Abstract
Hepatic encephalopathy (HE) has been related to gut bacteria and inflammation in the setting of intestinal barrier dysfunction. We aimed to link the gut microbiome with cognition and inflammation in HE using a systems biology approach. Multitag pyrosequencing (MTPS) was performed on stool of cirrhotics and age-matched controls. Cirrhotics with/without HE underwent cognitive testing, inflammatory cytokines, and endotoxin analysis. Patients with HE were compared with those without HE using a correlation-network analysis. A select group of patients with HE (n = 7) on lactulose underwent stool MTPS before and after lactulose withdrawal over 14 days. Twenty-five patients [17 HE (all on lactulose, 6 also on rifaximin) and 8 without HE, age 56 ± 6 yr, model for end-stage liver disease score 16 ± 6] and ten controls were included. Fecal microbiota in cirrhotics were significantly different (higher Enterobacteriaceae, Alcaligeneceae, and Fusobacteriaceae and lower Ruminococcaceae and Lachnospiraceae) compared with controls. We found altered flora (higher Veillonellaceae), poor cognition, endotoxemia, and inflammation (IL-6, TNF-α, IL-2, and IL-13) in HE compared with cirrhotics without HE. In the cirrhosis group, Alcaligeneceae and Porphyromonadaceae were positively correlated with cognitive impairment. Fusobacteriaceae, Veillonellaceae, and Enterobacteriaceae were positively and Ruminococcaceae negatively related to inflammation. Network-analysis comparison showed robust correlations (all P < 1E-5) only in the HE group between the microbiome, cognition, and IL-23, IL-2, and IL-13. Lactulose withdrawal did not change the microbiome significantly beyond Fecalibacterium reduction. We concluded that cirrhosis, especially when complicated with HE, is associated with significant alterations in the stool microbiome compared with healthy individuals. Specific bacterial families (Alcaligeneceae, Porphyromonadaceae, Enterobacteriaceae) are strongly associated with cognition and inflammation in HE.
Keywords: cirrhosis, inflammation, systems biology, multitag pyrosequencing
cirrhosis is often complicated by hepatic encephalopathy (HE), a condition characterized by cognitive impairment and poor survival (2, 8). There is evidence that pathogenic abnormalities in HE are related to the gut flora and their byproducts such as ammonia and endotoxin in the setting of intestinal barrier dysfunction and systemic inflammation (14, 35, 36, 44). Patients with cirrhosis also have widespread derangements of their immune response, which can potentiate insults such as sepsis and result in HE (36, 43). The present treatments for HE rely on manipulation of the gut flora; however, their mechanisms of action as well as prediction of resistance to therapy are not clear (2). In addition, the characterization of gut flora in prior HE studies has been limited by the use of culture-based techniques that do not identify the majority of the intestinal bacteria (23). Because the pathogenesis of HE likely spans several metabolic processes, we proposed that a systems biology approach could be useful to identify novel functional hypothesis and new therapeutic targets for HE. Specifically, we used correlation-network analysis to correlate features within each treatment group to dissect out functionality in the system (27). This analysis provides potential clues to the functionality of the system, leading the way to hypothesis-driven experimental research.
The aims of this study were 1) to link the gut microbiome with cognition and inflammation in cirrhotic patients with and without HE using a systems biology approach, 2) to identify differences in the microbiome of healthy controls and cirrhotic patients, and 3) to define the effect of lactulose withdrawal on microbiome of cirrhotic patients. The a priori hypothesis was that the gut microbiome composition would be correlated with cognition and inflammation in cirrhotic patients with HE and that this association would be different from those who have never developed HE.
MATERIALS AND METHODS
Patients with cirrhosis and healthy age-matched controls were recruited after providing a written informed consent. We only included controls without liver disease and those who were not taking medications apart from those for hypertension, hyperlipidemia, or gastroesophageal reflux disease. In the case of cirrhotic patients, we excluded those with a current infection (defined by elevated white blood cell count, clinical suspicion, or fever), who had experienced variceal bleeding within the last 4 wk, who were on gut-absorbable antibiotic therapy, or had alcohol or illicit drug intake within 3 mo (checked by drug and alcohol screens). The data collected from their medical record were model for end-stage liver disease (MELD) score, etiology of cirrhosis, complications of cirrhosis in the past, and present medication use. Patients in the no HE group had never had an episode of HE and were not on any therapy for it. Patients in the HE group had suffered at least one HE episode within the last 3 mo and were currently controlled on lactulose alone or lactulose with rifaximin. We did not include patients during an acute HE episode because those patients are often hospitalized, are on systemic antibiotic therapy, and are not able to give consent or perform cognitive testing.
All subjects underwent a mini-mental status exam, and only those scoring above 25 were included in the full study (11). Participants then underwent a 24-h dietary recall. Subsequently, a recommended cognitive battery consisting of the following tests was administered to the cirrhotic patients: 1) psychometric HE score (PHES), 2) block design test (BDT: subjects are required to replicate standardized designs with given blocks in a timed manner. The score is based on the designs correctly copied), and 3) inhibitory control test [ICT, this is a 15-min computerized test. Subjects are instructed to respond to alternating presentations of X and Y on the screen (targets) while inhibiting response when X and Y are not alternating (lures)] (3, 41). The PHES consists of five tests: number connection test-A/B (subjects are asked to “join the dots” between numbers or numbers and alphabets in a timed fashion, and the number of seconds required is the outcome), digit symbol test (DST: subjects are required to copy corresponding figures from a given list within 2 min, and the number correctly copied is the result), line drawing test [LDTt (time) and LDTe (errors): subjects are required to trace a line between two parallel lines and balance between speed and accuracy. Time required and the number of times the subject's line strays beyond the marked lines (errors) are recorded], and serial dotting test (subjects are asked to dot the center of a group of blank circles, and the time required is the outcome). The PHES is a validated battery for cognitive dysfunction in cirrhosis and tests for psychomotor speed, visuomotor coordination, attention, and set shifting (32). The BDT tests for visuomotor coordination. The ICT is a validated computerized test of attention, psychomotor speed, response inhibition, and working memory. A high score on BDT, DST, and ICT targets and a low score on the rest of the tests indicate good cognitive performance.
Cirrhotic patients also underwent serum collection for inflammatory cytokines testing for innate immunity (IL-1b, IL-6, TNF-α), Th1 response (IFN-γ and IL-2), Th2 response (IL-4, IL-10, IL-13), Th17 response (IL-17 and IL-23), and endotoxin. These were analyzed in duplicate by multiplex bead-based sandwich ELISA and Limulus Amebocyte Lysate assay for endotoxin using published techniques by Assay Gate, Ijamsville, MD (4, 17, 45).
Prospective study.
A selected group of seven cirrhotic patients in the HE group currently only on lactulose (who were also included in the cross-sectional study) were systematically withdrawn from therapy. Their diet was controlled over the study period. Their stool microflora was analyzed while on lactulose and then 14 days following discontinuation of lactulose therapy. Day 14 was chosen because prior culture-based studies have shown a change in fecal flora after lactulose initiation within that time frame (31).
Interrogation of the microbiome.
Stool was collected and DNA extracted for microbiome analysis within 24 h of collection from patients and controls (29). Stool (0.2 mg) was suspended in buffer argininosuccinate lyase to which 0.75-g 0.1-mm zirconia/silica beads (Biospec Products) were added, and cells were disrupted twice at 4,800 revolution/min for 1 min. Tubes were placed in a 90°C water bath for 10 min, and the remainder of the protocol was followed according to the manufacturer. We first routinely use length heterogeneity PCR (LH-PCR) fingerprinting of the 16S rRNA to rapidly survey our samples and standardize the community amplification. We then interrogated the microbial taxa associated with the gut fecal microbiome using multitag pyrosequencing (MTPS). This technique allows the rapid sequencing of multiple samples at one time, yielding thousands of sequence reads per sample (12).
Microbiome community fingerprinting.
LH-PCR was done to standardize the community analysis as previously published (21). Briefly, total genomic DNA was extracted from tissue using Bio101 kit from MP Biomedicals per the manufacturer's instructions. About 10 ng of extracted DNA was amplified by PCR using a fluorescently labeled forward primer 27F [5′- (6FAM) AGAGTTTGATCCTGGCTCA G-3′] and unlabeled reverse primer 355R′ (5′-GCTGCCTCCCGTAGGAGT-3′). Both primers are universal primers for bacteria (22). The LH-PCR products were diluted according to their intensity on agarose gel electrophoresis and mixed with ILS-600 size standards (Promega) and HiDi Formamide (Applied Biosystems). The diluted samples were then separated on an ABI 3130xl fluorescent capillary sequencer (Applied Biosystems) and processed using the Genemapper software package (Applied Biosystems). Normalized peak areas were calculated using a custom PERL script, and operational taxonomic units constituting less than 1% of the total community from each sample were eliminated from the analysis to remove the variable low-abundance components within the communities.
MTPS.
We employed the MTPS process to characterize the microbiome from the fecal samples. Specifically, we have generated a set of 96 emulsion PCR fusion primers that contain the 454 emulsion PCR linkers on the 27F and 355R primers and a different eight-base “barcode” between the A adapter and 27F primer. Thus each fecal sample was amplified with unique barcoded forward 16S rRNA primers, and then up to 96 samples were pooled and subjected to emulsion PCR and pyrosequenced using a GS-FLX pyrosequencer (Roche). Data from each pooled sample were “deconvoluted” by sorting the sequences into bins based on the barcodes using custom PERL scripts. Thus we were able to normalize each sample by the total number of reads from each barcode. We have noted that ligating tagged primers to PCR amplicons distorts the abundances of the communities, and thus it is critical to incorporate the tags during the original amplification step (12). Several groups have employed various barcoding strategies to analyze multiple samples, and this strategy is now well accepted (38).
RDP10 analysis.
We identified the taxa present in each sample using the Bayesian analysis tool in Version 10 of the Ribosomal Database Project (RDP10). The abundances of the bacterial identifications were then normalized using a custom PERL script, and taxa present at >1% of the community were tabulated. We chose this cutoff because of our a priori assumption that taxa present in <1% of the community vary between individuals and have minimal contribution to the functionality of that community and that 2,000 reads per sample will only reliably identify community components that are >1% in abundance (13).
This study was approved by the Institutional Review Boards of the McGuire VA Medical Center and the Virginia Commonwealth University Medical Center in Richmond.
Statistical analysis.
Clinical and microbiome features of controls were compared with patients with cirrhosis with Metastats using the P value and the false discovery rate (q value) for nonnormal distributions (42). A principal coordinate analysis was also used to show differences between the two groups. Only taxa with average abundances >1%, P values <0.05, and low q values (i.e., low risk of false discovery) were considered significant.
The cirrhosis group was divided into those with and without HE and were compared. Data from the significant variables between HE and non-HE groups were combined in a mutivariate ANOVA model. Within patients with HE, comparison was made between those on lactulose alone to those with lactulose and rifaximin. Microbiome abundance comparisons between groups were made at a family level using nonparametric tests. A comparison was performed between patients on and withdrawn from lactulose therapy using the Wilcoxon matched-pair signed rank tests. All values are presented as means ± SD unless mentioned otherwise.
Correlation-network models.
Groups were divided into HE or no HE, and they were analyzed separately. The microbiome features along with the presence of HE, cirrhosis severity, serum markers, and cognitive function tests were correlated using a Pearson's correlation function and then filtered for correlations >0.90. These correlates were calculated using a custom R module, and the correlations and corresponding attributes were imported into Cytoscape for visualization of the network models (34). We then compared the network topology of the two disease classes, HE and no HE, to identify which subnetworks were present in one and not the other, giving us clues on system functionality. It is assumed that correlations present in one treatment group that are missing in another not only differentiate the groups but indicate potential clues to the functionality of the system, leading the way to hypothesis-driven experimental research.
RESULTS
Twenty-five cirrhotic patients (MELD score 16 ± 6) and ten healthy controls were included. HE was present in 17 patients (68%; 11 were on lactulose alone, 6 were on both lactulose and rifaximin). None of the patients with HE were on rifaximin alone. All patients who were on rifaximin were started on it because of difficulties in tolerating lactulose alone. The mean age of the controls was 54 ± 5 yr with 6 men and 4 women and 60% Caucasian, 30% African-American, and 10% Hispanic ethnicity. The ages of cirrhotics with and without HE (56 ± 3 and 55 ± 5 yr) as well as the sex (16 men in HE and 7 men in no HE group) were similar to controls. The ethnicity distribution in the HE group (64% Caucasian, 29% African-American, and 6% Hispanic) and without HE group (63% Caucasian, 25% African-American, and 13% Hispanic) was statistically similar to controls. There was no significant difference in the body mass index between the groups (controls 25 ± 3, cirrhotics with HE 26 ± 5, and those without HE 25 ± 3).
All patients and controls were nonvegetarians and had similar dietary intake and constituents on recall before sample collection (mean intake 2,470 Kcal and 16% protein intake). Patients had been abstinent of alcohol and illicit drugs for at least 3 mo, confirmed by serum alcohol and urine drug screens. At the time of sample collection, none of the subjects had systemic infections as evidenced by normal white blood cell counts, normal body temperature, and physical examination unremarkable for infections. The majority of patients and none of the controls were on proton pump inhibitor (PPI) therapy (92%) (Table 1). Thirteen (52%) had alcoholic liver disease; the remainder had hepatitis C (40%) or cryptogenic cirrhosis (8%); eight (32%) had both alcoholic and hepatitis C virus disease. All patients in the HE group had residual cognitive impairment or minimal HE at the time of the testing (5, 30).
Table 1.
Features of patients with and without hepatic encephalopathy
| HE (n = 17) | No HE (n = 8) | P value | |
|---|---|---|---|
| Alcoholic etiology | 58% | 37% | 0.41 |
| Prior variceal bleeding | 23% | 0% | 0.07 |
| Prior SBP | 0% | 0% | 1.0 |
| Renal insufficiency | 0% | 18% | 0.09 |
| Clinically evident ascites | 29% | 47% | 0.65 |
| Median daily bowel movements | 2 | 1 | 0.02 |
| Proton pump inhibitor therapy | 94% | 86% | 0.51 |
| MELD score | 17 ± 6 | 12 ± 5 | 0.048 |
| Venous ammonia | 52 ± 28 | 31 ± 21 | 0.148 |
| WBC count, /mm3 | 5.2 ± 2 | 5 ± 3 | 0.33 |
| Endotoxin | 0.27 ± 0.24 | 0.059 ± 0.012 | 0.002 |
| IL-1b, pg/ml | 6.2 ± 11.1 | 1.07 ± 0.55 | 0.06 |
| IFN-γ, pg/ml | 11.3 ± 26.6 | 1.6 ± 1.8 | 0.148 |
| IL-10, pg/ml | 8.21 ± 8.70 | 2.9 ± 1.5 | 0.022 |
| IL-23, pg/ml | 1842 ± 4873 | 317 ± 359 | 0.205 |
| IL-17, pg/ml | 32.1 ± 81.3 | 4.53 ± 5.27 | 0.107 |
| IL-6, pg/ml | 67.8 ± 72.2 | 9.3 ± 7.8 | 0.004 |
| IL-2, pg/ml | 48 ± 91 | 2.7 ± 2 | 0.04 |
| TNF-α, pg/ml | 7.01 ± 4.09 | 4.33 ± 2.33 | 0.05 |
| IL-13, pg/ml | 32.0 ± 17.2 | 0.80 ± 0.02 | 0.0001 |
Applicable values are means ± SD. There were significant differences at baseline between those with and without HE with respect to endotoxemia and inflammation, all of which were significantly worse in patients with hepatic encephalopathy (HE). As expected, patients with HE had a significantly higher model for end-stage liver disease (MELD) score and a higher number of daily bowel movements because they were on lactulose.
SBP, spontaneous bacterial peritonitis; WBC, white blood cells.
Cross-sectional microbial analysis between controls and patients with cirrhosis.
There were significant differences in stool microbiome between cirrhotic patients and controls (Fig. 1, Table 2). There was a significantly higher abundance of Lachnospiraceae and Ruminococceae in the control group, whereas Enterobacteriaceae, Fusobacteriaceae, Alcaligenaceae, Lactobacillaceae, and Leuconostocaceae were significantly lower in the controls compared with cirrhotic patients. These differences persisted and widened when controls were compared with patients with and without HE (Tables 3 and 4). The differences existed for Leuconostocaceae, Clostridiales_Incertae Sedis XIV, Fusobacteriaceae, Lachnospiraceae, and Ruminococcaceae in both groups of cirrhotic patients (with or without HE). Interestingly, however, the HE group differed from controls on several additional bacterial families compared with cirrhotics without HE in that they had a significantly higher concentration of Enterobacteriaceae, Alcaligenaceae, Lactobacilaceae, and Streptococcaceae.
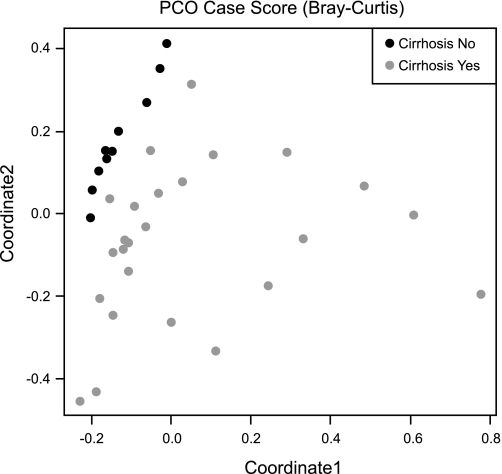
Fig. 1.
Principal coordinate analysis of the fecal microbiome of controls and cirrhotic patients. This graph shows the variation in fecal microbiome plotted on a principal coordinate analysis plot. Points that are closer to each other are similar with respect to their stool microbiota. The healthy controls represented by the black dots are clustered together, whereas the cirrhotic patients represented by the gray dots are distant from the controls. This indicates a difference in the stool microbiome of healthy controls compared to cirrhotic patients.
Table 2.
Differences in bacterial abundances between controls and cirrhotic patients
| Control |
Cirrhosis |
|||||
|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | P value | Q value | |
| Leuconostocaceae | 0.00 | 0.00 | 2.02 | 0.70 | 0.0009 | 0.009 |
| Clostridium Incertae sedis XIV | 7.35 | 1.59 | 1.08 | 0.18 | 0.0009 | 0.008 |
| Lachnospiraceae | 23.44 | 2.24 | 10.40 | 2.60 | 0.0009 | 0.008 |
| Ruminococcaceae | 17.72 | 1.89 | 6.75 | 1.28 | 0.001 | 0.008 |
| Enterobacteriaceae | 0.00 | 0.00 | 7.60 | 2.89 | 0.001 | 0.008 |
| Fusobacteriaceae | 0.00 | 0.00 | 1.80 | 1.06 | 0.0059 | 0.0408 |
| Alcaligenaceae | 0.89 | 0.34 | 2.76 | 0.73 | 0.032 | 0.1723 |
A comparison between controls and cirrhotic patients' microbial flora was performed using Metastats, and only significantly different and values with around 1% are shown; the rest were nonsignificant. Q value indicates that the false discovery rate and that a lower value is generally preferred to avoid a false positive result. There was a significantly higher abundance of Lachnospiraceae and Ruminococceae in the control group, whereas Enterobacteriaceae, Fusobacteriaceae, Alcaligenaceae, Lactobacillaceae, and Leuconostocaceae were significantly lower in the controls compared to patients with cirrhosis. Incertae sedis: uncertain placement.
Table 3.
Differences in bacterial abundances between controls and cirrhotic patients with HE
| Name | Control Mean | Control SE | HE Mean | HE SE | P value | Q value |
|---|---|---|---|---|---|---|
| Leuconostocaceae | 0.00 | 0.00 | 2.19 | 1.08 | 0.0009 | 0.007 |
| Clostridiales_Incertae Sedis XIV | 7.35 | 1.59 | 0.99 | 0.21 | 0.0009 | 0.007 |
| Ruminococcaceae | 17.72 | 1.89 | 5.68 | 1.42 | 0.0009 | 0.007 |
| Lachnospiraceae | 23.44 | 2.24 | 9.54 | 3.72 | 0.0039 | 0.022 |
| Enterobacteriaceae | 0.00 | 0.00 | 10.02 | 4.13 | 0.0049 | 0.026 |
| Streptococcaceae | 0.62 | 0.26 | 4.05 | 1.98 | 0.0239 | 0.099 |
| Fusobacteriaceae | 0.00 | 0.00 | 1.36 | 0.95 | 0.0369 | 0.146 |
| Alcaligenaceae | 0.89 | 0.34 | 2.61 | 0.74 | 0.048 | 0.169 |
This table shows the differences between bacterial abundances in stool of controls with patients with HE; only those bacteria whose abundances were >1% and were significantly different between groups are shown. There was a significantly higher abundance of Enterobacteriaceae, Fusobacteriaceae, Leuconostocaceae, Streptococcaceae, and Alcaligenaceae in patients with HE, whereas the rest of the bacteria listed were lower in the HE group.
Table 4.
Differences in bacterial abundances between controls and cirrhotic patients without HE
| Control Mean | Control SE | No HE Mean | No HE SE | P value | Q value | |
|---|---|---|---|---|---|---|
| Leuconostocaceae | 0.00 | 0.00 | 1.69 | 0.95 | 0.0001 | 0.001 |
| Clostridiales_Incertae Sedis XIV | 7.35 | 1.59 | 1.29 | 0.35 | 0.0001 | 0.001 |
| Fusobacteriaceae | 0.00 | 0.00 | 2.75 | 2.75 | 0.0001 | 0.001 |
| Lachnospiraceae | 23.44 | 2.245 | 12.08 | 2.47 | 0.003 | 0.002 |
| Ruminococcaceae | 17.72 | 1.89 | 9.04 | 2.62 | 0.019 | 0.008 |
This table shows the differences between bacterial abundances in stool of controls with cirrhotic patients without HE; only those bacteria whose abundances were >1% and were significantly different between groups are shown. There was a significantly higher abundance of Fusobacteriaceae and Leuconostocaceae in cirrhotic patients without HE, whereas the rest of the bacteria listed were lower in the cirrhotic no HE group.
Comparison within the HE group.
There was no significant difference between the clinical, inflammatory, or cognitive profile between HE patients on lactulose alone compared with those on lactulose and rifaximin (Table 5). Additionally, no differences in the microbiome components were identified using classic multivariate analysis. Specifically the normalized abundances at the family level of Alcaligenaceae (12.6 ± 9% vs. 10 ± 13%, P = 0.8), Enterobacteriaceae (26 ± 40% vs. 24 ± 40%, P = 0.7), Bacteroidaceae (39 ± 35% vs. 58 ± 39%, P = 0.35), Porphyromonadaceae (14 ± 21% vs. 8 ± 15%, P = 0.23), Prevotellaceae (18 ± 25% vs. 10 ± 19%, P = 0.45), Veillonellaceae (15 ± 13% vs. 15 ± 17%, P = 0.8), Ruminococcaceae (17 ± 22% vs. 17 ± 19%, P = 0.6), Streptococcaceae (4 ± 10 vs. 2 ± 3, P = 0.55), and Lactobacillaceae (2 ± 1% vs. 1 ± 1%, P = 0.42) between the two groups were not statistically significant.
Table 5.
Comparison within the HE group
| On Lactulose Alone (n = 11) | On Lactulose and Rifaximin (n = 6) | P value | |
|---|---|---|---|
| MELD score | 16.5 ± 7.6 | 18.2 ± 3.3 | 0.53 |
| Venous Ammonia | 52.8 ± 26.3 | 36.3 ± 32.6 | 0.23 |
| Endotoxin | 0.21 ± 0.21 | 0.41 ± 0.26 | 0.13 |
| IL-6, pg/ml | 47.8 ± 56.4 | 108.0 ± 94.3 | 0.20 |
| IL-2, pg/ml | 61 ± 108 | 21.6 ± 22.4 | 0.25 |
| TNF-α, pg/ml | 6.7 ± 4.2 | 7.7 ± 4.22 | 0.67 |
| IL-13, pg/ml | 31.9 ± 84.2 | 32.1 ± 49.7 | 0.99 |
| Number connection-A, s | 55.2 ± 34.1 | 79.4 ± 49.9 | 0.27 |
| Number connection-B, s | 189 ± 127 | 231 ± 105 | 0.51 |
| Digit symbol score | 36.5 ± 14.0 | 34.8 ± 12.3 | 0.82 |
| Block design, score | 24.9 ± 24.8 | 60.0 ± 53.4 | 0.23 |
| Serial dotting, s | 88.7 ± 29.2 | 124.2 ± 36.5 | 0.10 |
| Line drawing error score | 28.0 ± 18.6 | 43.0 ± 42.8 | 0.49 |
| Line drawing time, s | 106.9 ± 54.5 | 79.4 ± 72.1 | 0.48 |
| ICT targets, % | 90.6 ± 8.64 | 83.7 ± 21.0 | 0.52 |
| ICT lure number | 17.4 ± 10.8 | 19.6 ± 10.6 | 0.71 |
All applicable values are presented as means ± SD. There was no significant difference in any variable tested between patients with HE on lactulose alone compared to those on lactulose and rifaximin.
ICT, inhibitory control test.
Cross-sectional analysis within the cirrhosis group.
MELD was not correlated with endotoxin, inflammatory cytokines, or cognition. We also did not find any differences in the inflammatory cytokines or endotoxemia between cirrhotic patients of differing etiologies using ANOVA, possibly attributable to the sample size, dual etiologies, and probable effect of HE overwhelming the underlying etiologies (data not shown). Interestingly, MELD score was positively correlated with Enterobacteriaceae (r = 0.61, P = 0.001) and negatively with Ruminococcaceae (r = −0.38, P = 0.05) with a trend toward lower Prevotellaceae (r = −0.36, P = 0.056). Enterobacteriacae were also associated with TNF-α (r = 0.5, P = 0.03). Veillonellaceae and Fusobacteriaceae were also associated with worsening inflammation (IL-13, IL-6) and endotoxemia (P < 0.05). Ruminococcaceae, importantly, were negatively correlated with endotoxemia (r = −0.5, P = 0.02). The presence of Alcaligeneceae and Porphyromonadaceae was associated with poor cognition on individual tests (Table 6).
Table 6.
Correlation between poor cognitive performance and presence of Alcaligeneceae and Porphyromonadaceae in the entire group
|
Alcaligeneceae |
Porphyromonadaceae |
|||
|---|---|---|---|---|
| Cognitive Tests | R | P value | R | P value |
| Number connection-A, s | 0.68 | 0.001 | 0.63 | 0.002 |
| Number connection-B, s | 0.445 | 0.04 | 0.28 | 0.24 |
| Serial dotting, s | 0.52 | 0.018 | 0.41 | 0.05 |
| Line drawing error number | 0.58 | 0.009 | 0.59 | 0.008 |
| Line drawing time, s | 0.24 | 0.31 | 0.17 | 0.48 |
| ICT lure number | 0.26 | 0.26 | 0.29 | 0.19 |
| Digit symbol score | −0.63 | 0.003 | −0.46 | 0.04 |
| Block design score | −0.27 | 0.25 | −0.21 | 0.37 |
| ICT targets, % | −0.51 | 0.019 | −0.57 | 0.007 |
A high score on digit symbol, block design, and ICT targets indicates good cognition (i.e., lower value indicates poor cognition); for the rest of the tests, a high score indicates poor cognitive performance. Significant correlations are in bold text. Therefore, we found a significant correlation between impairment on most cognitive tests and relative abundance of Alcaligeneceae and Porphyromonadaceae.
Multivariate analysis of the HE and no HE groups.
HE patients, as expected had a higher MELD score and bowel movement frequency compared with those without HE (Table 1). The rate of PPI use and other complications of cirrhosis were not different between the groups. Although the major families were present in both sample classes, there were observable abundance differences in some of the taxa; there was a significantly higher abundance of Veillonellaceae in the HE group (14 ± 12% vs. 4 ± 9%, P = 0.046) compared with the no HE group. There were no significant differences in the other microbiome families between the HE and no HE groups. The multivariate ANOVA performed using Veillonellaceae, IL-13, IL-6, MELD score, and endotoxin demonstrated a P value of 0.002 using the Lawley-Hotelling test statistic of 2.25672 with an F statistic of 5.481.
Correlation-network analysis.
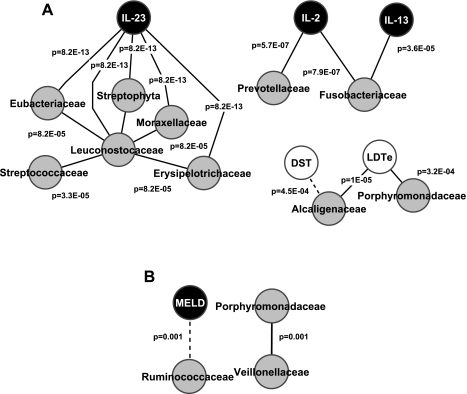
In patients with HE, in contrast to the multivariate analysis above, several significantly strong correlations were found between features within the HE group with the correlation coefficients (Fig. 2A). IL-23 was an important correlate with several bacterial families across different phyla, such as Leuconostocaceae, Eubacteriaceae, Erysipelotrichaceae, Moraxellaceae, Streptophyta, and Streptococcaceae within the HE group. All P values for this correlation were below 8.2E-05, indicating a highly robust linkage. The correlation of immune function with bacterial families was further illustrated by the highly significant correlation (P values <3.5E-0.5) between inflammatory cytokines IL-2 and IL-13 with Fusobacteriaceae and Prevotellaceae. The correlation between Porphyromonadacae and Alcaligenacae with poor performance on cognitive tests was observed in this group accompanied by very significant P values (P < 1E-05). In patients without HE, in sharp contrast, relatively few correlations that reached the stringent threshold we had set for this analysis were seen in patients without HE and markers of inflammation, cognition, and microbial families. MELD score was negatively correlated with Ruminococcaceae, whereas there was positive correlation between Porphyromonadaceae and Veillonellaceae (Fig. 2B). We did not find any significant correlations between inflammation and cognitive function that were abundant in the HE group correlation network.
Fig. 2.
Correlation-network analysis of cirrhotic patients with and without hepatic encephalopathy (HE). Only correlations with a coefficient >r = 0.90 are displayed. Gray nodes indicate microbiome families; white nodes indicate cognitive tests; and black nodes are serum inflammatory markers. A black line connecting nodes indicates positive correlation, and a dashed line indicates negative correlation >0.90. The P values for the correlations are displayed on the lines connecting the nodes in A and B. MELD, model for end-stage liver disease score; DST, digit symbol test; LDTe, line drawing test errors. A: patients with HE (n = 17) have a high number of significant correlations. There are significant positive correlations between IL-23 and several bacterial families. Prevotellaceae and Fusobacteriaceae are positively correlated with inflammation. Because a low score on DST and high one on LDTe indicate poor performance, Alcaligenaceae and Porphyromonadaceae were correlated with poor cognition. The P values for all these correlations are less than the 4th decimal place, indicating a very high significance. B: patients without HE have very few significant correlations (n = 8). There was a significant negative correlation between MELD score and Ruminococcaceae and a positive correlation between Veillonellaceae and Porphyromonadaceae.
Prospective study after lactulose withdrawal.
Seven male cirrhotic patients with HE (age 53 ± 8 yr) controlled on lactulose underwent a systematic withdrawal of lactulose. All patients had alcoholic liver disease, while five also had chronic hepatitis C. None of the patients had clinically recurrent HE at day 14. A significant (>1%) abundance was present for only 13 taxa at baseline on lactulose in those seven patients. These were mainly from the phylum Bacterioidetes (Bacteroides 35%, Prevotella 13%, Hallella 4%, Alistipes 3%, Parabacteroides spp. 2%, Porphyromonadaceae 1.7%) and Firmicutes (Faecalibacterium 6%, Lachnospira 5%, Roseburia 3%, Veillonella 2%, Dialister 2%, and Succinispira spp. 2%) with little contribution of Proteobacteria (Alcaligenaceae 2.6%, Hafnia 2%, and Sutterella spp. 1%) and none from Actinobacter spp. or Fusobacteria. There was <1% abundance on Lactobacillus spp., Clostridium spp., Streptococcus spp., Shigella spp., and Ruminococcus spp. After lactulose withdrawal, a decrease in Faecalibacterium spp. (abundance on lactulose 6% to 1% postwithdrawal, P = 0.026) and a trend toward decrease in Veillonella spp. (2% to 0%, P = 0.07) were seen. No other significant relative abundance change was identified, including Porphyromonadacae and Alcaligenacae.
DISCUSSION
This study demonstrates that a systems biology approach (correlation-network analysis) can be used to identify key linkages between the microbiome, inflammatory milieu, endotoxemia, and cognition in patients with HE. The IL-23 system was highly correlated with several bacterial families in patients within the HE group, and there was a direct correlation between cognition, Porphyromonadaceae, and Alcaligeneceae. We found significant differences between the microbial flora of age-matched healthy controls to the cirrhotic population with a higher degree of difference in patients with HE. The study showed that there was no significant difference in the stool flora between patients with HE on lactulose compared with those additionally on rifaximin. The results also indicate that a systematic withdrawal of lactulose therapy had minimal effect on the gut microflora abundance.
To date, it has been difficult to identify significant differences between control and disease groups using classic multivariate approaches (27). Specifically, the composition of the human gut microbiome has been shown to vary significantly between individuals, and this is a fundamental problem in associating the microbiome with diseases (12). Furthermore, most microbial abundance matrices derived from sequence data are both sparse and nonparametric. A microbial ecological interpretation of these issues is that different phylogenetic taxa play the same functional role in the complex nonlinear interactions between the human host and gut microbiome. It should be noted that these interactions are not static but form a nonlinear complex dynamic network that further confounds classic multivariate analysis methods. Unlike these methods, correlation network analysis allows the interrogation of these nonlinear dynamics to correlate phylogenetically defined taxa with function and disease phenotype. This was leveraged in our study where we found that HE was significantly correlated with microbiome components and inflammatory cytokines.
We found a significant difference in the bacterial composition of patients with cirrhosis compared with healthy controls that intensified when the cirrhotic group was divided into HE and those without HE. Ruminococcaceae and Clostridium incertae sedis XIV were overrepresented in controls similar to prior studies in inflammatory bowel disease and cirrhosis (7, 18, 19, 25, 46). The findings are also similar to those published by Chen et al. (7) in cirrhotic patients despite differences in cirrhosis etiology and diet (7). However, their study did not evaluate HE specifically, and they did not perform a systems biology analysis.
Alcaligenecaeae and Enterobacteriaceae were among the bacterial taxa that were differentially detected in cirrhotics with HE compared with controls but not different between controls and cirrhotics without HE. Increased Alcaligenaceae abundance was significantly associated with poor cognitive performance, whereas Enterobacteriaceae were associated with worsening inflammation and MELD score in the cirrhosis group. The correlation between the MELD score, HE, and Enterobacteriaceae is not surprising because these bacteria are responsible for most of the life-threatening infections associated with advanced cirrhosis (35, 37). Also, the negative correlation of Ruminococcaceae with endotoxemia and MELD score and reduction in this class in cirrhotics overall could indicate a protective role.
The striking finding was the direct correlation between specific bacterial taxa and cognitive function. To our knowledge bacterial taxa have not been previously related to cognitive and inflammatory markers in cirrhosis using culture-independent techniques. Porphyromonadaceae and Alcaligeneceae were associated with poor cognition in almost all tests (Table 6). It is unlikely that this is merely a manifestation of worsening liver disease because the MELD score was not significantly correlated either with cognitive performance or with these bacteria. Alcaligeneceae are Proteobacteria that are typically associated with opportunistic infections; interestingly they degrade urea to produce ammonia, which may explain part of this association (28). Porphyromonas are gram-negative anaerobes, whose fecal presence may be attributed to the deficient stomach acid and bile barrier function in cirrhosis (6, 33, 40). Interestingly in animal studies, gut microbial colonization with specific bacteria has been shown to influence neuronal circuitry involved in motor control and behavior (9, 15). The correlation of these bacterial families with cognition in humans is a novel finding that needs further study.
We confirmed the proinflammatory milieu and endotoxemia in patients with HE (36) and further demonstrated that specific microbial families, Enterobacteriacae, Veillonellaceae, and Fusobacteriaceae, were associated with inflammation (44). Specifically, in patients with HE, inflammatory markers IL-23, IL-1b, IL-2, IL-4, and IL-13 were highly correlated with gut microbiome components, possibly indicating a synergy between inflammation and cognition with microbiome changes (20, 26). It is interesting that IL-13, which, in addition to being an inflammatory mediator with IL-4, also mediates allergic reactions, would be increased in cirrhotic patients with HE. Although none of our patients had an allergic diatheses, the increased IL-13 concentration may also represent its profibrotic potential and the widespread immunomodulatory disturbances that are prevalent in cirrhosis (24). We did not find a difference in the inflammatory cytokines across etiologies, which is likely attributable to the limited sample size and patients with dual etiologies. The IL-23/IL-17 pathway is triggered by exposure to infectious agents in the intestine, which releases a cascade of proinflammatory cytokines (10). IL-23 functions as a stimulant of IL-17 production, and its role in inflammatory bowel disease has been well described (1, 16). The correlation between IL-23 and several bacterial families indicates that IL-23/IL-17 cytokine pathway may be an important mechanism behind intestinal inflammation in HE and cirrhosis.
Strengthening these correlations was the minimal effect that lactulose withdrawal had on the stool flora composition after 14 days; this replicates prior nonculture-based experience with lactulose in healthy individuals (39). We did not replicate prior culture-based studies in which lactulose therapy resulted in with higher lactobacillus or reduction in Escherichia coli and Staphyloccoci after symbiotic supplementation (23, 31). Our results are probably different because of the increased depth of the interrogation of the microbial community by MTPS rather than culture methodology. It is, however, possible that lactulose may act through change in bacterial functionality rather than change in abundances, which were measured in this study. These results suggest that these microbial abundances are reflective of HE and cirrhosis rather than just lactulose therapy.
The correlation-network model only indicates the coordinate changes in the features without implying causality, which is a limitation of this and all cross-sectional studies. The study is also limited by the small sample size; however, the correlations seen in the analysis were highly significant (P values <0.001). Network modeling indicates that robust analysis can be obtained with small sample sizes if the correlations are high as reflected in the high P values for the correlations in this study. In addition, age, socioeconomic status, medication use, cirrhosis complications, recent alcohol use, and dietary habits were similar between the groups. The current sample size did not allow for etiology-based differentiation in patients with cirrhosis, including inflammatory cytokines, and further research needs to be performed to elucidate those differences. We also could not study the effect of PPIs because most cirrhotic patients seen in our clinic are on PPI therapy, which could limit the generalizability of these results. This study was designed as a proof-of-concept experiment for generation of hypotheses for further definitive studies linking the cognitive impairment in cirrhosis with inflammation and the gut microbiome.
Collectively our data indicate that the gut microbiome components are significantly different between healthy controls and cirrhotic patients, especially those with HE, and are associated with cognition in cirrhosis. Additionally, markers of the Th17 and innate immune response were associated with Alcaligeneceae, Porphyromonadaceae, and Enterobacteriaceae in patients with HE. The IL-17/IL-23 pathway forms an important inflammatory link in this association. We anticipate that these findings could be a starting point for designing novel hypothesis-driven research and therapies such as targeted prebiotics and probiotics aimed at enhancing cognition through modulation of these microbiome components.
GRANTS
This work was partly supported by grant U01AT004428 from the National Center for Complementary and Alternative Medicine, grant RO1AA020203 from the National Institute on Alcohol Abuse and Alcoholism, the American College of Gastroenterology Junior Faculty Development Award, and the McGuire Research Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: J.S.B., D.M.H., and P.M.G. conception and design of research; J.S.B., P.B.H., L.R.T., S.S., M.S., and P.M.G. analyzed data; J.S.B., P.B.H., and L.R.T. interpreted results of experiments; J.S.B. prepared figures; J.S.B. and P.M.G. drafted manuscript; J.S.B., J.M.R., P.B.H., L.R.T., D.M.H., S.S., and P.M.G. edited and revised manuscript; J.S.B., P.B.H., and P.M.G. approved final version of manuscript; J.M.R., S.S., M.S., and P.M.G. performed experiments.
ACKNOWLEDGMENTS
We acknowledge Debulon Bell, R. N. and Melanie White for research coordination.
REFERENCES
- 1. Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, Powrie F. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity 33: 279– 288, 2010. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bajaj JS. Review article: the modern management of hepatic encephalopathy. Aliment Pharmacol Ther 31: 537– 547, 2010. . [DOI] [PubMed] [Google Scholar]
- 3. Bajaj JS, Hafeezullah M, Franco J, Varma RR, Hoffmann RG, Knox JF, Hischke D, Hammeke TA, Pinkerton SD, Saeian K. Inhibitory control test for the diagnosis of minimal hepatic encephalopathy. Gastroenterology 135: 1591– 1600, 2008. . [DOI] [PubMed] [Google Scholar]
- 4. Bajaj JS, Heuman DM, Wade JB, Gibson DP, Saeian K, Wegelin JA, Hafeezullah M, Bell DE, Sterling RK, Stravitz RT, Fuchs M, Luketic V, Sanyal AJ. Rifaximin improves driving simulator performance in a randomized trial of patients with minimal hepatic encephalopathy. Gastroenterology 140: 478– 487, 2011. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bajaj JS, Schubert CM, Heuman DM, Wade JB, Gibson DP, Topaz A, Saeian K, Hafeezullah M, Bell DE, Sterling RK, Stravitz RT, Luketic V, White MB, Sanyal AJ. Persistence of cognitive impairment after resolution of overt hepatic encephalopathy. Gastroenterology 138: 2332– 2340, 2010. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bajaj JS, Zadvornova Y, Heuman DM, Hafeezullah M, Hoffmann RG, Sanyal AJ, Saeian K. Association of proton pump inhibitor therapy with spontaneous bacterial peritonitis in cirrhotic patients with ascites. Am J Gastroenterol 104: 1130– 1134, 2009. . [DOI] [PubMed] [Google Scholar]
- 7. Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 54: 562– 572, 2011. . [DOI] [PubMed] [Google Scholar]
- 8. Cordoba J. New assessment of hepatic encephalopathy. J Hepatol 54: 1030– 1040, 2011. . [DOI] [PubMed] [Google Scholar]
- 9. Cryan JF, O'Mahony SM. The microbiome-gut-brain axis: from bowel to behavior. Neurogastroenterol Motil 23: 187– 192, 2011. . [DOI] [PubMed] [Google Scholar]
- 10. D'Elios MM, Del Prete G, Amedei A. Targeting IL-23 in human diseases. Expert Opin Ther Targets 14: 759– 774, 2010. . [DOI] [PubMed] [Google Scholar]
- 11. Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 12: 189– 198, 1975. . [DOI] [PubMed] [Google Scholar]
- 12. Gillevet P, Sikaroodi M, Keshavarzian A, Mutlu EA. Quantitative assessment of the human gut microbiome using multitag pyrosequencing. Chem Biodivers 7: 1065– 1075, 2010. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hamady M, Knight R. Tools, techniques, and challenges Microbial community profiling for human microbiome projects. Genome Res 19: 1141– 1152, 2009. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haussinger D, Schliess F. Pathogenetic mechanisms of hepatic encephalopathy. Gut 57: 1156– 1165, 2008. . [DOI] [PubMed] [Google Scholar]
- 15. Heijtz RD, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA 108: 3047– 3052, 2011. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huber S, Flavell RA. Checks and balances: IL-23 in the intestine. Immunity 33: 150– 152, 2010. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jin JJ, Kim HD, Maxwell JA, Li L, Fukuchi K. Toll-like receptor 4-dependent upregulation of cytokines in a transgenic mouse model of Alzheimer's disease (Abstract). J Neuroinflammation 5: 23, 2008. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut 60: 631– 637, 2011. . [DOI] [PubMed] [Google Scholar]
- 19. Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M, McSweeney CS. Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis 16: 2034– 2042, 2010. . [DOI] [PubMed] [Google Scholar]
- 20. Keshavarzian A, Holmes EW, Patel M, Iber F, Fields JZ, Pethkar S. Leaky gut in alcoholic cirrhosis: a possible mechanism for alcohol-induced liver damage. Am J Gastroenterol 94: 200– 207, 1999. . [DOI] [PubMed] [Google Scholar]
- 21. Komanduri S, Gillevet PM, Sikaroodi M, Mutlu E, Keshavarzian A. Dysbiosis in pouchitis: evidence of unique microfloral patterns in pouch inflammation. Clin Gastroenterol Hepatol 5: 352– 360, 2007. . [DOI] [PubMed] [Google Scholar]
- 22. Lane DJ. 16s/23s rRNA sequencing. In: Nucleic Acid Techniques in Bacterial Systematics, edited by Goodfellow M. West Sussex, England: John Wiley & Sons, 1991, p. 115– 175 . [Google Scholar]
- 23. Liu Q, Duan ZP, Ha DK, Bengmark S, Kurtovic J, Riordan SM. Synbiotic modulation of gut flora: effect on minimal hepatic encephalopathy in patients with cirrhosis. Hepatology 39: 1441– 1449, 2004. . [DOI] [PubMed] [Google Scholar]
- 24. Marra F, Annunziato F. Immunomodulation: a new approach to the therapy of cirrhosis? Gut 59: 868– 869, 2010. . [DOI] [PubMed] [Google Scholar]
- 25. Mondot S, Kang S, Furet JP, Aguirre de Carcer D, McSweeney C, Morrison M, Marteau P, Dore J, Leclerc M. Highlighting new phylogenetic specificities of Crohn's disease microbiota. Inflamm Bowel Dis 17: 185– 192, 2011. . [DOI] [PubMed] [Google Scholar]
- 26. Mutlu E, Keshavarzian A, Engen P, Forsyth CB, Sikaroodi M, Gillevet P. Intestinal dysbiosis: a possible mechanism of alcohol-induced endotoxemia and alcoholic steatohepatitis in rats. Alcohol Clin Exp Res 33: 1836– 1846, 2009. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Naqvi A, Rangwala H, Keshavarzian A, Gillevet P. Network-based modeling of the human gut microbiome. Chem Biodivers 7: 1040– 1050, 2010. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Obata T, Goto Y, Kunisawa J, Sato S, Sakamoto M, Setoyama H, Matsuki T, Nonaka K, Shibata N, Gohda M, Kagiyama Y, Nochi T, Yuki Y, Fukuyama Y, Mukai A, Shinzaki S, Fujihashi K, Sasakawa C, Iijima H, Goto M, Umesaki Y, Benno Y, Kiyono H. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc Natl Acad Sci USA 107: 7419– 7424, 2010. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ridlon JM, McGarr SE, Hylemon PB. Development of methods for the detection and quantification of 7alpha-dehydroxylating clostridia, Desulfovibrio vulgaris, Methanobrevibacter smithii, and Lactobacillus plantarum in human feces. Clin Chim Acta 357: 55– 64, 2005. . [DOI] [PubMed] [Google Scholar]
- 30. Riggio O, Ridola L, Pasquale C, Nardelli S, Pentassuglio I, Moscucci F, Merli M. Evidence of persistent cognitive impairment after resolution of overt hepatic encephalopathy. Clin Gastroenterol Hepatol 9: 181– 183, 2011. . [DOI] [PubMed] [Google Scholar]
- 31. Riggio O, Varriale M, Testore GP, Di Rosa R, Di Rosa E, Merli M, Romiti A, Candiani C, Capocaccia L. Effect of lactitol and lactulose administration on the fecal flora in cirrhotic patients. J Clin Gastroenterol 12: 433– 436, 1990. . [DOI] [PubMed] [Google Scholar]
- 32. Romero Gomez M, Cordoba J, Jover R, del Olmo J, Fernandez A, Flavia M, Company L, Poveda MJ, Felipo V. Normality tables in the Spanish population for psychometric tests used in the diagnosis of minimal hepatic encephalopathy. Med Clin (Barc) 127: 246– 249, 2006. . [DOI] [PubMed] [Google Scholar]
- 33. Savarino V, Mela GS, Zentilin P, Mansi C, Mele MR, Vigneri S, Cutela P, Vassallo A, Dallorto E, Celle G. Evaluation of 24-hour gastric acidity in patients with hepatic cirrhosis. J Hepatol 25: 152– 157, 1996. . [DOI] [PubMed] [Google Scholar]
- 34. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498– 2504, 2003. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shawcross DL, Sharifi Y, Canavan JB, Yeoman AD, Abeles RD, Taylor NJ, Auzinger G, Bernal W, Wendon JA. Infection and systemic inflammation, not ammonia, are associated with Grade 3/4 hepatic encephalopathy, but not mortality in cirrhosis. J Hepatol 54: 640– 649, 2011. . [DOI] [PubMed] [Google Scholar]
- 36. Shawcross DL, Wright G, Olde Damink SW, Jalan R. Role of ammonia and inflammation in minimal hepatic encephalopathy. Metab Brain Dis 22: 125– 138, 2007. . [DOI] [PubMed] [Google Scholar]
- 37. Tandon P, Garcia-Tsao G. Bacterial infections, sepsis, and multiorgan failure in cirrhosis. Semin Liver Dis 28: 26– 42, 2008. . [DOI] [PubMed] [Google Scholar]
- 38. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature 457: 480– 484, 2009. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vanhoutte T, De Preter V, De Brandt E, Verbeke K, Swings J, Huys G. Molecular monitoring of the fecal microbiota of healthy human subjects during administration of lactulose and Saccharomyces boulardii. Appl Environ Microbiol 72: 5990– 5997, 2006. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vlahcevic ZR, Buhac I, Bell CC, Jr, Swell L. Abnormal metabolism of secondary bile acids in patients with cirrhosis. Gut 11: 420– 422, 1970. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weissenborn K, Ennen JC, Schomerus H, Ruckert N, Hecker H. Neuropsychological characterization of hepatic encephalopathy. J Hepatol 34: 768– 773, 2001. . [DOI] [PubMed] [Google Scholar]
- 42. White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5: e1000352, 2009. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wong F, Bernardi M, Balk R, Christman B, Moreau R, Garcia-Tsao G, Patch D, Soriano G, Hoefs J, Navasa M. Sepsis in cirrhosis: report on the 7th meeting of the International Ascites Club. Gut 54: 718– 725, 2005. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wright G, Jalan R. Ammonia and inflammation in the pathogenesis of hepatic encephalopathy: Pandora's box? Hepatology 46: 291– 294, 2007. . [DOI] [PubMed] [Google Scholar]
- 45. Wu S, Yin R, Ernest R, Li Y, Zhelyabovska O, Luo J, Yang Y, Yang Q. Liver X receptors are negative regulators of cardiac hypertrophy via suppressing NF-kappaB signalling. Cardiovasc Res 84: 119– 126, 2009. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zoetendal EG, Ben-Amor K, Harmsen HJ, Schut F, Akkermans AD, de Vos WM. Quantification of uncultured Ruminococcus obeum-like bacteria in human fecal samples by fluorescent in situ hybridization and flow cytometry using 16S rRNA-targeted probes. Appl Environ Microbiol 68: 4225– 4232, 2002. . [DOI] [PMC free article] [PubMed] [Google Scholar]