Abstract
Background
SEL1L gene product is implicated in the endoplasmic reticulum (ER)-associated protein degradation and Unfolded Protein Response pathways. This gene and associated miRNAs have been indicated as predictive and prognostic markers of pancreatic cancer.
Aim
Explore the role of SEL1L in colorectal cancer (CRC) progression.
Methods
SEL1L expression was analysed immunohistochemically in 153 adenomas and 71 CRCs from African American and North Italian patients. The distribution of stained cells was determined by computing median and inter quartile range. The receiver operating characteristics plot was used as discriminate power of SEL1L expression, CRC diagnosis and the effects on patient survival.
Results
SEL1L was low in normal mucosa and confined to few scattered cells at the base crypt of the villi and in the foveolar glandular compartment. The highest levels were in Paneth cells within the lysosomes. The enterocytic progenitor cells and mature enterocytes showed less cytoplasmic staining. In CRCs, SEL1L expression significantly correlated with the progression from adenoma to carcinoma (P = 0.0001) being stronger in well-to-moderately differentiated cancers. No correlation was found with other clinicopathological characteristics or ethnicity.
Conclusions
SEL1L expression is a potential CRC tissue biomarker since its expression is significantly higher in adenoma cells with respect to normal mucosa. The levels of expression decrease sensibly in undifferentiated CRC cancers. Interestingly, Paneth cells contain high levels of SEL1L protein that could indicate pre-neoplastic mucosa undergoing neoplastic transformation. Since SEL1L’s major function lies within ER stress and active ERAD response, it may identify CRCs with differentiated secretory phenotype and acute cellular stress.
Keywords: SEL1L expression, Colorectal cancers, Paneth’s cells
Introduction
During carcinogenesis and tumor progression, transformed cells are exposed to a full spectrum of factors that determine structural alterations of secretory proteins [1]. Consistent with this, increasing evidence indicates that the unfolded protein response (UPR), a critical pathway required in the maintenance of cellular homeostasis, is activated in several human cancer types and in animal tumor model systems [2, 3]. This may be of critical clinical relevance, as it has been shown that the UPR may confer resistance to antitumor drugs [2–4] and may also become a target for cancer treatment. During UPR, the proteins targeted by the ER-associated protein degradation (ERAD) pathway [1] are selected by a quality control system within the ER lumen and destroyed by the cytoplasmic ubiquitin–proteasome system (UPS). UPS degradation requires substrate transport from the ER to the cytoplasm through the formation of a multiprotein complex involving E3 ligases, ubiquitination, dislocation and extraction of the misfolded proteins from the ER membrane to the cytosol, to be degraded by the proteasome or to be secreted. SEL1L is a key component of the ERAD machinery and, in association with the E3 ligase HRD1, clears several unfolded or orphan substrates [5–8]. In addition to HRD1, SEL1L interacts with several components of the dislocation machinery such as the erlectins OS-9 and XTP3-b, both implicated in substrate recognition; derlin-1, previously reported to protect human breast cancer cells from ER stress induced apoptosis [9]. Previous results indicate that SEL1L expression down-modulates breast and pancreatic carcinoma cell aggressiveness both in vivo and in vitro [10, 11], associates with the transition to severe dysplasia in esophageal carcinogenesis, being lost in aggressive esophageal cancers [12], and becomes consistently expressed in the initial stages of prostate and lung cancers [13, 14]. However, no data has yet been reported on the role of SEL1L protein nor of ERAD/UPR in colorectal cancer (CRC), a key model of human epithelial carcinogenesis. In this study we analyzed SEL1L expression in adenomas, carcinomas and matched adjacent normal tissue obtained from African–American and Italian affected patients. We here report on the significant activation of SEL1L in colonic cells undergoing transition from adenoma to carcinoma, being higher in better differentiated relative to poorly differentiated tumors.
Materials and Methods
Colorectal Cases
Archival formalin-fixed paraffin-embedded colorectal biopsies were obtained from African–American patients undergoing colonoscopy at Howard University Hospital, Washington DC, USA, and from Multimedica Hospital, Milan, Italy. Retrospective paraffin embedded surgical tissues were used for TMA analysis, and cores were obtained from pathology archival blocks. For western blot (WB) analysis fresh surgical tissue from Italian patients was used. Informed consent was obtained from the patients and the study was approved by the respective Institutional Ethical Board. Clinical data collected for each case included race, gender, associated past medical history, medication use, and family history of colorectal cancer. The two series of cases consisted of: 134 adenomas, 59 (55 moderately to well differentiated and 4 poorly differentiated) carcinomas and 57 matched adjacent normal tissue obtained from African Americans and 32 (19 adenomas, 13 adenocarcinoma) from the Italian group. The numbers of male patients in the adenoma and CRC groups were 68/153 (44.4%) and 40/71 (56.3%) and the number of females were 85/153 (55.5%) and 31/71 (43.6%), respectively. Ninety-six percent (96%) of the tumors were moderately to well differentiated, and 4% were poorly differentiated. Patients were deemed eligible if colonoscopy resulted in a first diagnosis of adenoma or adenocarcinoma, and confirmed by histopathology. From the review of the medical records, clinical information was collected and the TNM status was recorded based upon the American Joint Committee on Cancer staging system [15]. For survival data collection, we defined the vital status (dead or alive) of 55 CRC cases in this study, based on two different sources: the latest inpatient medical record at Howard University Hospital and data from the Social Security Death Index website (http://ssdi.rootsweb.com). For each deceased case the date of death was recorded. In cases with no record of death in both sources, the latest update of Social Security Death Index website (10/1/2007) was recorded as the date of follow-up. No death data were available for the Italian patients in this study.
Protein Extraction and Western Blot Analysis
Surgical fragments of normal tissue mucosa (M), adenoma (A) and adenocarcinoma (T) were lysed in 10 mM Tris–HCl (pH7.4), 150 mM NaCl, 1%NP-40 and protease inhibitors (Pierce, Celbio, Pero, Italy). Protein concentrations were determined by the Bradford assay; samples were resolved on 10% SDS–polyacrylamide gels, blotted onto PVDF membranes (GE, Milan, Italy) and hybridized with monoclonal SEL1L antibody and developed with ECL (Genespin, Milan, Italy). Densitometric analysis of the WB results was determined using the Scion imaging program (http://www.scioncorp.com).
Tissue Microarrays and Immunohistochemical Analysis
Tissue microarrays (TMAs) were constructed using Galileo X-3500 (http://www.isenet.it) semi-automatic arrayer. Each TMA contained normal colorectal mucosa, adenoma, and adenocarcinoma cores based on a published protocol [16, 17]. Donor blocks were always sampled in duplicate. In total, 282 tissue cores were analyzed for SEL1L expression by immunohistochemistry on TMA sections, including 134 adenomas, 59 carcinomas, of which 55 were moderately to well differentiated and four poorly differentiated, and 57 matched adjacent normal colorectal mucosa samples for control comparisons. Sections (5 µm) were mounted on charged glass slides, deparaffinized with xylene three times for 20 min and rehydrated using graded ethanol. Antigen retrieval was performed by placing deparaffinized sections in a microwave oven in citrate buffer for 12 min. The slides were then treated with hydrogen peroxide to quench endogenous peroxidase activity, followed by incubation with primary and secondary antibodies and subsequently, streptavidin–biotin complex, streptavidin-peroxidase and substrate-chromogen solution using the Envision system according to the manufacturers’ protocol (DAKO). Slides were then counterstained with hematoxylin, rinsed with ethanol, dried and visualized by light microscopy. TMA slides to which no primary antibody had been added were used as negative controls. As controls, SEL1L-positive prostate adenocarcinoma, liver, spleen, tonsil, and kidney tissues were included in the TMA. All immunohistochemistry reagents, except the SEL1L antibody [11], were purchased from DAKO (Carpinteria, CA). Slides were read by two pathologists (EL; WG). Percentages of positive cells (cytoplasmic staining) were recorded for each sample by approximating the area occupied by immunoreactive cells (average of 50 cells per filed, total of five filed) within the stained sections. The intensity of staining was also evaluated for more precise scoring.
Identification of Paneth’s Cells and Immunoelectron Microscopy
Localization of SEL1L in the crypt to villous axis was investigated on small intestinal mucosa samples, where the normal intestinal architecture is better developed than in the colorectal mucosa, involving the entire crypt to villous axis. Paneth’s cells were identified by light microscopy immunostaining using an antibody to lysosome, widely considered a specific marker. For ultrastructural immunocytochemistry, two samples of human duodenal mucosa were fixed for 2 h at 4°C in a mixture of 2% paraformaldehyde and 2% glutaraldehyde in 0.05 M pH 7.3 cacodylate buffer, post-fixed in 1% osmium tetroxide for 1 h at room temperature, dehydrated in ethanol and embedded in Epon-Araldite. After incubation with ovoalbumin 1% in TBS 0.05 M pH 7.4 for 5 min at room temperature, thin sections were pre-treated with 5% metaperiodate for 30 min, then incubated for 24 h at 4°C with the anti-SEL1L antibody diluted 1:10, then 1:20 gold-tagged goat anti-mouse (EY Laboratories, San Mateo, CA) for 1 h at room temperature and finally counterstained with uranyl acetate and lead citrate. The sections were examined with a Philips Morgagni electron microscope.
Statistical Analysis
Distribution for percentage of stained cells was studied by computing median and inter-quartile range (IQR) due to skewed distribution. The nonparametric Kruskal–Wallis test was then applied to verify whether the percentage of stained cells differed between normal, adenoma, and CRC samples. P values less than 0.05 were considered to be statistically significant. The discriminative power of SEL1L for CRC diagnosis was then computed by receiver operating characteristics (ROC) plot. In this curve SEL1L expression in cancer cells was compared to the non-cancer (normal and adenoma) tissue from the same patient. Survival was computed for patients and log rank test was used to test the effect of SEL1L expression on median survival.
Results
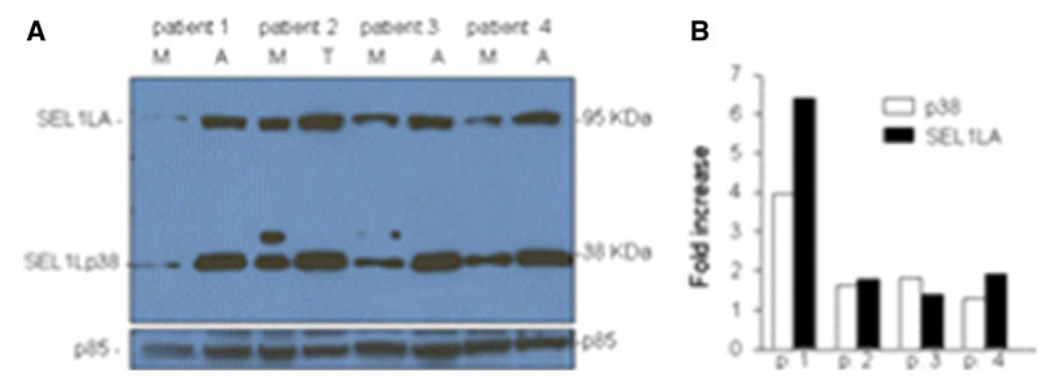
Western blot analysis was performed on adenoma (A), adenocarcinoma (T) and matched normal (M) mucosa from CRC patients. Protein lysates from fresh tissue fragments extracted from four Italian surgical fragments were probed with SEL1L monoclonal antibody. The levels of SEL1LA, the ER-resident protein, and of p38, the secreted variant, were higher in adenoma (A) and adenocarcinoma (T) cells with respect to the normal mucosa (M) of the same patient (Fig. 1a, lanes: patient 1-A, 2-T, 3-A and 4-A). Unexpectedly, a second protein of about 42 kDa was present in the normal mucosa of patients 2 and 3; the nature of this form is presently under investigation. The housekeeping p85 protein was used to normalize the loading of the protein on the filter and its intensity was used to determine the fold increase (Fig. 1b) of the two SEL1L proteins in adenoma or neoplastic cells over normal mucosa. The results, even if obtained on a too small number of cases, indicate SEL1L and its p38 variant activation in adenomas and carcinoma cells with respect to the normal mucosa of the same patient. These findings will be validated on a larger number of samples.
Fig. 1.
Western blot analysis for SEL1LA of the p38 SEL1L protein lysates obtained from normal tissue mucosa, adenoma and adenocarcinoma from four CRC patients. a An increase of SEL1LA, the ER-resident form of SEL1L, in the adenoma (A) and adenocarcinoma (T) cells when compared to normal mucosa (M) was noted in all four patients (a, lanes: patient 1-A, patient 2-T, patient 3-M and patient 4-A). In addition, a remarkable increase of the p38 SEL1L variant was seen in the neoplastic protein extracts. b Fold of SEL1L proteins increase in the neoplastic versus the normal mucosa cells, the levels were normalized toward the housekeeping p85 protein. Occasionally the p38 variant of SEL1L showed the presence of a post-translationally modified form
SEL1L Expression in CRC Progression
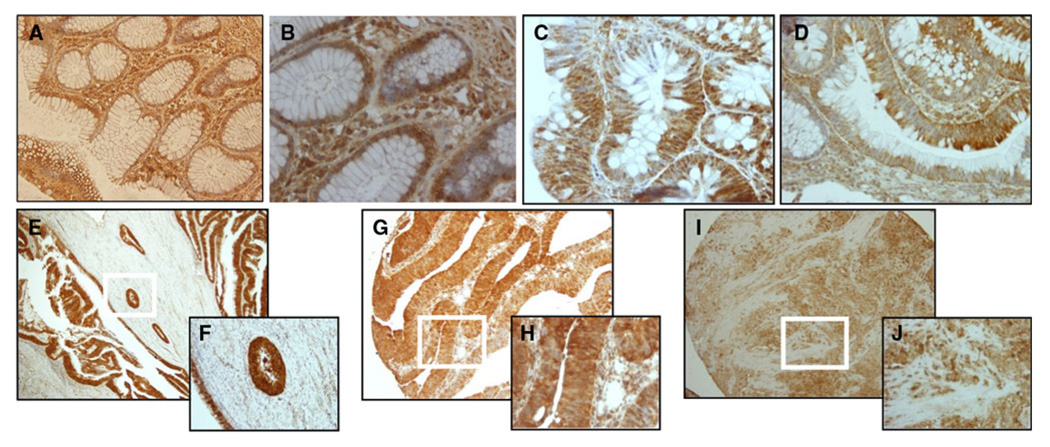
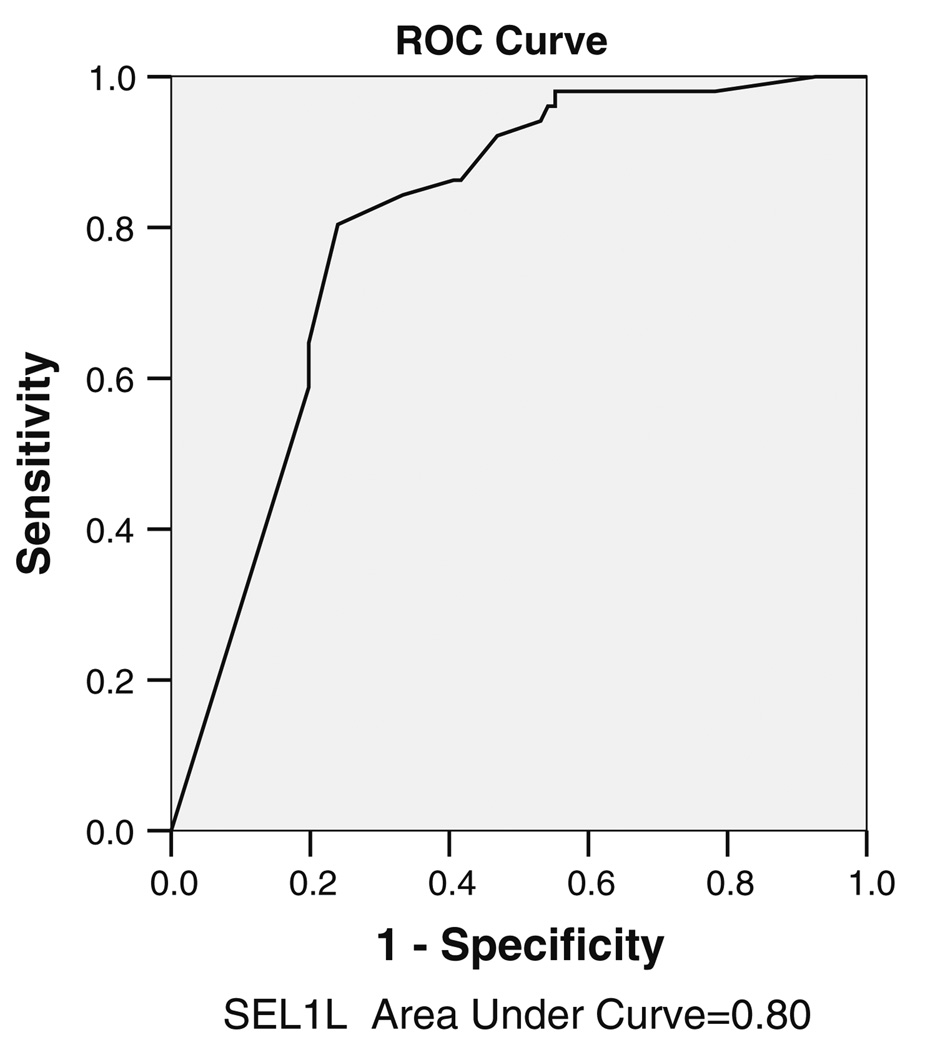
We analyzed the expression levels of SEL1L in endoscopic biopsies from 153 colorectal adenomas and 72 CRCs, plus 57 matched adjacent normal colorectal mucosa biopsies selected from African–American patients undergoing colonoscopy at Howard University Hospital, Washington DC, USA, and from Multimedica Hospital, Milan, Italy. Control samples were selected from the respective ethnic backgrounds. In general, SEL1L expression in normal biopsies is very low, if any, in the cytoplasm of mature colonocytes at the surface of colonic glands, and high in colonocyte progenitors at the proliferative zone of normal crypts (Fig. 2a, b). In marked contrast, strongly positive nuclear and cytoplasmic staining of epithelial cells was observed in adenomas and in moderately to well differentiated carcinomas (Fig. 2 c–h); while poorly differentiated carcinomas were low levels (Fig. 2i, j). The median percentages of SEL1L expression in cancer and adenoma (down into adjacent crypts, top-down) growth tissues were 100% and 20%, respectively (P = 0.0001). We determined the distribution of step-wise expression of SEL1L markers in well, moderately and poorly differentiated colorectal cancers (Table 1). SEL1L expression didn’t increase in poorly differentiated (median 90) versus well (median 92) and moderately differentiated (median 100) significantly (P = 0.5).The expression in the normal (proliferative zone, bottom-up process) was 100%. In addition, we studied the discriminating power of SEL1L in distinguishing between cancer and non-cancer diagnosis by ROC, in which the sensitivity and specificity of SEL1L expression was assessed (Fig. 3). The area under curve for SEL1L was 0.80. In this graph a SEL1L of 77% (cut-off value) had the best combination of sensitivity (84%) and specificity (67%) to diagnose the cancer versus non-cancer samples. The correlation coefficient test showed that SEL1L may have a meaningful and significant effect in progression of colon cancer.
Fig. 2.
a–j SEL1L expression in CRC progression. a, b Left, Normal colon with decreasing staining ascending the crypt 20X. Right, Normal colon base of crypt (proliferative zone) with cytoplasmic staining, 40X. c, d Left, Cytoplasmic staining in adenoma, 40X. Right, Nuclear staining in adenoma, 40X. e, f Left, Well-differentiated carcinoma, 10X. Right, 40X. g, h Moderately differentiated carcinoma, 10X. Right, 40X. i, j Left, Poorly differentiated carcinoma 10X. Right, 40X
Table 1.
Median and inter-quartile range of SEL1 expression (% of stained cell) in colorectal cancer samples
| Variable | Median (IQR) | P value |
|---|---|---|
| Sex | ||
| Male (n = 123) | 75 (20–100) | 0.77 |
| Female (n = 126) | 80 (15–100) | |
| Age | ||
| ≤60 (n = 107) | 60 (15–100) | 0.24 |
| >60 (n = 142) | 90 (20–100) | |
| Location | ||
| Proximal (n = 118) | 75 (10–100) | 0.49 |
| Distal (n = 60) | 60 (10–100) | |
| Stage | ||
| 0,1 (n = 14) | 97 (90–100) | 0.34 |
| 2 (n = 20) | 100 (95–100) | |
| 3,4 (n = 23) | 100 (83–100) | |
| Differentiation | ||
| Well (n = 8) | 92 (90–100) | 0.47 |
| Moderate (n = 46) | 100 (90–100) | |
| Poor (n = 3) | 90 |
IQR interquartile range
Fig. 3.
ROC curve for SEL1L as biomarker for colorectal cancer diagnosis
SEL1L Expression and Tumor Stage
In general, SEL1L immunostaining did not show statistically significant differences in relation to age and sex of the patients or anatomic location of the tumor. Of 59 CRC cases available for analysis, the number of cases with stage I, II, III, and IV disease were 14 (24.5%), 20 (35.1%), 21 (36.8%), and 2 (3.5%), respectively, while staging data were missing for the remaining two cases. Median expression of SEL1L was 97, 100, 100, and 100% for stages I–IV, respectively (P = 0.34, Table 1).
SEL1L Expression Lacks Correlation with Survival
Survival curves were calculated using the Kaplan–Meier method. Follow-up was completed in 55 cancer cases (93%) and, among them, 25 (46%) died in a follow-up period up to 123.4 months. The median follow-up time was 27.7 months. The median survival (95%CI) for CRC cases was 61.2 (range 24.5–97.8) months. Survival was 78% at 1 year, 66% at 2 years, 52% at 5 years and 17% at 10 years (Table 2). No significant difference in survival rate could be associated with regard to SEL1L immunostaining (Table 3).
Table 2.
Median (95% CI) survival time (in months) by demographic factors
| Factor | Median survival (95% CI) | P for log rank |
|---|---|---|
| Sex | ||
| Male | 79.5 (26.9–132.2) | 0.13 |
| Female | 41.3 (29.0–63.6) | |
| Age | ||
| <60 | 107.0 (89.9–124.2) | 0.001 |
| ≥60 | 40.6 (25.8–55.4) | |
| Tumor location | ||
| Right | 37.5 (19.8–55.2) | 0.38 |
| Left | 91.9 (0.0–197.4) | |
| Stage | ||
| 0 and 1 | 44.2 (38.2–50.2) | 0.72 |
| 2 | 73.8 (0.0–155.3) | |
| 3, 4 | 37.5 (13.0–62.0) |
CI confidence interval
Table 3.
Median (95% CI) survival time (in months) compared within SEL1L expression
| SEL1L expression |
N | Median | |||
|---|---|---|---|---|---|
| Estimate | Std. error |
95% Confidence interval (CI) |
|||
| Lower bound |
Upper bound |
||||
| ≤80% | 7 | 61.15 | 22.93 | 16.20 | 106.09 |
| >80% | 40 | 91.80 | 36.94 | 19.39 | 164.22 |
| Overall | 47 | 61.15 | 17.53 | 26.78 | 95.52 |
P for Log rank test = 0.64
Topographical Localization of SEL1L Expression
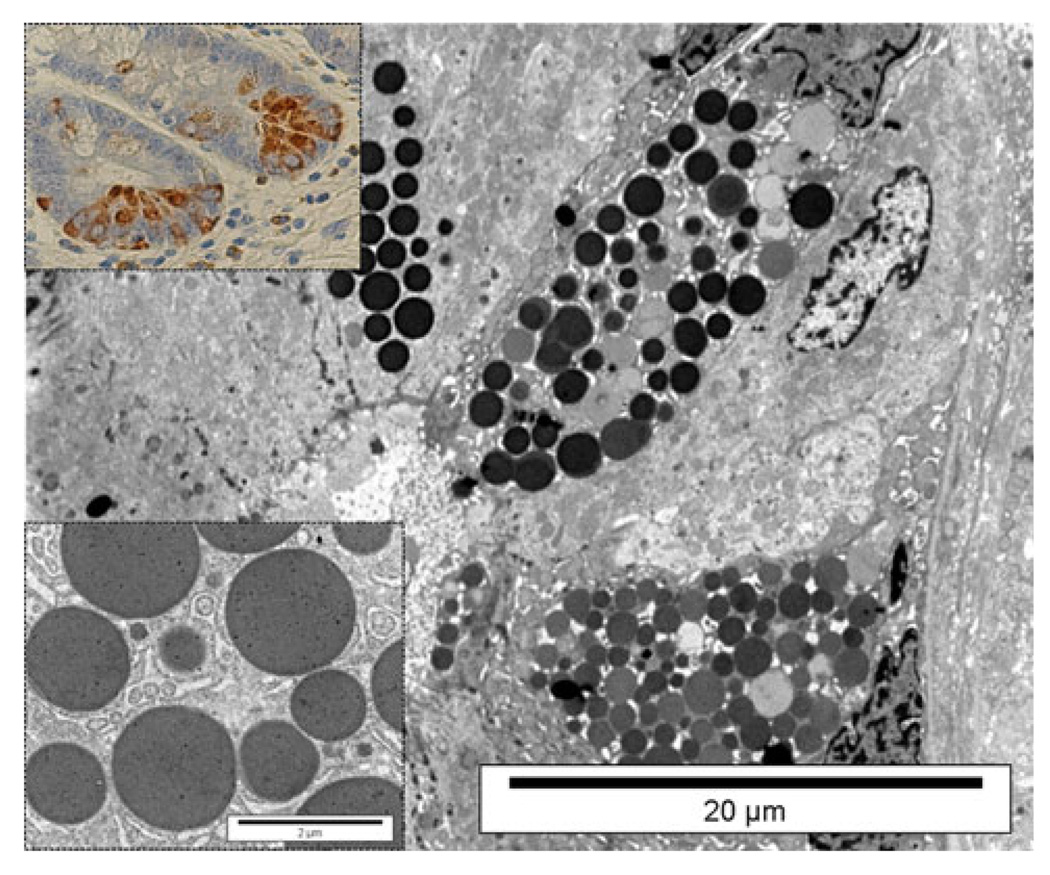
In normal duodenal mucosa SEL1L immunostaining appeared to be intense in a restricted number of cells located at the base of the crypts. These cells showed an eosiniphilic and granulated cytoplasm and were identified as Paneth’s cells based on positive immunoreaction for lysosomes (Fig. 4). Immunoelectronmicroscopic analysis confirmed the identification of these cells as Paneth’s cells. At the ultrastructural level, SEL1L immunoreactivity appeared to be distributed in the cytoplasm, and was particularly concentrated in secretory granules. The result shows high levels of SEL1L protein in Paneth’s cells, confirming the localization of the protein in secretory granules as previously reported [2, 18].
Fig. 4.
SEL1L immunoreactivity in intestinal mucosa Paneth’s cells both at optical (right bottom inset) and ultrastructural level. The electron microscopy shows that the immunoreactivity is concentrated in the secretory granules, as best shown in right top inset by the immunogold technique
Discussion
SEL1L is an ER-resident protein playing a fundamental role in the endoplasmic reticulum associated degradation (ERAD) pathway, acting on the disposal of unfolded proteins [18–22]. The role of SEL1L in cancer is not at all clear but it physiologically acts as a “gate keeper” to ensure that proper protein folding takes place before being transported to their functional destination. Previous results indicated that SEL1L expression is reduced in breast and pancreatic carcinomas. Tumor cell aggressiveness decreased both in vivo and in vitro [10, 11, 23] during the initial stages of esophageal, prostate and lung cancer with respect to SEL1L expression [12–14]. Moreover, the variant SEL1L genotype rs1235998 plays a role in modifying age of diagnosis of pancreatic adenocarcinoma in Caucasian non-smokers [24], and the overexpression of three miRNAs, negative regulators of SEL1L expression, could serve as useful biomarkers of this cancer [24]. However, no studies have dealt with the impact of SEL1L in CRC progression, in intestinal epithelial renewal nor on the migration of differentiated cells from the crypts to the epithelial luminal surface. CRC has been determined to arise from the crypt stem cells and the detail cascade of molecular events behind it is still not fully understood [25]. Here, we investigated the expression of SEL1L in the enterocytic differentiation and in the CRC epithelial neoplastic progression in a limited number of African–American and North Italian colorectal samples. We show that SEL1L is expressed in the proliferative zone of the non-neoplastic colonic mucosa, being particularly strong in Paneth’s cells, as its topographical localization which actively synthesizes secretory defensins and lysozymes, but less in mature enterocytes; SEL1L is mainly expressed in adenomas and well to moderately differentiated adenocarcinomas in agreement with previous observations in esophageal, prostate and lung cancers, with neoplastic progression having increasing protein expression while poorly differentiated tumors show negligible amounts [12–14]. The SEL1L expression is enhanced in those cells undergoing transition, consistent with the progression from adenoma to carcinoma. Paneth’s cells show remarkable levels of SEL1L protein and this may depend on the high secretary nature of these cells as well as to indicate that the pre-neoplastic mucosa may be undergoing oncogenic transformation.
No clear correlation was found between SEL1L expression levels, biomolecular/pathological/clinical parameters nor with overall survival contrary to what was reported for breast cancer [11]. This may be explained by shorter survival expectancy of colorectal cancer patients compared to breast cancer subjects. The expression analysis performed on three adenomas, one adenocarcinoma and relative normal mucosa revealed the up-modulation of the ER-resident SEL1LA protein and the p38 variant. This variant may be involved in cancer-related secretory pathways [2]. SEL1L is activated during the early phases of tumor formation and may indicate the activation of the UPR pathway. UPR is a cytoprotective signal transduction pathway activated in a large variety of human tumor types [3, 4, 6]; it has been reported that its activation alters the chemosensitivity to a variety of antitumor drugs [6, 8, 26]. Thus, the UPR pathway appears to be fundamental not only for cancer development, invasiveness and metastasis but also for resistance to chemotherapy and a target for pharmacological interventions. It has been established that the proteasome is a therapeutic target for a wide variety of cancers [5, 7, 26]. Within CRCs, SEL1L expression could become an identifier of that cell population with differentiated secretory phenotype, acute ER stress and active ERAD response. Given that prolonged or acute ER stress may overwhelm cytoprotective responses and induce cell death, such tumors could be specifically targeted by drugs aggravating ER stress or interfering with ERAD. The results presented here pave the way to investigate the sequence of events occurring as the cells turn from adenoma to carcinoma and the role of ER-stress and UPR pathway in conferring chemosensitivity or resistance to conventional antitumor drugs.
This study confirms previous findings on different tumor types, in essence, SEL1L is activated in the early stage of transformation, when likely a number of DNA mutations and rearrangements are taking place in cells. During this stage, many pathways including the ER-stress and UPR are activated in order to meet the changes. As the cells turn into carcinomas, the need for these pathways and SEL1L diminishes, indicating that another mechanism such as post-translational modification including very little protein refolding capacity may take place. SEL1L is a fundamental member of the UPR pathway, a cytoprotective signal transduction pathway activated in a large variety of human tumor types [3, 4, 6]. It has been reported that UPR activation alters the chemosensitivity to a variety of antitumor drugs [6, 8, 26]. Thus, the UPR pathway appears to be fundamental not only for cancer development, invasiveness and metastasis but also for resistance to chemotherapy and a target for pharmacological interventions. It has been established that the proteasome is a therapeutic target for a wide range of cancers [5, 7]. In this study we show that most normal/healthy colonic epithelial cells and surrounding stroma express detectable levels of SEL1L; the majority of dysplastic cells within adenoma contain high levels of protein; contrary to the poorly differentiated cells. SEL1L is up-regulated in well-differentiated CRCs containing a high fraction of transformed cells that progress toward a differentiated secretory phenotype; then it would be important to precisely locate SEL1L expression in the context of normal intestinal epithelial differentiation, as it could shed some light on the intriguing pattern of expression of this gene in epithelial carcinogenesis. Within CRCs, SEL1L expression could become an identifier of the cell population with differentiated secretory phenotype, acute ER stress and active ERAD response and could be a future a marker for targeted therapy.
Acknowledgments
This work was supported in part by Grants #CA102681 and CA90890 funded by the National Cancer Institute, NIH, by the RCMI, by the Italian Government MIUR-FIRB grant nRBIP064CRT to IB, by Italian Government MIUR 60% grants to RMC for 2008 and 2009 and by a grant from “Associazione Italiana per la Ricerca sul Cancro” (AIRC) to AM. Dr. A. Morgano is a PhD student in the Oncology program at G. d’Annunzio University, Chieti, Italy.
Footnotes
Conflict of Interest None
Contributor Information
Hassan Ashktorab, Email: hashktorab@howard.edu, Department of Medicine and Cancer Center, Howard University College of Medicine, 2041 Georgia Avenue, NW, Washington, DC 20060, USA; Department of Pathology, Howard University College of Medicine, Washington, DC 20060, USA.
William Green, Department of Medicine and Cancer Center, Howard University College of Medicine, 2041 Georgia Avenue, NW, Washington, DC 20060, USA; Department of Pathology, Howard University College of Medicine, Washington, DC 20060, USA.
Giovanna Finzi, Department of Pathology, Ospedale di Circolo, Varese, Italy.
Fausto Sessa, Department of Human Morphology, University of Insubria, Varese, Italy; Department of Pathology, Multimedica IRCCS, Milan, Italy.
Mehdi Nouraie, Department of Medicine and Cancer Center, Howard University College of Medicine, 2041 Georgia Avenue, NW, Washington, DC 20060, USA; Department of Pathology, Howard University College of Medicine, Washington, DC 20060, USA.
Edward L. Lee, Department of Medicine and Cancer Center, Howard University College of Medicine, 2041 Georgia Avenue, NW, Washington, DC 20060, USA Department of Pathology, Howard University College of Medicine, Washington, DC 20060, USA.
Annalisa Morgano, Department of Oncology and Neuroscience, “G. d’Annunzio” University, Chieti, Italy; Center of Excellence on Ageing, G. d’Annunzio University Foundation, Chieti-Pescara, Italy; Laboratory of Lipid Metabolism and Cancer, Department of Translational Pharmacology (DTP), Consorzio Mario Negri Sud, S. Maria Imbaro, Chieti, Italy.
Antonio Moschetta, Laboratory of Lipid Metabolism and Cancer, Department of Translational Pharmacology (DTP), Consorzio Mario Negri Sud, S. Maria Imbaro, Chieti, Italy.
Monica Cattaneo, Institute of Genetics and Biomedical Research-National Research Council, Via Fantoli 16/15, Milano, Italy.
Renato Mariani-Costantini, Department of Oncology and Neuroscience, “G. d’Annunzio” University, Chieti, Italy; Center of Excellence on Ageing, G. d’Annunzio University Foundation, Chieti-Pescara, Italy.
Hassan Brim, Department of Medicine and Cancer Center, Howard University College of Medicine, 2041 Georgia Avenue, NW, Washington, DC 20060, USA; Department of Pathology, Howard University College of Medicine, Washington, DC 20060, USA.
Ida Biunno, Email: ida.biunno@itb.cnr.it, Institute of Genetics and Biomedical Research-National Research Council, Via Fantoli 16/15, Milano, Italy.
References
- 1.Vembar SS, Brodsky JL. One step at a time: Endoplasmic reticulum-associated degradation. Natl Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cattaneo M, Lotti LV, Martino S, et al. Secretion of novel SEL1L endogenous variants is promoted by ER stress/UPR via endosomes and shed vesicles in human cancer cells. PLoS One. 2011;6:e17206. doi: 10.1371/journal.pone.0017206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma J, Stampfer MJ, Giovannucci E, et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 1997;57:1098–1102. [PubMed] [Google Scholar]
- 4.Ma Y, Hendershot LM. ER chaperone functions during normal and stress conditions. J Chem Neuroanat. 2004;28:51–65. doi: 10.1016/j.jchemneu.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Orlowski RZ, Zeger EL. Targeting the proteasome as a therapeutic strategy against haematological malignancies. Expert Opin Investig Drugs. 2006;15:117–130. doi: 10.1517/13543784.15.2.117. [DOI] [PubMed] [Google Scholar]
- 6.Scriven P, Brown NJ, Pockley AG, Wyld L. The unfolded protein response and cancer: A brighter future unfolding? J Mol Med. 2007;85:331–341. doi: 10.1007/s00109-006-0150-5. [DOI] [PubMed] [Google Scholar]
- 7.Voorhees PM, Dees EC, O’Neil B, Orlowski RZ. The proteasome as a target for cancer therapy. Clin Cancer Res. 2003;9:6316–6325. [PubMed] [Google Scholar]
- 8.Zhang F, Hamanaka RB, Bobrovnikova-Marjon E, et al. Ribosomal stress couples the unfolded protein response to p53-dependent cell cycle arrest. J Biol Chem. 2006;281:30036–30045. doi: 10.1074/jbc.M604674200. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Hua H, Ran Y, et al. Derlin-1 is overexpressed in human breast carcinoma and protects cancer cells from endoplasmic reticulum stress-induced apoptosis. Breast Cancer Res. 2008;10:R7. doi: 10.1186/bcr1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cattaneo M, Orlandini S, Beghelli S, et al. SEL1L expression in pancreatic adenocarcinoma parallels SMAD4 expression and delays tumor growth in vitro and in vivo. Oncogene. 2003;22:6359–6368. doi: 10.1038/sj.onc.1206665. [DOI] [PubMed] [Google Scholar]
- 11.Orlandi R, Cattaneo M, Troglio F, et al. SEL1L expression decreases breast tumor cell aggressiveness in vivo and in vitro. Cancer Res. 2002;62:567–574. [PubMed] [Google Scholar]
- 12.Granelli P, Cattaneo M, Ferrero S, et al. SEL1L and squamous cell carcinoma of the esophagus. Clin Cancer Res. 2004;10:5857–5861. doi: 10.1158/1078-0432.CCR-04-0075. [DOI] [PubMed] [Google Scholar]
- 13.Barberis MC, Roz E, Biunno I. SEL1L expression in prostatic intraepithelial neoplasia and adenocarcinoma: An immunohistochemical study. Histopathology. 2006;48:614–616. doi: 10.1111/j.1365-2559.2005.02274.x. [DOI] [PubMed] [Google Scholar]
- 14.Ferrero S, Falleni M, Cattaneo M, et al. SEL1L expression in non-small cell lung cancer. Hum Pathol. 2006;37:505–512. doi: 10.1016/j.humpath.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Greene FL, Sobin LH. A worldwide approach to the TNM staging system: Collaborative efforts of the AJCC and UICC. J Surg Oncol. 2009;99:269–272. doi: 10.1002/jso.21237. [DOI] [PubMed] [Google Scholar]
- 16.Ashktorab H, Belgrave K, Hosseinkhah F, et al. Global histone H4 acetylation and HDAC2 expression in colon adenoma and carcinoma. Dig Dis Sci. 2009;54:2109–2117. doi: 10.1007/s10620-008-0601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hewitt SM. The application of tissue microarrays in the validation of microarray results. Methods Enzymol. 2006;410:400–415. doi: 10.1016/S0076-6879(06)10020-8. [DOI] [PubMed] [Google Scholar]
- 18.Cattaneo M, Lotti LV, Martino S, et al. Functional characterization of two secreted SEL1L isoforms capable of exporting unassembled substrate. J Biol Chem. 2009;284:11405–11415. doi: 10.1074/jbc.M805408200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cattaneo M, Otsu M, Fagioli C, et al. SEL1L andHRD1 are involved in the degradation of unassembled secretory Ig-mu chains. J Cell Physiol. 2008;215:794–802. doi: 10.1002/jcp.21364. [DOI] [PubMed] [Google Scholar]
- 20.Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci USA. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller B, Lilley BN, Ploegh HL. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol. 2006;175:261–270. doi: 10.1083/jcb.200605196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cattaneo M, Fontanella E, Canton C, Delia D, Biunno I. SEL1L affects human pancreatic cancer cell cycle and invasiveness through modulation of PTEN and genes related to cell-matrix interactions. Neoplasia. 2005;7:1030–1038. doi: 10.1593/neo.05451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q, Chen J, Mai B, et al. A single-nucleotide polymorphism in tumor suppressor gene SEL1L as a predictive and prognostic marker for pancreatic ductal adenocarcinoma in caucasians. Mol Carcinog. 2011 doi: 10.1002/mc.20808. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 26.Fribley AM, Evenchik B, Zeng Q, et al. Proteasome inhibitor PS-341 induces apoptosis in cisplatin-resistant squamous cell carcinoma cells by induction of Noxa. J Biol Chem. 2006;281:31440–31447. doi: 10.1074/jbc.M604356200. [DOI] [PubMed] [Google Scholar]