Abstract
Sox9 is an high-mobility group box transcription factor that is expressed in the stem cell zone of the small intestine and colon. We have previously used a Sox9EGFP mouse model to demonstrate that discrete levels of Sox9 expression mark small intestine epithelial stem cells that form crypt/villus-like structures in a three-dimensional culture system (Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Am J Physiol Gastrointest Liver Physiol 296: G1108–G1118, 2009; Gracz AD, Ramalingam S, Magness ST. Am J Physiol Gastrointest Liver Physiol 298: G590–G600, 2010). In the present study, we hypothesized that discrete levels of Sox9 expression would also mark colonic epithelial stem cells (CESCs). Using the Sox9EGFP mouse model, we show that lower levels of Sox9 mark cells in the transit-amplifying progenitor cell zone, while higher levels of Sox9 mark cells in the colonic crypt base. Furthermore, we demonstrate that variable SOX9 levels persist in cells of colonic adenomas from mice and humans. Cells expressing lower Sox9 levels demonstrate gene expression profiles consistent with more differentiated populations, and cells expressing higher Sox9 levels are consistent with less differentiated populations. When placed in culture, cells expressing the highest levels of Sox9 formed “colonoids,” which are defined as bodies of cultured colonic epithelial cells that possess multiple cryptlike structures and a pseudolumen. Cells expressing the highest levels of Sox9 also demonstrate multipotency and self-renewal in vitro, indicating functional stemness. These data suggest a dose-dependent role for Sox9 in normal CESCs and cells comprising colon tumors. Furthermore, distinct Sox9 levels represent a new biomarker to study CESC and progenitor biology in physiological and disease states.
Keywords: SOX factors, colon epithelium culture, stem cell
for decades, the general region of the intestinal epithelial stem cell (IESC) niche in the small intestine and colon has been recognized to exist in the crypts (5, 31); however, the lack of specific biomarkers that can be used to identify and isolate IESCs has been a challenge to the study of IESCs during physiological states, disease, and injury. Recent studies have identified genes that exhibit restricted expression in the IESC zones of the small intestine, suggesting that they might serve as stem cell biomarkers (12, 26, 37, 39, 41). In vivo genetic lineage tracing studies in mice have demonstrated that the G protein-coupled receptor Lgr5, cell surface marker CD133, polycomb ring finger oncogene Bmi1, and telomerase (Tert) mark a population of IESCs in the crypt base of the small intestine. Additionally, the cell surface marker CD24 has been shown to be useful for fluorescent-activated cell sorting (FACS) enrichment of a small intestine IESC population that possesses stem cell characteristics, which are defined by multipotency and self-renewal capacity in vitro (15, 44). Other studies in the small intestine have correlated gene expression patterns of Ascl2, Olfm4, Musashi1, and Dcamkl1 to IESC zone (17, 22, 30, 42, 43); however, it is still unclear whether these genes mark cells that possess functional stem cell properties based on in vivo lineage tracing or in vitro culture of FACS isolated cells.
Our laboratory has previously demonstrated that distinct Sox9 expression levels are useful biomarkers in the identification and isolation of different small intestine epithelial cell lineages, including IESCs (15). Using a Sox9EGFP mouse model in which enhanced green fluorescent protein (EGFP) is driven by large regulatory regions of the Sox9 5′-flanking regions, we demonstrated that “high” Sox9EGFP levels mark chromogranin A-positive cells, which are consistent with enteroendocrine lineages; “low” levels mark IESCs, and “sublow” levels mark transit-amplifying progenitor cells (10, 15). Furthermore, we demonstrated functional stem cell characteristics of Sox9EGFPlow cells by introducing these cells into a culture system that facilitates formation of crypt/villus-like structures from a single cell (15). Subsequently, genetic lineage tracing has shown Sox9 to mark multipotent IESCs in vivo (13).
While considerable efforts have been made to define the genetic signature of small intestine epithelial cells (2, 9, 10, 15, 17, 18, 25, 26, 30, 37, 41–44), fewer studies have focused on identifying genetic biomarkers of colonic epithelial stem cells (CESCs) (11, 13, 36, 39). The concept of two populations of CESCs has been postulated: one is a slowly dividing label-retaining population, and the other is an actively dividing high-turnover population (19). Two genetic lineage-tracing studies describe Lgr5 and Sox9 as biomarkers of small IESCs and CESCs (13, 39). Two other candidate biomarkers, β1-integrin and Phlda1 (Pleckstrin homology-like domain family A member 1), localize to the stem cell zone in the colon (11, 36). Phlda1 demonstrates a restricted expression pattern to the base of the crypt and appears to be consistent with Lgr5-expressing cells (36). FACS-isolated β1-integrin cells demonstrated the ability to form colonies in culture; however, at the time of this study, culture conditions did not permit testing of whether the β1-integrin cells were multipotent and self-renewing (11). It is currently unknown whether any of these putative or established stem cell biomarkers demonstrate overlapping expression patterns with a label-retaining or high-turnover CESC population.
SOX9 exhibits a broader crypt expression pattern in the colonic crypts compared with the crypts of the small intestines (4, 10); however, like the small intestine, different levels of Sox9EGFP (and SOX9) are regionally restricted to the progenitor and stem cell zones. Therefore, we hypothesized that distinct levels of Sox9 could serve as useful biomarkers for CESCs and progenitor populations in the colon. This study describes the cellular and molecular characterization of CESCs that express Sox9. We quantify different levels of Sox9 in single cells of the colonic crypt base and in cells of colon tumors from both mice and humans using confocal microscopy and computational image analysis. We assess the stemness of colonic epithelial cells exhibiting different Sox9 expression levels. Additionally, we define culture conditions that support colonoid formation from single CESCs. These studies provide a strong framework for identification of other novel markers of CESCs and facilitate investigation into the behavior of CESCs in disease models of the colon.
EXPERIMENTAL PROCEDURES
Mice/genotyping.
The Sox9EGFP mouse line was originally generated as part of the GENSAT Brain Atlas Project (14) and contains genomic integration of a modified bacterial artificial chromosome (BAC) (RP32–140D18) with ∼75.5-kb upstream and ∼151-kb downstream sequence to Sox9. Frozen Sox9EGFP mouse embryos were obtained from the Mutant Mouse Regional Resource Center (University of California-Davis) and reconstituted by transfer into foster mice. All mice are on the outbred CD-1 strain and were maintained as heterozygotes on the CD-1 genetic background. Mice breed normally and live to adulthood with no overt phenotypes due to the transgene. At ∼10 days postnatal, tail snips were viewed under an epifluorescent microscope fitted with filters for EGFP visualization. A high level of EGFP fluorescence compared with transgene negative control mice was scored as positive for the Sox9EGFP transgene.
Epithelial cell dissociation/FACS.
The distal colons from three freshly killed 6- to 8-wk-old Sox9EGFP mice were removed, flushed of luminal contents with ice-cold Hanks' balanced salt solution (HBSS), and incubated at 37°C for 1 h in 5 ml of colonic epithelium dissociation medium I [CEDM I; Dulbecco's modified eagle medium-high glucose (DMEM-H) containing 1 U/ml penicillin, 1 μg/ml streptomycin, 2.9 μg/ml l-glutamine, 1% fetal bovine serum (FBS), 5 mM calcium chloride, 150 U/ml collagenase XI (Sigma, St. Louis, MO), and 0.03 U/ml dispase (BD Biosciences, Franklin Lakes, NJ)]. The tissues were washed once in 50 ml DMEM-H, and once in 50 ml PBS, and then transferred to CEDM II [CEDM II: PBS containing 60 mM EDTA, 3 mM dithiothreitol, 10 μM anoikis inhibitor Y27632 (Sigma, St. Louis, MO)], and incubated on ice for 20 min. Tissues were removed and transferred to CEDM III (CEDM III: PBS containing 60 mM EDTA, and 10 μM Y27632) for 10 min at 37°C. Next, the tissue was shaken vigorously for 1 min in CEDM III. The remnant tissue (muscularis) was then removed from CEDM III, and dissociated crypts were pelleted at 820 g at 4°C for 5 min. The crypts were washed in 10 ml ice-cold Dulbecco's phosphate-buffered saline (GIBCO), pelleted at 1,280 g, and incubated at 37°C for 10 min in CEDM IV [CEDM IV: HBSS (GIBCO) containing 1.35 U/ml dispase and 10 μM Y27632]. Crypts were shaken vigorously every 2 min to further dissociation into single cells. FBS was then added to 10% (vol/vol), and the suspension was sequentially filtered through a 70-μm filter and then a 40-μm filter. The final single-cell suspension was pelleted at 1,280 g at 4°C for 5 min and washed thrice in 10 ml of ice-cold DMEM-H containing 10% FBS, 1 U/ml penicillin, 1 μg/ml streptomycin, 2.9 μg/ml l-glutamine, and 10 μM Y27632. Between each wash step, the cells were pelleted at 1,280 g for 5 min at 4°C. The cell pellet was resuspended in 2 ml of colonoid culture medium [CCM; advanced DMEM-F12, N2, B27 supplements, 1 U/ml penicillin, 1 μg/ml streptomycin, 2.9 μg/ml l-glutamine (GIBCO), 10 μM HEPES buffer (Invitrogen), supplemented with 10 μM Y27632, and 25 μg/ml DNase I before FACS].
Sox9EGFP forward-side scatter gating was used to exclude 99.8% of all dead cells and lymphocytes. Both forward scatter and side scatter height-area plots were used for doublet discrimination to maximize efficient single-cell sorting. Sort gates for populations 1, 2, and 3 were based on the natural peaks that were present on the univariate EGFP histogram. Cells were collected in ice-cold CCM supplemented with 10 μM Y27632 (Sigma, Saint Louis, MO) and kept on ice throughout the sort.
cDNA preparation/real-time PCR analysis.
Total RNA was isolated from 0.25–0.5 × 104 cells (populations 1, 2, and 3) using RNAqueous Micro Kit 9 (Ambion, Austin, TX), according to the manufacturer's protocols. cDNA was generated using High Capacity cDNA kit (Applied Biosystems, Pleasanton, CA). Time from tissue harvest to RNA extraction was kept in between 3.5–4 h to control the quality of RNA. Real-time PCR was conducted for each sample in triplicate on ∼1/250 of the total amount of cDNA generated. Taqman probes for each gene (18S, HS99999901; Atoh1, Mm00476035_s1; ChgA Mm00514341_m1; Lgr5, Mm00438890_m1; Notch1, Mm00435245_m1; Sox9, Mm00448840_m1; Tff3, Mm00495590_m1) were used in real-time PCR reactions, according to the manufacturer's protocols. 18S ribosomal RNA was amplified and used as the internal control gene for sample comparison. ΔΔCT values were calculated to obtain fold changes for sample comparison (29). Due to mosaic Sox9EGFP expression that is unique to the colonic epithelium, we normalized fold changes to population 1, which exhibited the lowest levels of Sox9EGFP expression. All data points represent means ± SE from three separate experiments; statistical analysis was conducted by one-way ANOVA and post hoc Tukey-Kramer pairwise tests. A P value of <0.05 was considered statistically significant.
CESC culturing/intestinal myofibroblasts isolation.
E16.5 embryonic intestinal myofibroblasts (MFs) were isolated from a transgenic mouse model in which the red fluorescent reporter gene, dsRED is driven by the smooth muscle actin (SMA) promoter (21). A CD-1 female was time-mated with a SMA-DsRED male mouse. Embryos were collected at 16.5 days postcoitus and phenotyped for expression of SMAdsRed by visualization under fluorescent microscopy. Intestines were dissected from SMAdsRed-positive embryos with the aid of a dissecting microscope and rinsed in sterile, ice-cold PBS to remove any adherent nonintestinal tissues. Special care was taken to assure no embryonic liver remained in the cell prep. Tissue was minced into ∼1-mm2 pieces and transferred to a 50-ml conical containing 5 ml DMEM-H (GIBCO) with 0.3 U/ml dispase (Collaborative Biomedical) and 300 U/ml collagenase I (Sigma). The tissue slurry was rotated for 30 min at room temperature before 10 ml of DMEM-H +10% FBS (Gemini) was added. The slurry was then pipetted vigorously 50 times to mechanically dissociate any remaining clumps of cells. The cells were centrifuged for 5 min at 460 g, resuspended in DMEM-H with 10% FBS, and plated in a 10-cm culture dish. Cells underwent three passages to obtain a pure population of dsRED-positive MFs before use in colonoid culture. Flow cytometry demonstrated that >98% of cells were dsRED-positive.
Ten thousand sorted cells from each gate (1, 2, or 3) were individually pelleted at 3,000 g for 5 min, the media was carefully removed, and 3 μl of media were added to the cell pellet, which was resuspended by pipetting. The cells were added to 50 μl of ice-cold growth factor reduced Matrigel (BD, Franklin Lakes, NJ) supplemented with 1 μg/ml of mR-Spondin1 (R&D Systems, Minneapolis, MN), 1 μM Jagged-1 (AnaSpec, San Jose, CA), 100 ng/ml Noggin (Peprotech, Rocky Hill, NJ), 80 ng/ml Wnt3a (R&D Systems, Minneapolis, MN), and 50 ng/ml epidermal growth factor (EGF) (R&D Systems) and thoroughly mixed by pipetting. The cell/Matrigel mixture was plated on a Transwell insert designed for a 24-well plate format. The Matrigel was allowed to polymerize for 30 min at 37°C. The Transwells were placed into the 24-well plate, which contained E18.5 MFs that were 30–50% confluent. Two-hundred and fifty microliters of CCM with 10 μM Y27632 were added to the well containing the MFs, and 200 μl of CCM were added to the Transwell insert (500-μl total volume). Growth factors were replenished every 2 days by diluting the factors (R-spondin1, Wnt3a, Noggin, EGF) in a volume of growth medium, such that 10 μl represented a 50× solution of concentrations described above. Growth medium containing R-spondin, Wnt3a, Noggin, and EGF at 1× concentrations was completely changed every 4 days.
For passaging experiments, colonoids were isolated from Matrigel with a pipette and transferred to advanced DMEM/F12 with 10 μM Y27632 in 2.0 ml conical tubes. Single cells were obtained by addition of dispase (0.3 U/ml), incubation for 20 min at 37°C, and combined with trituration 20× through a 10-ml pipette connected to a mechanical pipettor. If cells were not dissociated to satisfaction, another 20 triturations were performed. Cells were pelleted and washed twice in advanced DMEM/F12, containing 10% FBS and 10 μM Y27632. Cells were washed once in advanced DMEM/F12, pelleted at 3,000 g, and replated in Matrigel, as described above.
Immunostaining/microscopy.
For tissue preparation, small intestines were dissected from adult Sox9EGFP mice (>8 wk of age), and luminal contents were flushed out with PBS, followed immediately by a single flush with freshly prepared 4% paraformaldehyde. The colon was opened along the proximal-distal axis and fixed for an additional 14–18 h at 4°C. The tissues were then prepared for cryosectioning by immersion in 30% sucrose solution for at least 24 h at 4°C. Tissues were then embedded in optimal cutting temperature (OCT) medium and frozen on dry ice. The OCT blocks were stored at −80°C until cryosectioning. Thin sections (8–10 μm) were cut on a cryostat and placed on positively charged microscope slides for staining and microscopy. This tissue preparation technique is critical for preserving the EGFP fluorescence. Human colon tumor samples were obtained from US Biomax (Rockville, MD). Twenty-four cases were contained on a tissue microarray (TMA; serial no. CO482). Paraffin was removed from the TMA using Histoclear, and immunostaining was conducted as described for mouse tissues.
For immunostaining, the sections were washed twice in PBS to remove OCT, followed by incubation in blocking medium (5% normal goat or donkey serum), in PBS-0.3% Triton X-100 for at least 30 min at room temperature (21–25°C). Primary antibodies were applied to the tissue sections in antibody-staining solution (1% normal goat serum, in PBS-0.3% Triton X-100). Dilutions were as follows: α-lysozyme (rabbit, 1:1,000, Diagnostics Biosystems, Pleasanton, CA, RP 028), α-mucin 2 (rabbit; 1:100, Santa Cruz Biotechnology, Santa Cruz, CA, sc-15334), α-SOX9 (rabbit, 1:1,000, Chemicon, Temecula, CA, AB5535), α-substance P (rat; 1:500, Chemicon, MAB356), α-chromogranin A (rabbit; 1:500, Immunostar, 20086), and α-sucrase isomaltase (goat; 1:100, Santa Cruz Biotechnology, sc-27603). All secondary antibodies (α-rabbit-Cy3, Sigma, St. Louis, MO, C2306; α-rabbit-Alexa fluor 488, Molecular Probes, Eugene, OR, Z-25302; α-rat-Alexa fluor 555, Molecular Probes, Z-25305; α-goat-Cy3) were used at a 1:500 dilution in staining buffer. Ki67 immunostaining required special conditions. Tissue sections were cut as described above and were antigen retrieved using Reveal Buffer (Biocare Medical) for 10 s at 120°C in a pressure cooker. Slides were allowed to cool at room temperature for 20 min and washed 3× in Tris buffer (TB; 0.05 M Tris·HCl; pH 7.6) for 2 min each. Ki67 (DAKO) was diluted 1:100 in DAKO diluent (DAKO) and incubated in a humidified chamber for 2 h at room temperature. The Ki67 antibody solution was removed, and the tissues were washed 3× in TB for 2 min each. Biotinylated anti-rat-IgG was diluted 1:100 in DAKO diluent and applied to the tissue sections for 1 h at room temperature in a humidified chamber. The secondary antibody was removed, and the tissue sections were washed 3× in TB, as described above. For Ki67 detection, Alexa-fluor 555 streptavidin was diluted 1:250 in DAKO diluent and incubated with tissues for 1 h at room temperature in a humidified chamber. The slides were then washed 3×, as described above, and mounted using aqueous mounting medium. All nuclei were stained with Draq5 (1:10,000, Biostatus, San Diego, CA, BOS-889–001). Background staining was negligible, as determined by nonspecific IgG staining.
Day-to-day development of colonoids was tracked using light microscopy. Epifluorescence images were captured on an Olympus IX70 fitted with an Olympus digital camera. Objective lenses used were ×20 and ×40, with numerical apertures of 0.55 and 1.40, respectively. All confocal images represent ×200 (1.0-μm optical sections), ×400 (1.0-μm optical sections), or ×630 (1.0-μm optical sections). Objective lenses for the confocal images had a numerical aperture of 0.1.
Image analysis.
Images for quantifying individual cell fluorescence intensities were obtained using a Zeiss LSM 510 confocal microscope at ×400 optical zoom with a 1.0-μm optical section. The red, green, and blue images were stacked in the image analysis program, FIJI (NIH ImageJ). To measure the level of SOX9 immunostaining in individual cells, an oval was superimposed on the nucleus (blue channel), and the “measure” feature was used to sequentially determine the average fluorescence intensity (or pixel intensity) of the red and green wavelengths. The base of the crypt was defined as cell position 1, increasing to the right and decreasing to the left. Measurement data were then transferred into Microsoft Excel for statistical analysis. For each image, average fluorescence intensities were loaded into the analysis application “R” (R Foundation for Statistical Computing, Vienna, Austria), and the kernel density estimate was computed and plotted using the “kernel” function with Gaussian kernel and bandwidth set to 4.
RESULTS
Sox9 expression levels vary between cells in the normal colonic epithelium.
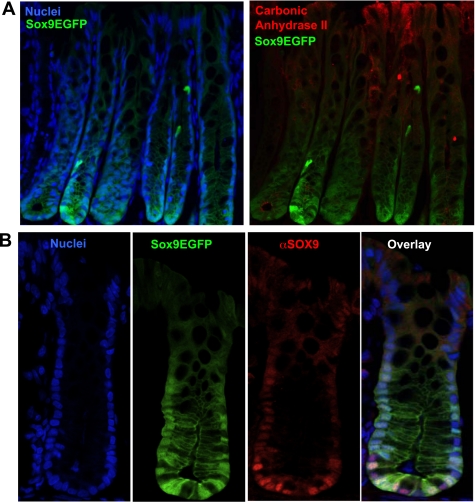
Sox9 is expressed at different levels in the small intestine epithelium, facilitating the histological identification and FACS isolation of IESCs (10, 15). In the colon, Sox9EGFP is primarily expressed in the lower one-half of the crypt and is regionally separated from the more differentiated absorptive colonocytes, which express carbonic anhydrase II (Fig. 1A). To determine whether Sox9 was expressed at different levels in the colonic epithelium, we used a Sox9EGFP reporter gene mouse or immunostaining for endogenous SOX9 to visualize Sox9 expression (Fig. 1B). The data qualitatively suggest that Sox9EGFP (and SOX9) are expressed at variable levels, with higher Sox9EGFP expression in the crypt base and lower Sox9EGFP expression in cells positioned closer to the luminal surface (Fig. 1A).
Fig. 1.
Variations in Sox9EGFP/SOX9 expression levels in the base of colonic crypts. A, left: a gradient of Sox9EGFP and SOX9 cellular expression exists in colonic crypts. Increased Sox9EGFP and SOX9 levels localize to the crypt base, whereas lower levels localize toward the lumen. Right: expression of Sox9EGFP is exclusive of the absorptive colonocyte biomarker, carbonic anhydrase II (CAII). Images are ×200 original magnification. B: SOX9 and Sox9EGFP are expressed at different levels in the crypt base, and the Sox9EGFP transgene recapitulates endogenous SOX9 expression. Images are ×630 original magnification.
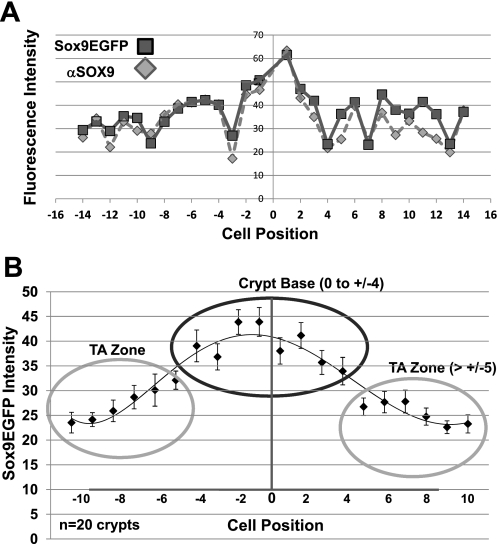
To quantitatively assess whether cells in the crypt base expressed higher levels of Sox9EGFP and SOX9, image analysis software was used to analyze 1.0-μm confocal optical sections of Sox9EGFP colon that had been stained for endogenous SOX9 (Fig. 2). The nuclei of individual cells in the images were digitally marked (independent of Sox9 expression), and then Sox9EGFP and endogenous SOX9 levels in the marked nuclear regions of corresponding images were quantified. Comparisons were conducted on a cell-by-cell basis using the conventional crypt cell position method, where position “zero” is at the base of the crypt, cells to the left of zero are sequentially designated by negative numbers, and cells to the right of zero are sequentially designated as positive numbers. Single crypt analysis shows that endogenous SOX9 levels trend with Sox9EGFP levels both at the single cell level and by location (Fig. 2A). Analyses of 20 crypts show that higher levels of Sox9EGFP and SOX9 are expressed toward the base of the crypt, and lower levels are expressed toward the luminal surface (Fig. 2B). The nonparametric Wilcoxon rank sum test supports the hypothesis that the distribution of Sox9EGFP intensities is significantly higher in the crypt base from cell positions −4 to +4. Together, these data indicate that Sox9EGFP/SOX9 expression in colonic crypt cells is indeed variable, with higher levels in the crypt base cells (from positions −4 to +4) and lower levels in cells toward the lumen (positions higher than −4/+4), suggesting that different Sox9 levels could serve as useful biomarkers for distinct and localized cell lineages.
Fig. 2.
Confocal and computational image analysis demonstrates higher Sox9EGFP/SOX9 expression levels in the colonic crypt base. A: single-cell image analysis. Sox9EGFP and SOX9 immunofluorescence intensities were measured on a cell-by-cell basis and plotted as a function of cell position. Spearman rank correlation test indicated Sox9EGFP intensities correlated with endogenous SOX9 fluorescence intensities by a significant dependence between endogenous SOX9 and enhanced green fluorescent protein (EGFP) intensity distributions (rs = 0.69 with P < 2.2 × 10−16). B: Sox9EGFP levels form distinct subgroups. Fluorescence intensities were averaged across 20 crypts for each cell position. When organized by location into two subgroups at the crypt base (positions −4 to +4; solid circle) and higher (positions +/−5 to +/−10; shaded circle), the distribution of Sox9EGFP intensities was significantly higher in the crypt base group compared with the TA (transit amplifying) zone group (Wilcoxon rank sum test, P = 1.214 × 10−6 for EGFP, P = 3.482 × 10−8 for SOX9).
Colonic tumor cells exhibit variable SOX9- or Sox9EGFP-expression levels.
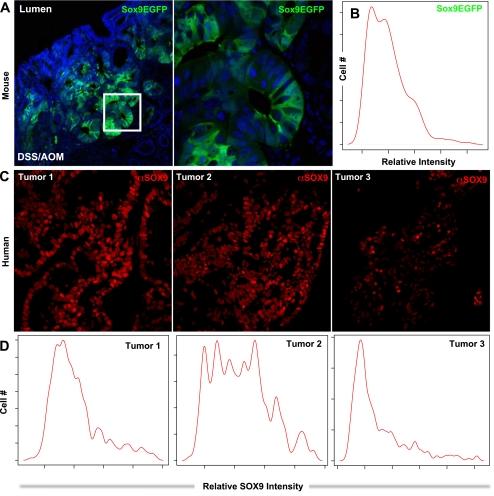
SOX9 is expressed in colonic adenomas in mice and humans (3, 4, 20, 27, 33); however, Sox9 expression levels in individual cells have not been examined. To determine whether differential expression of Sox9EGFP persisted in murine adenomas, we treated Sox9EGFP mice with azoxymethane (AOM)/dextran sodium sulfate (DSS) and examined the levels of Sox9EGFP in developed tumors. Confocal images of adenomas visually show positive Sox9EGFP expression, while closer examination demonstrates differential Sox9EGFP expression levels in cells of the tumor (Fig. 3A). Statistical kernel density estimation of the distribution of Sox9EGFP is multimodal, indicating multiple levels of expression exist (Fig. 3B). To determine whether differential levels of SOX9 were expressed in primary human tumors, we assessed SOX9 expression levels in individual cells from TMAs containing primary tumor biopsies (Fig. 3C). Again, statistical kernel density estimation of the distribution of SOX9 expression is multimodal, indicating all three human tumors show multiple, differential levels of expression (Fig. 3D).
Fig. 3.
Differential Sox9EGFP/SOX9 expression levels persist in cells of azoxymethane (AOM)/dextran sodium sulfate (DSS) tumors in mice and in cells of primary human colon tumors. A: variable Sox9EGFP expression levels are preserved in AOM/DSS adenomas. Region depicted in white box is magnified in right panel. Images represent 1-μm optical sections imaged at ×200 (left) and ×400 (right) original magnification. B: Sox9EGFP fluorescence was quantified at the single-cell level using the image analysis strategy described in Fig. 2. Multiple peaks in the histogram indicate a multimodal distribution of different Sox9EGFP expression levels. C: three primary colon tumors from separate individuals were immunostained for SOX9. The data visually demonstrate variable levels of SOX9 in tumor cells. D: single-cell quantification of SOX9 expression in colon tumors demonstrates multimodal SOX9 expression and also different ratios of lower and higher SOX9-expressing cells between tumors from different individuals.
Sox9 is expressed in undifferentiated proliferating cells and a subset of mucin 2 expressing cells.
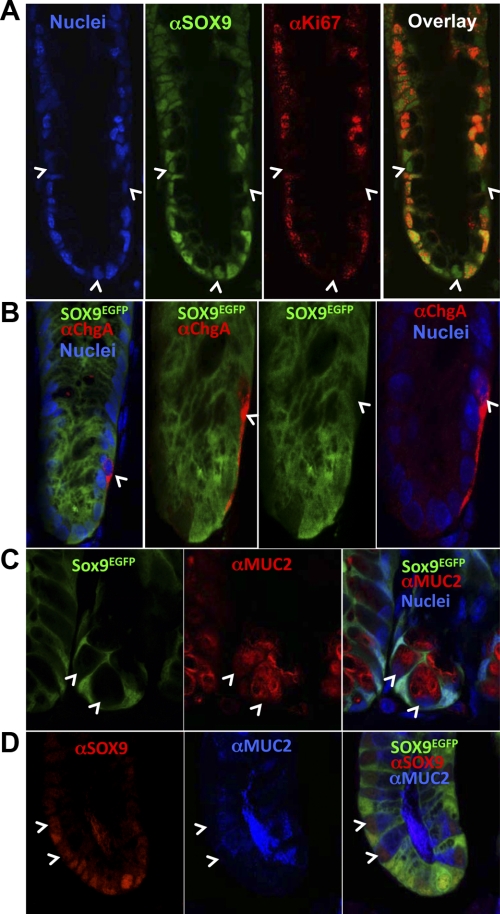
Like most SOX9-expressing cells in the small intestine crypts, a majority of the SOX9-expressing cells in colonic crypts also express the Ki67 proliferation marker (Fig. 4A). We identified a minority subset of SOX9-expressing cells that did not colocalize with the Ki67 proliferation marker (Fig. 4A; white arrows). The basal nuclei and large mucous globules in the cytoplasm suggest these nonproliferating cells are goblet cells (6). To determine the lineage identity of Sox9EGFP-expressing cells, we stained Sox9EGFP colonic tissue for goblet and enteroendocrine cell biomarkers. Immunostaining demonstrates that enteroendocrine cells (chromogranin A+; Fig. 4B) and goblet cells (MUC2+; Fig. 4C) do not express detectable levels of Sox9EGFP. Although the absence of endogenous SOX9 expression is consistent with the absence of Sox9EGFP transgene in enteroendocrine cells (data not shown), a subset of MUC2+/Sox9EGFP-goblet cells do express endogenous SOX9 (Fig. 4D). Thus, in the colonic epithelium, SOX9 is expressed in CESCs/progenitors and a subset of goblet cells. Despite the inconsistent expression pattern of the Sox9EGFP transgene and endogenous SOX9 in the crypt-based MUC2+ populations, the transgene faithfully recapitulates the endogenous gene in the proliferating stem/progenitor populations. The lack of Sox9EGFP expression in the crypt-based MUC2+ cells was exploited as a method to FACS exclude MUC2+ cells from putative CESC populations.
Fig. 4.
Sox9EGFP is expressed in proliferating cells and a subset of mucin 2 (MUC2+) cells. A: SOX9 colocalizes with proliferative marker Ki67, except in select cells closer to the crypt base (white arrows). B: Sox9EGFP is not expressed in chromogranin A+ (ChgA) cells. C: Sox9EGFP−/MUC2+ cells are intercalated between Sox9EGFP+/MUC2− cells (white arrows). D: crypt-based Sox9EGFP−/MUC2+ cells express SOX9 (red nuclei). Images represent 1-μm optical sections imaged at ×400 (A and D) and ×1,260 (B and C) original magnification.
Increasing expression levels of Sox9 in colonic epithelial cells correlate with increased stemness.
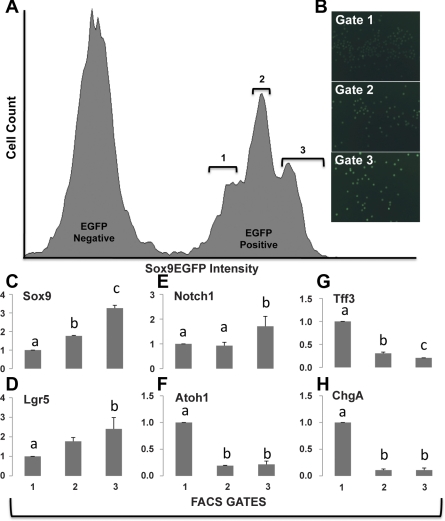
In the small intestine epithelium, Sox9EGFP is expressed at different levels in the base of the crypts, and distinct levels of EGFP fluorescence facilitate differential identification and FACS isolation of IESCs from transit amplifying progenitors and enteroendocrine cells (15). To assess whether different levels of Sox9EGFP expression observed in the colonic epithelium could be used in a similar manner to isolate CESCs, flow cytometry was used to analyze the Sox9EGFP levels in dissociated colonic epithelial cells (Fig. 5). The results show three different Sox9EGFP-expressing populations, which are defined by three distinct EGFP peak intensities (Fig. 5A). “Population 1” was defined by cells expressing the lowest levels of Sox9EGFP, “population 2” by intermediate Sox9EGFP levels, and “population 3” by cells expressing the highest levels of Sox9EGFP.
Fig. 5.
Three different Sox9EGFP-expressing populations can be identified by flow cytometery, and higher levels of Sox9EGFP correlate with increased stemness. A: univariate flow cytometry histogram demonstrates a trimodal distribution of Sox9EGFP intensity. B: postsort analysis of cells sorted using gates 1, 2, and 3 validate fluorescent-activated cell sorting (FACS) based on increasing EGFP intensity. Images are ×200 original magnification with identical exposure time and gain. Semiquantitative RT-PCR conducted on populations 1, 2, and 3 showed significant enrichment of Sox9 in population 3 (C), Lgr5, a known colonic epithelial stem cells (CESC) marker, in population 3 (D), and Notch1 in population 3 (E), whereas Atoh1, a marker associated with secretory progenitors, was enriched in population 1 (F). Enrichment of differentiated cell markers Tff3 (G) and ChgA (H) was observed in population 1. Statistical comparisons were performed using one-way ANOVA with post hoc Tukey-Kramer pairwise comparisons; tests were considered significant if P < 0.05. a,b,c The different letters above each bar represent data points that are statistically different from each other.
To characterize Sox9EGFP-expressing populations at the transcript level, FACS was used to differentially isolate the three Sox9EGFP-expressing populations, and lineage identity was assessed by real-time RT-PCR analysis of each population. Increasing endogenous Sox9 mRNA expression correlated with increasing Sox9EGFP expression validating the FACS (Fig. 5, B and C). Previous studies using genetic lineage tracing have identified that Lgr5 marks a CESC population in vivo (39). Population 3, which is most enriched for Sox9 mRNA, also demonstrates significant enrichment of Lgr5 mRNA, compared with population 1, suggesting that higher levels of Sox9EGFP correlate with increased “stemness” in the colon (Fig. 5D). Notch pathway activation has been demonstrated to be essential for maintaining stemness (16, 35, 40). When the Notch pathway is downregulated, there is an increase in Atoh1 expression, which results in secretory lineage specification in transit amplifying progenitor cells. The data demonstrate a modest but significant decrease in Notch1 mRNA in the population 1, which expresses the lowest level of Sox9, and more notably a significant increase in secretory progenitor cell marker, Atoh1, mRNA in population 1. These data indicate that lower levels of Sox9EGFP mark cells consistent with transit amplifying progenitor cells (Fig. 5, E and F). Additionally, there was increased mRNA expression associated with goblet cells (Tff3) and enteroendocrine cells (chromogranin A) in cells that expressed the lowest levels of Sox9EGFP (Fig. 5, G and H). It is noteworthy that neither goblet cells nor enteroendocrine cells are observed to express Sox9EGFP by fluorescence microscopy; however, the high sensitivity of EGFP detection by flow cytometry is likely detecting small amounts of EGFP, even in cells that have downregulated Sox9EGFP and upregulated Muc2 and ChgA upon differentiation. Taken together, these data strongly suggest that higher levels of Sox9EGFP/Sox9 mark CESCs, and lower levels mark differentiating progenitors.
Colonic epithelial cells expressing the highest level of Sox9EGFP by flow cytometry form colonoids in culture.
Recent advances in culturing techniques for small intestine IESCs have revolutionized single-cell analysis of putative IESC populations that are initially identified and FACS isolated based on their gene expression signatures. It should be stated that a true clonogenic assay in which validated single cells are FACSed into individual wells has not been developed for IESCs, but, rather, single cells are typically FACS isolated and plated together as putative singlets into a three-dimensional culture system. Sato et al. (39) first described the culture system that supported the development of crypt/villus-like structures from single IESCs in the absence of mesenchymal support cells. These culture conditions provide essential signaling that is usually supplied by the nonepithelial cells in vivo, namely, a laminin-enriched extracellular matrix, EGF; Wnt-pathway agonist, R-Spondin1; Notch-receptor agonist, Jagged; and bone morphogenic protein antagonist, Noggin (39). A subsequent study from the same group indicated that a “support cell” is likely required to produce mitogens and morphogens to FACS-isolated IESCs, which would promote survival and growth of single IESCs in culture. In the absence of a more precise clonogenic assay, we utilized the strategy developed by Sato et al. (39) to assess the stemness of Sox9EGFP-expressing colonocytes.
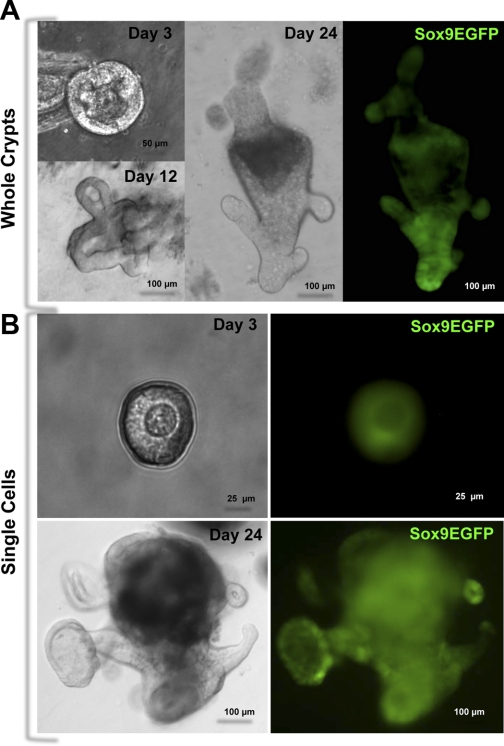
To test whether distinct Sox9 levels correlated with functional stemness, we FACS isolated single Sox9EGFP cells from populations 1, 2, and 3 and introduced these cells into the culture conditions that support the development of small intestine organoids. Forty-eight hours postplating, no viable cells remained, based on absence of Sox9EGFP expression and morphological characteristics consistent with widespread apoptosis. Recent studies indicate that a Paneth cell-derived WNT3a source is critical for the high-efficiency development of small intestine organoids from single IESCs (38). Paneth cells are not present in the colonic epithelium; however, we reasoned that a “Paneth cell equivalent” might be providing WNT3a in vivo. The conditions that support generation of organoids from IESCs were modified to include exogenously supplied recombinant WNT3a, as well as an underlying layer of MFs derived from embryonic murine intestine. Pericryptal MFs have been implicated in the support of epithelial growth and development by secreting mitogens and morphogens in the IESC zones (23, 32). These new conditions supported development of mature colonoids, which formed from isolated whole crypts (Fig. 6A) and FACS-isolated Sox9EGFP cells from population 3 (Fig. 6B). Early development of colonospheres, a colonoid precursor defined as a multicellular sphere (with no crypt-like structures) possessing a pseudolumen, developed from populations 1, 2, and 3 at an incidence of 0.3, 0.6, and 1.1%, respectively, between 2 and 3 days postplating. However, by day 7, all colonospheres from populations 1 and 2 ceased further development and died, while all colonospheres from population 3 progressed development to colonoid stage by day 12. Colonoid development persisted through day 24, at which point the colonoids were mechanically dissociated and replated. Each colonoid at day 12 derived from FACS-isolated CESCs produced three independent colonoids upon passaging, indicating symmetric division and self-renewal capacity of population 3 CECSs (data not shown).
Fig. 6.
High Sox9EGFP expression can be used as a FACS parameter to isolate CESCs that form colonoids in culture. Colonoids are defined as complex crypt-like bodies that develop from whole crypts or single CESCs. Colonoids are composed of colonic epithelial cells surrounding a pseudolumen and do not contain a mesenchymal cellular component. By 3 days in culture, both whole colonic crypts (A) and single CESCs (B) first develop into colonospheres that are composed of epithelial cells that have not formed crypt-like structures but contain a pseudolumen. Colonospheres progress to colonoids by 24 days in culture. Images were captured at ×200 original magnification.
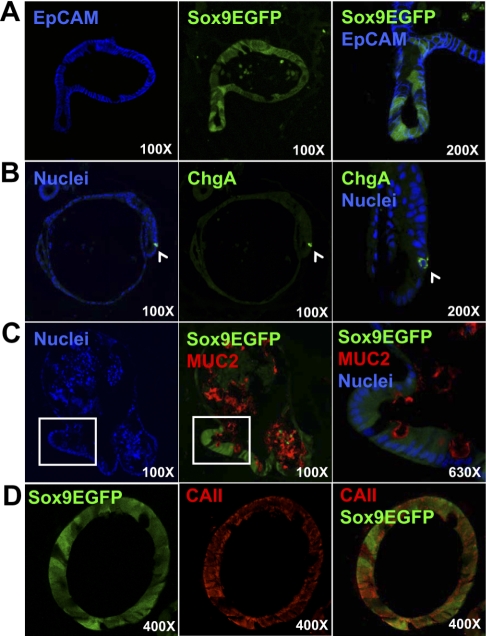
To assess the multipotency of population 3 CECS, colonoids were sectioned and immunostained for markers of postmitotic lineages. Sox9EGFP expression persisted in colonoids and showed variable expression levels similar to those observed in vivo (Fig. 7). Epithelial cell adhesion molecule staining demonstrated that all cells were epithelial in nature (Fig. 7A). Chromogranin A and MUC2 expression indicated that population 3 CESCs were competent to generate enteroendocrine (CHGA+) and goblet cells (MUC2+) (Fig. 7, B and C). MUC2 staining in the pseudolumen of the colonoids also indicates that the cells derived from single CESCs are polarized and functional (Fig. 7C; middle). Carbonic anhydrase II expression is consistent with absorptive colonocytes that localize to more luminal regions in vivo (Fig. 7D; see Fig. 1A). Together these data indicate that population 3 CESCs characterized by the highest Sox9 levels are multipotent and self-renewing.
Fig. 7.
Sox9EGFP CECSs that form colonoids are multipotent. A: epithelial cell adhesion molecule (EpCAM) staining confirms that colonoid cells are epithelial. ChgA (white arrow; B); MUC2 (white box indicates magnified region; C, right); and CAII (D) immunostaining indicate Sox9EGFP CESCs are competent to generate enteroendocrine, goblet, and absorptive colonocytes, respectively. Images represent 1-μm optical sections. Magnifications are indicated in each panel.
DISCUSSION
Despite recent and rapid advances in the identification of stem cell biomarkers across nearly all tissue types, precise cell-specific gene expression levels and the phenotypic consequences of these levels remain an understudied metric of mechanisms regulating stem cell maintenance and differentiation. Technologies that enable detection and monitoring of a gene's expression are becoming more robust, and the threshold of these detection methods is sufficient to enable quantification of gene expression in single cells. We propose that precise gene expression levels, and not simply the “on/off” qualitative parameter of gene expression, may be a more robust and critical identifier of cellular phenotype than previously appreciated.
In past studies, we have demonstrated this concept by using differential Sox9 expression at the cellular level to identify and isolate stem and progenitor cells from the small intestine epithelium (15). Other independent studies have used differential levels of Lgr5EGFP expression in the small intestine to demonstrate that “high” Lrg5EGFP-expressing cells are more stem-like, and “low” Lgr5EGFP-expressing cells are more consistent with transit amplifying progenitor cells (43). In the present study, we further validate this concept by showing that higher Sox9-expression levels mark cells that have gene expression signatures and functional characteristics of CESCs, while lower Sox9-expression levels are consistent with more differentiated cell types. Importantly, we also demonstrate that the presence of Sox9 expression in colonic epithelial cells is not, in and of itself, a sufficient indicator of stemness, as progenitor cells and subsets of goblet cells also express Sox9. These findings indicate that, while in vivo lineage tracing remains a powerful assay of stemness, careful analysis of cell-specific gene expression levels and the phenotypes associated with these levels are necessary criteria when defining and validating stem cell biomarkers.
We have demonstrated that colonic epithelial cells expressing the highest levels of Sox9EGFP also coexpress the highest levels of Lgr5, suggesting that these cells have functional stem cell qualities. Recent breakthroughs in culturing of isolated IESCs from the small intestine provided us with a framework to identify conditions that would support colonoid growth (15, 38, 39). The conditions described by Sato et al. (38, 39) for culture of small intestine IESCs did not support growth of colonoids from CESCs. We identified that additional requirements of WNT3a, in combination with critical unidentified diffusible factors from an underlying MF layer, were essential to support generation of colonoids from single cells. WNT3a is a secreted signaling protein that binds to receptors of the Frizzled family and has critical developmental roles in proliferation control and morphogenesis (24). Additionally, WNT3a has been shown to promote tissue-specific stem cell proliferation and maintenance in vitro (28, 34, 45). A recent study suggests that Paneth cells in the small intestine supply WNT3a to the small intestine IESCs and are required for maintenance of the IESC niche (38). Our studies demonstrate that WNT3a is essential for the growth of CESCs in culture, and we speculate that the WNT3a source in the colon could be generated from a similar cell type that is intimately associated with the CESCs. Our data suggest that this cell type is consistent with a Sox9-expressing goblet cell that is intimately connected to the CESCs in a manner similar to that of Paneth cells and IESCs in the small intestine. Moreover, both the crypt-based goblet cells and Paneth cells express Sox9, indicating a conserved role for this transcription factor in differentiated cell types that comprise the stem cell zone.
Interestingly, the Sox9EGFP transgene is not expressed in either MUC2+ cells in the base of the colonic crypt or Paneth cells in the small intestine, despite the expression of endogenous SOX9 in these cells types (10, 15). The Sox9EGFP transgene is randomly integrated into the genome; thus the disparity between the Sox9EGFP transgene expression and endogenous SOX9 expression could be due to genome position effects. It is also possible that the regulatory elements for Sox9 expression in crypt-based Paneth cells and goblet cell types are not contained in the BAC construct. If the latter is true, then efforts to identify the cis-regulatory elements required for lineage-specific Sox9 expression in Paneth cells and crypt-based goblet cell expression could be focused to genomic regions that flank the BAC ends. Identifying these regulatory elements could provide important insights into upstream signaling mechanisms that maintain Sox9 in cell types that support the IESC and CESC niche.
The ability to identify and isolate cancer cells based on their gene expression profiles and associated phenotypic characteristics has been central to the study of carcinogenesis (7, 46). Currently, the nature of how individual cells contribute to colon cancer growth and invasion remains controversial, as technological limitations have precluded the ability to precisely identify colon cancer stem cells. In this study and others, we have shown that Sox9EGFP levels vary between cell types with different lineage identities and functional stem cell characteristics in normal physiology (10, 15). A novel finding from our studies is that these variable levels are preserved in cells of AOM/DSS mouse colon tumors and primary human tumors. Based on the result from this study that higher levels of Sox9 expression correlate with increased stemness in physiological states, the quantitative assessment of the ratio of higher vs. lower Sox9-expressing cells within a tumor may provide an indication of cancer cell stemness. Further investigation into the levels of SOX9 in cancer cell stemness holds promise as a diagnostic tool to predict outcomes and direct therapeutic interventions.
Prevailing evidence suggests that Sox9 has anti-oncogenic properties, in that higher levels suppress proliferation and decrease Wnt signaling activity (4, 10). Interestingly, a new splice variant of SOX9 has been recently identified in human cancer cell lines and primary tumors that appear to exhibit oncogenic properties (1). The SOX9 splice variant (termed miniSox9) retains intron 2, producing in a premature stop codon and an aberrant COOH-terminus. The mutant protein retains the DNA binding domain, but lacks the transactivation domain. MiniSox9 inhibits the anti-oncogenic properties of full-length SOX9 and stimulates the canonical Wnt pathway, presumably increasing proliferative capacity and tumorgenicity (1). The miniSox9 study highlights the importance of characterizing the levels of Sox9 at the cellular level and also understanding how these Sox9 levels mechanistically confer cellular phenotype. For example, determining expression levels of full-length or miniSox9 in the same cell may identify subpopulations of cells within the colonic tumors that have differential capacity for proliferation and differentiation.
The Sox9EGFP mouse model will be a useful tool to study the role of differential Sox9-expression levels in cells of colon tumors. Currently, the Sox9EGFP mice are on a genetic background that is highly resistant to AOM/DSS tumor induction; thus the amount of tumor tissue obtained in this study was insufficient for gene expression analysis and culture assays at the single-cell level. On-going studies are underway to introduce the Sox9EGFP allele onto genetic backgrounds that are more amenable to colonic tumor induction. Based on our studies, we speculate that colonic adenoma cells expressing higher levels of Sox9 may exhibit slower division and more stem cell-like characteristics that have been hypothesized to be more resistant to chemotherapy and radiation, while lower levels of Sox9 would be consistent with more rapidly dividing progenitor-like cells that may be more responsive to standard cancer therapies (8). The concepts and image analysis strategies developed in this study will enable detailed analysis of SOX9 levels in human colonic adenomas and carcinomas and will likely establish rationale for screening tumor TMAs for different levels of oncogenic gene products and correlating cellular expression levels of these oncogenic proteins to clinical outcomes.
GRANTS
We acknowledge the University of North Carolina (UNC) Center for GI Biology and Disease Imaging and Histology COREs (P30-DK034987), the UNC Neuroscience Confocal Imaging Facility (P30-NS045892), the UNC Chapel Hill Mutant Mouse Regional Resource Center, and the UNC Flow Cytometry CORE. Funding was provided by the National Institutes of Health, K01-DK080181; the American Gastroenterological Association Research Scholar Award; the North Carolina Biotechnology Center; the UNC-Chapel Hill Center for Gastrointestinal Biology and Disease, P30-DK034987; and the UNC SPORE in GI Cancer, 5-P50-CA 106991.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: S.R., G.W.D., M.J.J., A.D.G., and S.T.M. conception and design of research; S.R., G.W.D., M.J.J., A.D.G., and S.T.M. performed experiments; S.R., G.W.D., M.J.J., A.D.G., and S.T.M. analyzed data; S.R., G.W.D., M.J.J., A.D.G., and S.T.M. interpreted results of experiments; S.R., G.W.D., M.J.J., A.D.G., and S.T.M. prepared figures; S.R., G.W.D., M.J.J., A.D.G., and S.T.M. drafted manuscript; S.R., G.W.D., M.J.J., A.D.G., and S.T.M. edited and revised manuscript; S.R., G.W.D., M.J.J., A.D.G., and S.T.M. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Ian Williamson for technical assistance, and Drs. Jim Simmons and P. Kay Lund for providing the protocol that we adapted for embryonic MF isolation and propagation. Additionally, we thank Dr. Susan Henning, Dr. Victoria Newton, Dr. Laurianne Van Landeghem, and Brent Puthoff for critical reading of the manuscript, and Dr. P. Kay Lund for useful discussions on terminology.
REFERENCES
- 1. Abdel-Samad R, Zalzali H, Rammah C, Giraud J, Naudin C, Dupasquier S, Poulat F, Boizet-Bonhoure B, Lumbroso S, Mouzat K, Bonnans C, Pignodel C, Raynaud P, Fort P, Quittau-Prevostel C, Blache P. MiniSOX9, a dominant-negative variant in colon cancer cells. Oncogene 30: 2493–2503, 2011 [DOI] [PubMed] [Google Scholar]
- 2. Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007, 2007 [DOI] [PubMed] [Google Scholar]
- 3. Bastide P, Darido C, Pannequin J, Kist R, Robine S, Marty-Double C, Bibeau F, Scherer G, Joubert D, Hollande F, Blache P, Jay P. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol 178: 635–648, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blache P, van de Wetering M, Duluc I, Domon C, Berta P, Freund JN, Clevers H, Jay P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol 166: 37–47, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cai WB, Roberts SA, Potten CS. The number of clonogenic cells in crypts in three regions of murine large intestine. Int J Radiat Biol 71: 573–579, 1997 [DOI] [PubMed] [Google Scholar]
- 6. Cheng H. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. II. Mucous cells. Am J Anat 141: 481–501, 1974 [DOI] [PubMed] [Google Scholar]
- 7. Choi D, Lee HW, Hur KY, Kim JJ, Park GS, Jang SH, Song YS, Jang KS, Paik SS. Cancer stem cell markers CD133 and CD24 correlate with invasiveness and differentiation in colorectal adenocarcinoma. World J Gastroenterol 15: 2258–2264, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med 17: 313–319, 2011 [DOI] [PubMed] [Google Scholar]
- 9. Dekaney CM, Rodriguez JM, Graul MC, Henning SJ. Isolation and characterization of a putative intestinal stem cell fraction from mouse jejunum. Gastroenterology 129: 1567–1580, 2005 [DOI] [PubMed] [Google Scholar]
- 10. Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol 296: G1108–G1118, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fujimoto K, Beauchamp RD, Whitehead RH. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology 123: 1941–1948, 2002 [DOI] [PubMed] [Google Scholar]
- 12. Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, Masui T, Kawaguchi M, Takaori K, Doi R, Nishi E, Kakinoki R, Deng JM, Behringer RR, Nakamura T, Uemoto S. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet 43: 34–41, 2011 [DOI] [PubMed] [Google Scholar]
- 13. Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, Masui T, Kawaguchi M, Takaori K, Doi R, Nishi E, Kakinoki R, Deng JM, Behringer RR, Nakamura T, Uemoto S. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet 43: 34–41, 2011 [DOI] [PubMed] [Google Scholar]
- 14. Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425: 917–925, 2003 [DOI] [PubMed] [Google Scholar]
- 15. Gracz AD, Ramalingam S, Magness ST. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol 298: G590–G600, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet 24: 36–44, 2000 [DOI] [PubMed] [Google Scholar]
- 17. Kayahara T, Sawada M, Takaishi S, Fukui H, Seno H, Fukuzawa H, Suzuki K, Hiai H, Kageyama R, Okano H, Chiba T. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett 535: 131–135, 2003 [DOI] [PubMed] [Google Scholar]
- 18. Levin TG, Powell AE, Davies PS, Silk AD, Dismuke AD, Anderson EC, Swain JR, Wong MH. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology 139: 2072–2082 e2075, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science 327: 542–545, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu B, Xu J, Lai M, Zhang H, Chen J. A transcriptome anatomy of human colorectal cancers. BMC Cancer 6: 40, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Magness ST, Bataller R, Yang L, Brenner DA. A dual reporter gene transgenic mouse demonstrates heterogeneity in hepatic fibrogenic cell populations. Hepatology 40: 1151–1159, 2004 [DOI] [PubMed] [Google Scholar]
- 22. May R, Riehl TE, Hunt C, Sureban SM, Anant S, Houchen CW. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells 26: 630–637, 2008 [DOI] [PubMed] [Google Scholar]
- 23. Mifflin RC, Pinchuk IV, Saada JI, Powell DW. Intestinal myofibroblasts: targets for stem cell therapy. Am J Physiol Gastrointest Liver Physiol 300: G684–G696, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mikels AJ, Nusse R. Wnts as ligands: processing, secretion and reception. Oncogene 25: 7461–7468, 2006 [DOI] [PubMed] [Google Scholar]
- 25. Montgomery RK, Breault DT. Small intestinal stem cell markers. J Anat 213: 52–58, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour-Awuah NY, Ambruzs DM, Fogli LK, Algra S, Breault DT. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A 108: 179–184, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mori-Akiyama Y, van den Born M, van Es JH, Hamilton SR, Adams HP, Zhang J, Clevers H, de Crombrugghe B. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology 133: 539–546, 2007 [DOI] [PubMed] [Google Scholar]
- 28. Nusse R. Wnt signaling and stem cell control. Cell Res 18: 523–527, 2008 [DOI] [PubMed] [Google Scholar]
- 29. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Potten CS, Booth C, Tudor GL, Booth D, Brady G, Hurley P, Ashton G, Clarke R, Sakakibara S, Okano H. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation 71: 28–41, 2003 [DOI] [PubMed] [Google Scholar]
- 31. Potten CS, Kellett M, Roberts SA, Rew DA, Wilson GD. Measurement of in vivo proliferation in human colorectal mucosa using bromodeoxyuridine. Gut 33: 71–78, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Powell DW, Mifflin RC, Valentich JD, Crowe SE, Saada JI, West AB. Myofibroblasts. II. Intestinal subepithelial myofibroblasts. Am J Physiol Cell Physiol 277: C183–C201, 1999 [DOI] [PubMed] [Google Scholar]
- 33. Ramocki NM, Wilkins HR, Magness ST, Simmons JG, Scull BP, Lee GH, McNaughton KK, Lund PK. Insulin receptor substrate-1 deficiency promotes apoptosis in the putative intestinal crypt stem cell region, limits Apc−/+ tumors, and regulates Sox9. Endocrinology 149: 261–267, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423: 409–414, 2003 [DOI] [PubMed] [Google Scholar]
- 35. Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, Strobl LJ, Honjo T, Clevers H, Radtke F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep 9: 377–383, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sakthianandeswaren A, Christie M, D'Andreti C, Tsui C, Jorissen RN, Li S, Fleming NI, Gibbs P, Lipton L, Malaterre J, Ramsay RG, Phesse TJ, Ernst M, Jeffery RE, Poulsom R, Leedham SJ, Segditsas S, Tomlinson IP, Bernhard OK, Simpson RJ, Walker F, Faux MC, Church N, Catimel B, Flanagan DJ, Vincan E, Sieber OM. PHLDA1 expression marks the putative epithelial stem cells and contributes to intestinal tumorigenesis. Cancer Res 71: 3709–3719, 2011 [DOI] [PubMed] [Google Scholar]
- 37. Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40: 915–920, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469: 415–418, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265, 2009 [DOI] [PubMed] [Google Scholar]
- 40. Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, Zoghbi HY. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 132: 2478–2488, 2007 [DOI] [PubMed] [Google Scholar]
- 41. Snippert HJ, van Es JH, van den Born M, Begthel H, Stange DE, Barker N, Clevers H. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology 136: 2187–2194 e2181, 2009 [DOI] [PubMed] [Google Scholar]
- 42. van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 137: 15–17, 2009 [DOI] [PubMed] [Google Scholar]
- 43. van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, van Es JH, Barker N, Peters PJ, van de Wetering M, Clevers H. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136: 903–912, 2009 [DOI] [PubMed] [Google Scholar]
- 44. von Furstenberg RJ, Gulati AS, Baxi A, Doherty JM, Stappenbeck TS, Gracz AD, Magness ST, Henning SJ. Sorting mouse jejunal epithelial cells with CD24 yields a population with characteristics of intestinal stem cells. Am J Physiol Gastrointest Liver Physiol 300: G409–G417, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, 3rd, Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423: 448–452, 2003 [DOI] [PubMed] [Google Scholar]
- 46. Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, Poppleton H, Zakharenko S, Ellison DW, Gilbertson RJ. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 457: 603–607, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]