: 10.1007/s00213-008-1300-y. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Ku BS, Yao HY, et al. The effects of curcumin on depressive-like behaviors in mice. Eur J Pharmacol. 2005;518:40–46. doi: 10.1016/j.ejphar.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Fu Y, Zheng S, Lin J, et al. Curcumin protects the rat liver from CCl4-caused injury and fibrogenesis by attenuating oxidative stress and suppressing inflammation. Mol Pharmacol. 2008;73:399–409. doi: 10.1124/mol.107.039818. [DOI] [PubMed] [Google Scholar]
- 11.Park EJ, Jeon CH, Ko G, et al. Protective effect of curcumin in rat liver injury induced by carbon tetrachloride. J Pharm Pharmacol. 2000;52:437–440. doi: 10.1211/0022357001774048. [DOI] [PubMed] [Google Scholar]
- 12.Donatus IA, Sardjoko, Vermeulen NP. Cytotoxic and cytoprotective activities of curcumin. Effects on paracetamol-induced cytotoxicity, lipid peroxidation and glutathione depletion in rat hepatocytes. Biochem Pharmacol. 1990;39:1869–1875. doi: 10.1016/0006-2952(90)90603-i. [DOI] [PubMed] [Google Scholar]
- 13.Kiso Y, Suzuki Y, Watanabe N, et al. Antihepatotoxic principles of Curcuma longa rhizomes. Planta Med. 1983;49:185–187. doi: 10.1055/s-2007-969845. [DOI] [PubMed] [Google Scholar]
- 14.Xu J, Fu Y, Chen A. Activation of peroxisome proliferator-activated receptor-gamma contributes to the inhibitory effects of curcumin on rat hepatic stellate cell growth. Am J Physiol Gastrointest Liver Physiol. 2003;285:G20–G30. doi: 10.1152/ajpgi.00474.2002. [DOI] [PubMed] [Google Scholar]
- 15.Zheng S, Chen A. Activation of PPARgamma is required for curcumin to induce apoptosis and to inhibit the expression of extracellular matrix genes in hepatic stellate cells in vitro. Biochem J. 2004;384(Part 1):149–157. doi: 10.1042/BJ20040928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidd PM. Bioavailability and activity of phytosome complexes from botanical polyphenols: the silymarin, curcumin, green tea, grape seed extracts. Altern Med Rev. 2009;14:226–246. [PubMed] [Google Scholar]
- 17.Marczylo TH, Verschoyle RD, Cooke DN, et al. Comparison of systemic availability of curcumin with that of curcumin formulated with phosphatidylcholine. Cancer Chemother Pharmacol. 2007;60:171–177. doi: 10.1007/s00280-006-0355-x. [DOI] [PubMed] [Google Scholar]
- 18.Pan MH, Huang TM, Lin JK. Biotransformation of curcumin through reduction and glucuronidation in mice. Drug Metab Dispos. 1999;27:486–494. [PubMed] [Google Scholar]
- 19.Sharma RA, Steward WP, Gescher AJ. Pharmacokinetics and pharmacodynamics of curcumin. Adv Exp Med Biol. 2007;595:453–470. doi: 10.1007/978-0-387-46401-5_20. [DOI] [PubMed] [Google Scholar]
- 20.Anand P, Kunnumakkara AB, Newman RA, et al. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007;4:807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 21.Garcea G, Berry DP, Jones DJ, et al. Consumption of the putative chemopreventive agent curcumin by cancer patients: assessment of curcumin levels in the colorectum and their pharmacodynamic consequences. Cancer Epidemiol Biomarkers Prev. 2005;14:120–125. [PubMed] [Google Scholar]
- 22.Bisht S, Mizuma M, Feldmann G, et al. Systemic administration of polymeric nanoparticle-encapsulated curcumin (NanoCurc) blocks tumor growth and metastases in preclinical models of pancreatic cancer. Mol Cancer Ther. 2010;9:2255–2264. doi: 10.1158/1535-7163.MCT-10-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcea G, Jones DJ, Singh R, et al. Detection of curcumin and its metabolites in hepatic tissue and portal blood of patients following oral administration. Br J Cancer. 2004;90:1011–1015. doi: 10.1038/sj.bjc.6601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhillon N, Aggarwal BB, Newman RA, et al. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res. 2008;14:4491–4499. doi: 10.1158/1078-0432.CCR-08-0024. [DOI] [PubMed] [Google Scholar]
- 25.Bisht S, Feldmann G, Koorstra JB, et al. In vivo characterization of a polymeric nanoparticle platform with potential oral drug delivery capabilities. Mol Cancer Ther. 2008;7:3878–3888. doi: 10.1158/1535-7163.MCT-08-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bisht S, Feldmann G, Soni S, et al. Polymeric nanoparticle-encapsulated curcumin (‘nanocurcumin’): a novel strategy for human cancer therapy. J Nanobiotechnology. 2007;5:3. doi: 10.1186/1477-3155-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders RA, Subudhi SK, Wang J, et al. Contribution of the lymphotoxin beta receptor to liver regeneration. J Immunol. 2005;175:1295–1300. doi: 10.4049/jimmunol.175.2.1295. [DOI] [PubMed] [Google Scholar]
- 28.Vogel S, Piantedosi R, Frank J, et al. An immortalized rat liver stellate cell line (HSC-T6): a new cell model for the study of retinoid metabolism in vitro. J Lipid Res. 2000;41:882–893. [PubMed] [Google Scholar]
- 29.Aggarwal BB, Sundaram C, Malani N, et al. Curcumin: the Indian solid gold. Adv Exp Med Biol. 2007;595:1–75. doi: 10.1007/978-0-387-46401-5_1. [DOI] [PubMed] [Google Scholar]
- 30.Bisht S, Maitra A. Systemic delivery of curcumin: 21st century solutions for an ancient conundrum. Curr Drug Discov Technol. 2009;6:192–199. doi: 10.2174/157016309789054933. [DOI] [PubMed] [Google Scholar]
- 31.Sudo K, Yamada Y, Moriwaki H, et al. Lack of tumor necrosis factor receptor type 1 inhibits liver fibrosis induced by carbon tetrachloride in mice. Cytokine. 2005;29:236–244. doi: 10.1016/j.cyto.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Adams DH, Ju C, Ramaiah SK, et al. Mechanisms of immune-mediated liver injury. Toxicol Sci. 2010;115:307–321. doi: 10.1093/toxsci/kfq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Diehl AM. Innate immunity in the liver. Curr Opin Gastroenterol. 2003;19:565–571. doi: 10.1097/00001574-200311000-00009. [DOI] [PubMed] [Google Scholar]
- 34.Szabo G, Mandrekar P, Dolganiuc A. Innate immune response and hepatic inflammation. Semin Liver Dis. 2007;27:339–350. doi: 10.1055/s-2007-991511. [DOI] [PubMed] [Google Scholar]
- 35.Marra F, Aleffi S, Galastri S, et al. Mononuclear cells in liver fibrosis. Semin Immunopathol. 2009;31:345–358. doi: 10.1007/s00281-009-0169-0. [DOI] [PubMed] [Google Scholar]
- 36.Ginsburg I, Koren E, Horani A, et al. Amelioration of hepatic fibrosis via Padma Hepaten is associated with altered natural killer T lymphocytes. Clin Exp Immunol. 2009;157:155–164. doi: 10.1111/j.1365-2249.2009.03936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishikawa S, Ikejima K, Yamagata H, et al. CD1d-restricted natural killer T cells contribute to hepatic inflammation and fibrogenesis in mice. J Hepatol. 2010 doi: 10.1016/j.jhep.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 38.Park O, Jeong WI, Wang L, et al. Diverse roles of invariant natural killer T cells in liver injury and fibrosis induced by carbon tetrachloride. Hepatology. 2009;49:1683–1694. doi: 10.1002/hep.22813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Safadi R, Zigmond E, Pappo O, et al. Amelioration of hepatic fibrosis via beta-glucosylceramide-mediated immune modulation is associated with altered CD8 and NKT lymphocyte distribution. Int Immunol. 2007;19:1021–1029. doi: 10.1093/intimm/dxm069. [DOI] [PubMed] [Google Scholar]
- 40.Melhem A, Muhanna N, Bishara A, et al. Anti-fibrotic activity of NK cells in experimental liver injury through killing of activated HSC. J Hepatol. 2006;45:60–71. doi: 10.1016/j.jhep.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 41.Radaeva S, Sun R, Jaruga B, et al. Natural killer cells ameliorate liver fibrosis by killing activated stellate cells in NKG2D-dependent and tumor necrosis factor-related apoptosis-inducing ligand-dependent manners. Gastroenterology. 2006;130:435–452. doi: 10.1053/j.gastro.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 42.Daley JM, Thomay AA, Connolly MD, et al. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J Leukocyte Biol. 2008;83:64–70. doi: 10.1189/jlb.0407247. [DOI] [PubMed] [Google Scholar]
- 43.Karlmark KR, Weiskirchen R, Zimmermann HW, et al. Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology. 2009;50:261–274. doi: 10.1002/hep.22950. [DOI] [PubMed] [Google Scholar]
- 44.Weiler-Normann C, Herkel J, Lohse AW. Mouse models of liver fibrosis. Z Gastroenterol. 2007;45:43–50. doi: 10.1055/s-2006-927387. [DOI] [PubMed] [Google Scholar]
- 45.Burk RF, Lane JM, Patel K. Relationship of oxygen and glutathione in protection against carbon tetrachloride-induced hepatic microsomal lipid peroxidation and covalent binding in the rat. Rationale for the use of hyperbaric oxygen to treat carbon tetrachloride ingestion. J Clin Invest. 1984;74:1996–2001. doi: 10.1172/JCI111621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burk RF, Patel K, Lane JM. Reduced glutathione protection against rat liver microsomal injury by carbon tetrachloride. Dependence on O2. Biochem J. 1983;215:441–445. doi: 10.1042/bj2150441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng S, Yumei F, Chen A. De novo synthesis of glutathione is a prerequisite for curcumin to inhibit hepatic stellate cell (HSC) activation. Free Radic Biol Med. 2007;43:444–453. doi: 10.1016/j.freeradbiomed.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedman SL. Molecular mechanisms of hepatic fibrosis and principles of therapy. J Gastroenterol. 1997;32:424–430. doi: 10.1007/BF02934504. [DOI] [PubMed] [Google Scholar]
- 49.Friedman SL, Roll FJ, Boyles J, et al. Hepatic lipocytes: the principal collagen-producing cells of normal rat liver. Proc Natl Acad Sci USA. 1985;82:8681–8685. doi: 10.1073/pnas.82.24.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawase T, Shiratori Y, Sugimoto T. Collagen production by rat liver fat-storing cells in primary culture. Exp Cell Biol. 1986;54:183–192. doi: 10.1159/000163355. [DOI] [PubMed] [Google Scholar]
- 51.Ramadori G, Knittel T, Odenthal M, et al. Synthesis of cellular fibronectin by rat liver fat-storing (Ito) cells: regulation by cytokines. Gastroenterology. 1992;103:1313–1321. doi: 10.1016/0016-5085(92)91522-6. [DOI] [PubMed] [Google Scholar]
- 52.Neubauer K, Knittel T, Armbrust T, et al. Accumulation and cellular localization of fibrinogen/fibrin during short-term and long-term rat liver injury. Gastroenterology. 1995;108:1124–1135. doi: 10.1016/0016-5085(95)90211-2. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Trevijano ER, Iraburu MJ, Fontana L, et al. Transforming growth factor beta1 induces the expression of alpha1(I) procollagen mRNA by a hydrogen peroxide-C/EBPbeta-dependent mechanism in rat hepatic stellate cells. Hepatology. 1999;29:960–970. doi: 10.1002/hep.510290346. [DOI] [PubMed] [Google Scholar]
- 54.Gressner AM. Transdifferentiation of hepatic stellate cells (Ito cells) to myofibroblasts: a key event in hepatic fibrogenesis. Kidney Int Suppl. 1996;54:S39–S45. [PubMed] [Google Scholar]
- 55.Galli A, Crabb D, Price D, et al. Peroxisome proliferator-activated receptor gamma transcriptional regulation is involved in platelet-derived growth factor-induced proliferation of human hepatic stellate cells. Hepatology. 2000;31:101–108. doi: 10.1002/hep.510310117. [DOI] [PubMed] [Google Scholar]
- 56.Marra F, Efsen E, Romanelli RG, et al. Ligands of peroxisome proliferator-activated receptor gamma modulate profibrogenic and proinflammatory actions in hepatic stellate cells. Gastroenterology. 2000;119:466–478. doi: 10.1053/gast.2000.9365. [DOI] [PubMed] [Google Scholar]
- 57.Miyahara T, Schrum L, Rippe R, et al. Peroxisome proliferator-activated receptors and hepatic stellate cell activation. J Biol Chem. 2000;275:35715–35722. doi: 10.1074/jbc.M006577200. [DOI] [PubMed] [Google Scholar]
- 58.Lin J, Zheng S, Chen A. Curcumin attenuates the effects of insulin on stimulating hepatic stellate cell activation by interrupting insulin signaling and attenuating oxidative stress. Lab Invest. 2009;89:1397–1409. doi: 10.1038/labinvest.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiao J, Friedman SL, Aloman C. Hepatic fibrosis. Curr Opin Gastroenterol. 2009;25:223–229. doi: 10.1097/mog.0b013e3283279668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Everhart J. Washington, DC: US Government; 2008. The Burden of Digestive Diseases in the United States, Vol. No. 09-6443. [Google Scholar]
- 61.van Vlerken LE, Amiji MM. Multi-functional polymeric nanoparticles for tumour-targeted drug delivery. Expert Opin Drug Deliv. 2006;3:205–216. doi: 10.1517/17425247.3.2.205. [DOI] [PubMed] [Google Scholar]
- 62.Vicent MJ, Duncan R. Polymer conjugates: nanosized medicines for treating cancer. Trends Biotechnol. 2006;24:39–47. doi: 10.1016/j.tibtech.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 63.Vasir JK, Labhasetwar V. Targeted drug delivery in cancer therapy. Technol Cancer Res Treat. 2005;4:363–374. doi: 10.1177/153303460500400405. [DOI] [PubMed] [Google Scholar]
- 64.Ganta S, Devalapally H, Amiji M. Curcumin enhances oral bioavailability and anti-tumor therapeutic efficacy of paclitaxel upon administration in nanoemulsion formulation. J Pharm Sci. 2010;99:4630–4641. doi: 10.1002/jps.22157. [DOI] [PubMed] [Google Scholar]
- 65.Appiah-Opong R, de Esch I, Commandeur JN, et al. Structure-activity relationships for the inhibition of recombinant human cytochromes P450 by curcumin analogues. Eur J Med Chem. 2008;43:1621–1631. doi: 10.1016/j.ejmech.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 66.Volak LP, Ghirmai S, Cashman JR, et al. Curcuminoids inhibit multiple human cytochromes P450, UDP-glucuronosyltransferase, and sulfotransferase enzymes, whereas piperine is a relatively selective CYP3A4 inhibitor. Drug Metab Dispos. 2008;36:1594–1605. doi: 10.1124/dmd.108.020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin J, Chen A. Activation of peroxisome proliferator-activated receptor-gamma by curcumin blocks the signaling pathways for PDGF and EGF in hepatic stellate cells. Lab Invest. 2008;88:529–540. doi: 10.1038/labinvest.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaeschke H. Reactive oxygen and mechanisms of inflammatory liver injury: Present concepts. J Gastroenterol Hepatol. 2011;26(Suppl 1):173–179. doi: 10.1111/j.1440-1746.2010.06592.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.