Abstract
With the use of molecular techniques, numerous studies have evaluated the composition of the intestinal microbiota in health and disease. However, it is of major interest to supplement this with a functional analysis of the microbiota. In this review, the different approaches that have been used to characterize microbial metabolites, yielding information on the functional end products of microbial metabolism, have been summarized. To analyze colonic microbial metabolites, the most conventional way is by application of a hypothesis-driven targeted approach, through quantification of selected metabolites from carbohydrate (e.g., short-chain fatty acids) and protein fermentation (e.g., p-cresol, phenol, ammonia, or H2S), secondary bile acids, or colonic enzymes. The application of stable isotope-labeled substrates can provide an elegant solution to study these metabolic pathways in vivo. On the other hand, a top-down approach can be followed by applying metabolite fingerprinting techniques based on 1H-NMR or mass spectrometric analysis. Quantification of known metabolites and characterization of metabolite patterns in urine, breath, plasma, and fecal samples can reveal new pathways and give insight into physiological regulatory processes of the colonic microbiota. In addition, specific metabolic profiles can function as a diagnostic tool for the identification of several gastrointestinal diseases, such as ulcerative colitis and Crohn's disease. Nevertheless, future research will have to evaluate the relevance of associations between metabolites and different disease states.
Keywords: fermentation, metabolites, metabolomics, microbiota, short-chain fatty acids
the human intestinal microbiota complements our physiology with functions that we have not had to develop on our own. In fact, the intestinal microbiota have a metabolic capacity that is comparable to that of the liver (83). The human colon contains an extremely complex and dynamic microbial ecosystem with high densities of living bacteria in concentrations of 1011-1012 cells/g of luminal contents belonging to more than 1,000 different species. In healthy adults, 80% of the identified fecal microbiota can be classified into three dominant phyla: Firmicutes, Bacteroidetes, and Actinobacteria, but there is substantial variation in the species composition between individuals (30, 101).
The intestinal microbiota plays an important role in human physiology. For example, the intestinal microbiota is responsible for the further metabolism and energy harvest from nondigested nutrients, is involved in the synthesis of vitamins such as B and K and metabolism of polyphenols, provides colonization resistance toward potential pathogens, is involved in the metabolism of bile acids, and stimulates the immune function of the host (74, 83).
Molecular approaches, mainly based on the 16S ribosomal RNA gene, have revolutionized the field of gut microbial ecology. Nowadays, the uncultured and complex microbial communities can be characterized with greater sensitivity by using high-throughput technologies, such as pyrosequencing (5) and phylogenetic microarrays (79), compared with former molecular fingerprinting methods, such as PCR-denaturing gradient gel electrophoresis (DGGE). Complementary quantitative technologies, such as fluorescence in situ hybridization (FISH) and real-time quantitative PCR, can be used to confirm shifts in particular groups or species (113).
In recent years an increasing amount of literature has demonstrated that several diseases are related to alterations in the intestinal microbiota (known as dysbiosis), such as irritable bowel syndrome (54), inflammatory bowel disease (93), diabetes (55), atopic diseases (76), cancer (85), and obesity (7). For example, a reduction in the abundance and diversity of Firmicutes is frequently associated with inflammatory bowel disease and irritable bowel syndrome (105, 113). These studies have mainly shown that differences in the composition of the intestinal microbiota are associated with disease.
Functional Capacity of the Microbiota
The human microbiota is characterized by a significant degree of functional redundancy, meaning that different bacteria can perform similar functions and metabolize the same substrate (60, 64). Therefore, not only the composition but also the functional capacity of the intestinal microbiota is highly important regarding the clinical end points.
Next-generation pyrosequencing can be applied to further evaluate the functional capacity of the colonic microbiota by creating a catalog of the genetic potential of the microbiota. However, it has to be realized that the detection of genes in a metagenomic library does not necessarily mean that these are functionally important (113). To gain better insight into the activity and functionality of the intestinal microbiota, other meta-“omics” approaches can be applied that use RNA, proteins, and metabolites as targets. In this review, we summarize the different approaches that have been used to quantify and characterize the metabolites produced by the microbiota, which yield information on the actual end products of metabolism. Colonic microbial fermentation results in the production of large amounts of different end products. The type and amount of these fermentation-derived metabolites largely depend on the composition of the microbiota, transit time, and the substrates available for fermentation (95). Some of these end products have been shown to be protective to the colonic epithelium, and others have proved to be proinflammatory or procarcinogenic metabolites (3, 48). By using knowledge of these specific metabolites, a hypothesis-driven targeted approach can be applied to evaluate changes in colonic metabolism following dietary interventions or during different disease states, for example, through quantification of selected metabolites from carbohydrate and protein fermentation, secondary bile acids, or colonic enzymes. On the other hand, a top-down approach can be followed by applying metabolite fingerprinting techniques based on 1H-nuclear magnetic resonance (NMR) or mass spectrometric analysis. By following this approach, novel metabolites and mechanisms can be identified that are involved in health and disease. This is, however, not an easy task, since the signals first have to be identified and their metabolic roles elucidated.
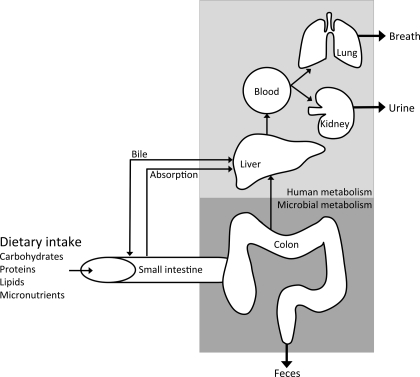
For analyses of metabolites as end products of intestinal metabolism in humans, we mainly rely on fecal samples or on breath (for example, hydrogen, methane, and carbon dioxide), urine, and plasma samples due to the relative inaccessibility of the colon to sample at different locations (Fig. 1) (50). In these human studies, an elegant solution to study metabolic pathways in vivo is the application of stable isotope tracers.
Fig. 1.
Simplified scheme of the metabolites produced by the interplay between the microbial and human metabolome. After passing through the small intestine, unabsorbed dietary substrates (mainly unabsorbed carbohydrates and proteins) along with other endogenous substrates (such as bile enzymes, mucus and exfoliated cells) reach the large intestine, where they are fermented by the intestinal microbiota. These microbial metabolites are excreted in feces or absorbed and transported to the liver. Subsequently, these absorbed metabolites are subject to further human metabolism and can be excreted in urine and breath (50).
Types of Fermentation
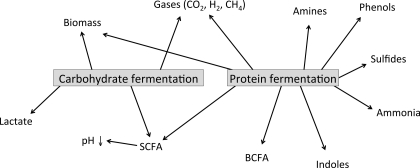
The colonic microbiota ferment endogenous host-derived substrates such as mucus, pancreatic enzymes, and exfoliated epithelial cells, as well as dietary components that escape digestion in the upper gastrointestinal tract. Two main types of colonic microbial fermentation can be distinguished, including saccharolytic fermentation of carbohydrate and proteolytic fermentation of protein (Fig. 2). In the proximal part of the colon, mainly saccharolytic fermentation takes place, since most microorganisms preferentially ferment carbohydrates and switch to protein fermentation when carbohydrate sources are depleted (75). The main products of carbohydrate metabolism are short-chain fatty acids (SCFA), mainly acetate, propionate, and butyrate, which have been shown to contribute to colonic health. SCFA provide energy to the colonic epithelial cells, decrease luminal pH, and improve mineral absorption. Furthermore, butyrate has been shown to possess an anti-inflammatory and anticarcinogenic potential (46, 63). Besides their contribution to gut health and maintenance, SCFA may provide further benefits for the systemic metabolism. For example, it has been shown that acetate and propionate affect hepatic lipid metabolism. Acetate is the primary substrate for cholesterol synthesis, whereas propionate can inhibit cholesterol synthesis (27, 110). Since the different SCFA may show distinct effects, not only the amount of SCFA produced but also their ratio is of importance with regard to these health effects. Furthermore, SCFA stimulate increased plasma levels of satiety hormones such as peptide YY (PYY), leptin, and glucagon-like peptide-1 (GLP-1) and may attenuate insulin resistance (1, 34). These effects may occur due to the fact that SCFA are ligands for G protein-coupled receptors (GPR) 41 and 43, expressed on adipocytes, enteroendocrine L-cells, and immune cells (88). In addition, recent studies have linked SCFA activation of GPR 43 to the suppression of colon cancer (94, 96). Proteolytic fermentation also leads not only to the production of SCFA (lower amounts than produced from carbohydrates) and branched-chain fatty acids but also to potentially toxic metabolites such as phenolic compounds, sulfur-containing compounds, amines, and ammonia (Fig. 2) (48). The toxicity of these protein fermentation metabolites has mainly been established in in vitro studies (6, 11, 58) and animal studies (4, 97). However, human epidemiological studies do not consistently support an association between protein intake and colorectal cancer (2) and inflammatory bowel disease (47). Unfortunately, actual protein fermentation metabolites were not determined in these studies. The different metabolites of protein fermentation and their potential involvement in colorectal cancer have previously been reviewed (48).
Fig. 2.
Different metabolites produced from colonic fermentation of carbohydrates and proteins. BCFA, branched-chain fatty acids; SCFA, short-chain fatty acids.
Targeted Approach
Quantification of carbohydrate fermentation.
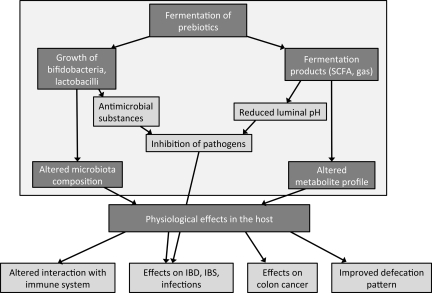
The beneficial effects of saccharolytic fermentation have mainly been ascribed to the production of SCFA (68, 91). Several studies have evaluated SCFA concentrations and profiles in patients with different diseases. For example, a lower butyrate-to-acetate ratio has been found in colonic luminal content of patients with adenomatous polyps or colon cancer compared with healthy controls (107), and increased amounts of SCFA were found in fecal samples from obese compared with lean individuals (89). Dietary interventions with prebiotics, such as inulin and oligofructose, defined as “nondigestible food ingredients that stimulate the growth and/or activity of bacteria in the digestive system which are beneficial to the health of the host,” aim to increase and prolong the saccharolytic fermentation toward the distal colon and thereby aim to reduce proteolytic fermentation (38) (Fig. 3). Increased SCFA production after addition of different prebiotics to the diet has previously been demonstrated by measuring fecal or plasma concentrations of SCFA (52, 73). However, the in situ production of SCFA is difficult to determine, since more than 95% of the produced SCFA are absorbed and metabolized by the host depending on the gastrointestinal transit time (59). To gain more insight into the actual SCFA production over time, the use of stable isotope tracers can be considered. A recent study with 13C-labeled barley has evaluated the kinetics of SCFA appearance in the systemic circulation in healthy volunteers. In this study, the pattern of SCFA appearing in the systemic circulation was different after consumption of a meal with dietary fiber (nonstarch polysaccharides) combined with resistant starch compared to a meal with dietary fiber alone (102). Further studies are necessary to address the significance of these different SCFA profiles with regard to health benefits. In another study, stable isotope tracers were applied to determine the production rate of acetate during colonic fermentation of lactulose in humans. Healthy volunteers received a primed continuous infusion of [1-13C]acetate followed by the ingestion of lactulose. The colonic acetate production was calculated from the reduction in the plasma [13C]acetate enrichment as a result of the colonic fermentation of lactulose (77). With the use of a dynamic in vitro model of the large intestine, the fermentation of 13C-labeled starch was evaluated by determination of the label incorporation into the microbial biomass and metabolites using stable isotope probing and NMR analysis (53). From the labeling pattern of microbial metabolites, it was concluded that cross-feeding between Ruminoccus bromii and Eubacterium rectale occurred, wherein R. bromii produced acetate, which was subsequently converted to butyrate by E. rectale.
Fig. 3.
Proposed mechanism of action of prebiotics. IBD, inflammatory bowel disease; IBS, irritable bowel syndrome.
Quantification of protein fermentation.
The degree of proteolytic fermentation can be determined by quantification of typical end products of protein fermentation (Fig. 2) (48). Some of these metabolites are reused as nitrogen source for bacterial growth, whereas others will be taken up by colonocytes and transported into the bloodstream. For instance, phenols and indoles are breakdown products of the aromatic acids tyrosine and tryptophan, respectively. Generally, once these compounds are produced, they enter the hepatic circulation to be detoxified in the liver and eventually excreted in urine. Since p-cresol and phenol are unique bacterial metabolites from protein fermentation that are not produced by human enzymes, these metabolites have been frequently used to assess the degree of proteolytic fermentation (25, 37). Urinary levels of p-cresol and phenol have shown to be increased during high protein intake (37) and decreased after oral supplementation with oligofructose-enriched inulin (OF-IN) (25).
In hemodialysis patients, p-cresol generation and p-cresyl sulfate serum concentrations were lowered after oral OF-IN administration (69). As renal function declines in these patients, substances that are either excreted or metabolized by the kidney accumulate in the circulation, resulting in increased levels of numerous molecules in blood (33). A number of these retained solutes originate from colonic bacterial protein metabolism (33). In addition, small intestinal assimilation of proteins is impaired in renal failure (10), resulting in an increased availability of proteins for fermentation in the colon (31). Accumulation of the protein fermentation metabolites in serum has been suggested to alter endothelial function (32, 70) and has been associated with increased mortality in hemodialysis patients (9). As a consequence, a dietary strategy with OF-IN that contributes to a lower generation of protein fermentation metabolites might constitute a significant improvement in the treatment of these patients. In addition to p-cresol and phenol, other protein fermentation metabolites, such as sulfur-containing metabolites, were decreased in fecal samples after incubation with OF-IN in vitro (22). Increased concentrations of sulfides have been associated with the pathogenesis of ulcerative colitis (UC). Hydrogen sulfide (H2S) has been found to provoke genomic DNA damage in colonic cancer cells (HT-29 cells) in concentrations similar to those found in the human colon (6). In addition to inducing DNA damage, sulfide impairs the oxidation of butyrate, the major energy substrate in colonocytes, by inhibition of cytochrome c oxidase (81). These observations are the basis for Roediger's “energy deficiency” hypothesis as a cause of UC (67, 82). Different studies have reported increased luminal concentrations of sulfide as well as high numbers of sulfide-reducing bacteria in patients with UC (84). This patient group may therefore also benefit from dietary interventions with prebiotics.
Colonic ammonia metabolism.
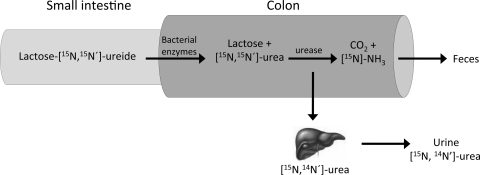
To evaluate the ammonia metabolism in the colon, the stable isotope-labeled biomarker lactose[15N,15N′]ureide has been validated (20, 36) (Fig. 4). Human enzymes are not able to hydrolyze the bond between the sugar moiety and urea, whereas this bond can be split by bacterial enzymes when it reaches the colon. Released [15N,15N′]urea is rapidly hydrolyzed to [15N]ammonia by ubiquitous bacterial urease. As such, lactose[15N,15N′]ureide is used as a vehicle to introduce a known amount of 15N, in the form of ammonia, to the colon. The formed [15N]ammonia can be either taken up by the colonic microbiota, followed by fecal excretion, or absorbed through the mucosa and renally excreted after conversion to [15N]urea in the liver. When microbial activity was stimulated after intake of a prebiotic, the urinary 15N excretion decreased, whereas the fecal 15N excretion increased (20, 36). In general, fermentable carbohydrates stimulate bacterial proliferation, which leads to incorporation of nitrogen (from ammonia and other sources) into bacterial cells and consequent excretion in feces (25, 26, 108). Thus a shift of nitrogen excretion from urine to feces can be explained by increased bacterial protein synthesis and a subsequent decrease in colonic absorption of nitrogen in the form of ammonia. The removal of ammonia from the colonic lumen might be considered a health benefit, since it may prevent ammonia from damaging colonocytes (87) and increased systemic levels of ammonia cause neurotoxic effects (106). Accumulation of ammonia in the bloodstream is associated with hepatic encephalopathy. Lactulose is frequently utilized in the treatment to reduce ammonia levels in these patients (90, 106).
Fig. 4.
Lactose[15N,15N′]ureide is resistant to digestion but is fermented by colonic bacterial enzymes into [15N,15N′]urea and subsequently [15N]ammonia (NH3). The formed [15N]ammonia can either be taken by the microbiota and excreted via the feces or be absorbed through the mucosa and renally excreted after conversion to [15N,14N′]urea in the liver. The proportionate recovery of 15N in feces or urine indicates the degree of colonic protein fermentation.
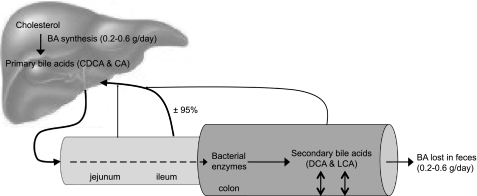
Secondary bile acids.
Bile acids are natural amphipathic detergents that assist the emulsification and absorption of lipids and fat-soluble vitamins. The human liver synthesizes the primary bile acids, cholic acid and chenodeoxycholic acid. Primary bile acids are secreted in bile from the gallbladder into the small intestine during digestion. They are then actively absorbed in the ileum and returned to the liver via the portal vein (Fig. 5). About 5% of bile salts escape this enterohepatic circulation and enter the colon, where they are subject to bacterial biotransformation reactions. When the primary bile acids reach the colon, they are deconjugated and successively undergo other enzymatic reactions, the most predominant being the dehydroxylation by bacterial 7α-dehydroxylase to form the secondary bile acids, primarily deoxycholic (DCA) and lithocholic acids (LCA) (8). Secondary bile acids have been hypothesized to be cocarcinogenic and tumor promoters (18, 112). It has been reported that patients with adenomatous polyps have a higher concentration of fecal bile acids and total secondary bile acids compared with control subjects (49). Furthermore, the ratios of the primary bile acids and their secondary bile acids are significantly lower in cancer patients compared with controls (17, 49). However, a large meta-analysis of 20 clinical trials and a total number of 1,226 individuals showed no difference between the fecal concentrations of secondary bile acids (DCA and LCA) in colorectal cancer patients compared with controls (98). It has been shown that dietary interventions are able to modulate fecal bile acid concentrations. For example, Boutron-Ruault et al. (15) have shown that ingestion of short-chain fructooligosaccharides decreased fecal concentrations of LCA. This decrease may be related to the increased production of SCFA, since SCFA decrease the colonic pH. A low pH inhibits dehydroxylation of bile acids and, consequently, conversion to secondary bile acids (19). This hypothesis is supported by the negative correlation between butyrate and acetate production and bile acid metabolism (100).
Fig. 5.
Enterohepatic circulation of bile acids. After each meal, gallbladder contraction empties bile acids into the intestinal tract. When passing through the intestinal tract, some bile acids are reabsorbed in the upper intestine by passive diffusion, but most bile acids (±95%) are reabsorbed in the ileum. Bile acids are transdiffused across the enterocyte to the basolateral membrane and excreted into portal blood circulation back to the liver. In the colon, secondary bile acids are formed by bacterial 7α-dehydroxylation of their primary bile acid precursor. Secondary bile acids may have a cytotoxic effect on the colonic epithelium. Bile acids lost in the feces (0.2–0.6 g/day) are replenished by de novo synthesis in the liver to maintain a constant bile acid pool. BA, bile acids; CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; LCA, lithocholic acid.
Bacterial enzyme activity.
Several bacterial enzyme activities have been studied in relation to their effect on host health. Bacterial β-glucosidase and β-glucuronidase hydrolyze the glycosidic bond of glycosides and glucuronide conjugates, respectively, to release aglycones. Glycosides in the gut originate from the plant material in the diet. Glucuronide conjugates are endogenously produced compounds that are metabolized in the liver and conjugated to glucuronic acid before being excreted by the liver via the bile into the small intestine (86). Since some aglycones have been reported to be potentially toxic or carcinogenic, elevated activity of these bacterial enzymes is hypothesized to be a risk factor for colon cancer (39, 43). On the other hand, anticarcinogenic effects of flavonoid aglycones also have been reported (29). Other examples of bacterial enzymes are azoreductase, nitrate reductase, nitroreductase, and sulfatases (39). Azoreductase leads to the production of amines, whereas nitrate reductase converts nitrate into nitrite and nitroreductase is involved in the formation of heterocyclic and aromatic nitro compounds (66). Sulfatases are involved in the degradation of mucosal glycans, such as colonic mucins (14). Activities of these bacterial enzymes are related to the composition of the microbiota. For example, lactobacilli and bifidobacteria produce low levels of β-glucuronidase but also low levels of azoreductase and nitroreductase, whereas strict anaerobes (Bacteroides sp., Eubacterium sp., and Clostridium sp.) produce high levels of these enzymes (14, 71). A number of animal and human intervention studies have investigated the ability of pre- and probiotics to modulate these bacterial enzyme activities in the colon. However, these studies have obtained conflicting results (23, 40–42, 45, 62, 65, 66, 92), and their significance for host health is often difficult to determine (23, 65).
Top-Down Approach/Metabolomics
New techniques such as metabolomics allow evaluation of the colonic metabolism by a top-down approach, bypassing the need for an a priori hypothesis. Metabolomics has been defined as “the quantitative measurement of the multiparametric metabolic responses of a living system to pathophysiological stimuli or genetic modification” (72).
Metabolomic analysis requires analytical technologies that are capable of detecting and quantifying the large number of metabolites in biological samples, such as feces (24, 35, 37, 103), intact tissue (12, 16), breath (80), blood (111), and urine (109). Analyses of these multiple biological samples complement each other, since they yield different information with regard to the origin of the metabolites. Certain breath and urinary metabolites result from cometabolism by the host and the intestinal microbiota, whereas fecal metabolites are primarily derived from the intestinal microbiota (Fig. 1).
Ideally, the sample preparation for metabolomic analysis should be minimal to enable the quantification of as many metabolites as possible. However, no single analytical technique can measure all the metabolites present in a particular sample due to the diverse chemical properties of metabolites and the limitations of each analytical technique (57). Commonly used analytical platforms are mass spectrometry (MS) (28) and NMR (13). Multivariate statistical techniques are required to determine which metabolites differ between samples from different groups. Commonly used multivariate discriminant tests in metabolomic studies include principal component analysis (PCA) and partial least-squares discriminant analysis (PLS-DA). PCA may be considered an unsupervised classification method, since the variation in the data is analyzed without a priori designation of samples into their classes. In contrast, PLS-DA is considered a supervised classification method, because the samples are designated into their classes for comparison. These different platforms to detect hundreds, or even thousands, of metabolites in one single analysis can be used to evaluate the differences in metabolite profiles between different diseases (51, 109) or different stages of disease (56) and to evaluate the effect of different (dietary) interventions (44, 57).
Williams et al. (109) used NMR spectroscopy to evaluate urinary metabolites in patients with UC and Crohn's disease compared with controls. PCA analysis revealed that each of the three groups clustered together and had different principal components, indicating different metabolite patterns in patients with the two distinct inflammatory bowel diseases and controls. After further analysis of the individual metabolites, hippurate levels were found to be lowest in Crohn's disease patients, whereas formate levels were higher and p-cresol sulfate levels were lower in Crohn's disease patients compared with UC patients and controls, reflecting inherent differences in intestinal microbes between cohorts (109). It remains to be determined whether these metabolites play a causative role in the development of inflammatory bowel diseases or are just innocent bystanders associated with the disease. Metabolic profiling of fecal samples using gas chromatography-MS (GC-MS) also demonstrated that the patterns of volatile organic compounds from feces of patients with UC, C. difficile, and C. jejuni infection were each significantly different (35). In another study in which metabolite patterns in fecal slurries were studied using GC-MS, incubation with OF-IN resulted in increased SCFA levels as expected. However, a more discriminatory factor for the clustering of the fecal slurries incubated with different doses of OF-IN was an increase in the concentration and number of esters (22). Increased acid production might be the origin of a higher presence of esters. The relevance of esters to health is yet unknown. Future studies are warranted and may also have to consider esters as a marker of saccharolytic activity. Incubation of fecal slurries with OF-IN was furthermore associated with a reduction in sulfur-containing compounds and phenols in a dose- and time-dependent way (22). Analysis of patterns of volatile organic compounds in exhaled breath has been proved to be useful to discriminate patients with chronic obstructive pulmonary disease (COPD) from healthy controls (99) and to discriminate asthmatic children from healthy subjects (21).
Microbial Metabolites Relevant to Health?
The top-down evaluation of microbial metabolites may lead to new insights into which components could be associated with different diseases and (dietary) interventions. Another future challenge remains to associate which microbial species or consortia are responsible for the production of distinct types of metabolites. Several recent studies have used a “transgenomic” approach to link gut microbiome and metabolic phenotype variation (61, 104). These approaches can offer new possibilities for future interventions. However, more knowledge on the clinical effects of a decrease or increase in certain metabolites and microbial species is warranted, because detected associations may not always be relevant. It remains unknown whether changes in metabolites are a cause or a consequence of the disease. Large prospective cohort studies are warranted to evaluate whether there exists a causal relationship between the production of different metabolites and the different diseases, their clinical features, or disease progression.
Metabolic profiles can also be used for diagnosis of different diseases or classification of patients. This is a promising noninvasive, rapid, and relatively inexpensive diagnostic tool. Before introduction into the clinical practice, the specificity and sensitivity needs to be further evaluated (35, 78).
Conclusions
Changes in colonic metabolism can be evaluated following a targeted approach or a top-down approach. The targeted approach may include quantification of known metabolites from carbohydrate fermentation (SCFA) or protein fermentation (such as p-cresol, phenol, ammonia, or H2S), levels of secondary bile acids, or bacterial enzyme activities. All these “biomarkers” have been identified in epidemiological or intervention studies and have been related to different pathologies, such as colorectal cancer, inflammatory bowel disease, or metabolic syndrome. However, at present, these biomarkers have been insufficiently validated in large prospective trials. With the emergence of metabolomic analyses, new insight into metabolic pathways in relation to health and disease will evolve. Furthermore, these specific metabolic profiles can function as a diagnostic tool for the identification of several diseases, such as ulcerative colitis and Crohn's disease. Until now, the described biomarkers of bacterially mediated colonic metabolism can only be regarded as intermediate end points. An important challenge for current and future research remains to relate changes in bacterial metabolism to human health.
GRANTS
V. De Preter is a postdoctoral fellow of the Fund for Scientific Research-Flanders (F.W.O. Vlaanderen, Belgium).
DISCLOSURES
All authors declare that they have nothing to disclose and that there is no potential conflict of interest.
AUTHOR CONTRIBUTIONS
Author contributions: H.M.H. conception and design of research; H.M.H. and K.V. prepared figures; H.M.H., V.D.P., and K.V. drafted manuscript; H.M.H., V.D.P., K.W., and K.V. edited and revised manuscript; H.M.H., V.D.P., K.W., and K.V. approved final version of manuscript; K.V. interpreted results of experiments.
REFERENCES
- 1. Al-Lahham SH, Roelofsen H, Priebe M, Weening D, Dijkstra M, Hoek A, Rezaee F, Venema K, Vonk RJ. Regulation of adipokine production in human adipose tissue by propionic acid. Eur J Clin Invest 40: 401–407, 2010 [DOI] [PubMed] [Google Scholar]
- 2. Alexander DD, Cushing CA, Lowe KA, Sceurman B, Roberts MA. Meta-analysis of animal fat or animal protein intake and colorectal cancer. Am J Clin Nutr 89: 1402–1409, 2009 [DOI] [PubMed] [Google Scholar]
- 3. Andoh A, Tsujikawa T, Fujiyama Y. Role of dietary fiber and short-chain fatty acids in the colon. Curr Pharm Des 9: 347–358, 2003 [DOI] [PubMed] [Google Scholar]
- 4. Andriamihaja M, Davila AM, Eklou-Lawson M, Petit N, Delpal S, Allek F, Blais A, Delteil C, Tome D, Blachier F. Colon luminal content and epithelial cell morphology are markedly modified in rats fed with a high-protein diet. Am J Physiol Gastrointest Liver Physiol 299: G1030–G1037, 2010 [DOI] [PubMed] [Google Scholar]
- 5. Armougom F, Raoult D. Use of pyrosequencing and DNA barcodes to monitor variations in Firmicutes and Bacteroidetes communities in the gut microbiota of obese humans. BMC Genomics 9: 576, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res 4: 9–14, 2006 [DOI] [PubMed] [Google Scholar]
- 7. Backhed F. Changes in intestinal microflora in obesity: cause or consequence? J Pediatr Gastroenterol Nutr 48, Suppl 2: S56–S57, 2009 [DOI] [PubMed] [Google Scholar]
- 8. Bajor A, Gillberg PG, Abrahamsson H. Bile acids: short and long term effects in the intestine. Scand J Gastroenterol 45: 645–664, 2010 [DOI] [PubMed] [Google Scholar]
- 9. Bammens B, Evenepoel P, Keuleers H, Verbeke K, Vanrenterghem Y. Free serum concentrations of the protein-bound retention solute p-cresol predict mortality in hemodialysis patients. Kidney Int 69: 1081–1087, 2006 [DOI] [PubMed] [Google Scholar]
- 10. Bammens B, Verbeke K, Vanrenterghem Y, Evenepoel P. Evidence for impaired assimilation of protein in chronic renal failure. Kidney Int 64: 2196–2203, 2003 [DOI] [PubMed] [Google Scholar]
- 11. Bartram HP, Scheppach W, Schmid H, Hofmann A, Dusel G, Richter F, Richter A, Kasper H. Proliferation of human colonic mucosa as an intermediate biomarker of carcinogenesis: effects of butyrate, deoxycholate, calcium, ammonia, and pH. Cancer Res 53: 3283–3288, 1993 [PubMed] [Google Scholar]
- 12. Beckonert O, Coen M, Keun HC, Wang Y, Ebbels TM, Holmes E, Lindon JC, Nicholson JK. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc 5: 1019–1032, 2010 [DOI] [PubMed] [Google Scholar]
- 13. Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, Nicholson JK. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc 2: 2692–2703, 2007 [DOI] [PubMed] [Google Scholar]
- 14. Benjdia A, Martens EC, Gordon JI, Berteau O. Sulfatases and a radical S-adenosyl-l-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron. J Biol Chem 286: 25973–25982, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boutron-Ruault MC, Marteau P, Lavergne-Slove A, Myara A, Gerhardt MF, Franchisseur C, Bornet F. Effects of a 3-mo consumption of short-chain fructo-oligosaccharides on parameters of colorectal carcinogenesis in patients with or without small or large colorectal adenomas. Nutr Cancer 53: 160–168, 2005 [DOI] [PubMed] [Google Scholar]
- 16. Chan EC, Koh PK, Mal M, Cheah PY, Eu KW, Backshall A, Cavill R, Nicholson JK, Keun HC. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J Proteome Res 8: 352–361, 2009 [DOI] [PubMed] [Google Scholar]
- 17. Chaplin MF. Bile acids, fibre and colon cancer: the story unfolds. J R Soc Health 118: 53–61, 1998 [DOI] [PubMed] [Google Scholar]
- 18. Cheng K, Raufman JP. Bile acid-induced proliferation of a human colon cancer cell line is mediated by transactivation of epidermal growth factor receptors. Biochem Pharmacol 70: 1035–1047, 2005 [DOI] [PubMed] [Google Scholar]
- 19. Christl SU, Bartram HP, Paul A, Kelber E, Scheppach W, Kasper H. Bile acid metabolism by colonic bacteria in continuous culture: effects of starch and pH. Ann Nutr Metab 41: 45–51, 1997 [DOI] [PubMed] [Google Scholar]
- 20. Cloetens L, De Preter V, De Loor H, Rutgeerts P, Verbeke K. Does the biomarker 15N-lactose ureide allow to estimate the site of fermentation of resistant starch? Eur J Nutr 47: 217–223, 2008 [DOI] [PubMed] [Google Scholar]
- 21. Dallinga JW, Robroeks CM, van Berkel JJ, Moonen EJ, Godschalk RW, Jobsis Q, Dompeling E, Wouters EF, van Schooten FJ. Volatile organic compounds in exhaled breath as a diagnostic tool for asthma in children. Clin Exp Allergy 40: 68–76, 2010 [DOI] [PubMed] [Google Scholar]
- 22. De Preter V, Falony G, Windey K, Hamer HM, De Vuyst L, Verbeke K. The prebiotic, oligofructose-enriched inulin modulates the faecal metabolite profile: an in vitro analysis. Mol Nutr Food Res 54: 1791–1801, 2010 [DOI] [PubMed] [Google Scholar]
- 23. De Preter V, Raemen H, Cloetens L, Houben E, Rutgeerts P, Verbeke K. Effect of dietary intervention with different pre- and probiotics on intestinal bacterial enzyme activities. Eur J Clin Nutr 62: 225–231, 2008 [DOI] [PubMed] [Google Scholar]
- 24. De Preter V, Van Staeyen G, Esser D, Rutgeerts P, Verbeke K. Development of a screening method to determine the pattern of fermentation metabolites in faecal samples using on-line purge-and-trap gas chromatographic-mass spectrometric analysis. J Chromatogr A 1216: 1476–1483, 2009 [DOI] [PubMed] [Google Scholar]
- 25. De Preter V, Vanhoutte T, Huys G, Swings J, De Vuyst L, Rutgeerts P, Verbeke K. Effects of Lactobacillus casei Shirota, Bifidobacterium breve, and oligofructose-enriched inulin on colonic nitrogen-protein metabolism in healthy humans. Am J Physiol Gastrointest Liver Physiol 292: G358–G368, 2007 [DOI] [PubMed] [Google Scholar]
- 26. De Preter V, Vanhoutte T, Huys G, Swings J, Rutgeerts P, Verbeke K. Effect of lactulose and Saccharomyces boulardii administration on the colonic urea-nitrogen metabolism and the bifidobacteria concentration in healthy human subjects. Aliment Pharmacol Ther 23: 963–974, 2006 [DOI] [PubMed] [Google Scholar]
- 27. Demigne C, Morand C, Levrat MA, Besson C, Moundras C, Remesy C. Effect of propionate on fatty acid and cholesterol synthesis and on acetate metabolism in isolated rat hepatocytes. Br J Nutr 74: 209–219, 1995 [DOI] [PubMed] [Google Scholar]
- 28. Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev 26: 51–78, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Di Cagno R, Mazzacane F, Rizzello CG, Vincentini O, Silano M, Giuliani G, De Angelis M, Gobbetti M. Synthesis of isoflavone aglycones and equol in soy milks fermented by food-related lactic acid bacteria and their effect on human intestinal Caco-2 cells. J Agric Food Chem 58: 10338–10346, 2010 [DOI] [PubMed] [Google Scholar]
- 30. Diamant M, Blaak EE, de Vos WM. Do nutrient-gut-microbiota interactions play a role in human obesity, insulin resistance and type 2 diabetes? Obes Rev 12: 272–281, 2011 [DOI] [PubMed] [Google Scholar]
- 31. Evenepoel P, Claus D, Geypens B, Hiele M, Geboes K, Rutgeerts P, Ghoos Y. Amount and fate of egg protein escaping assimilation in the small intestine of humans. Am J Physiol Gastrointest Liver Physiol 277: G935–G943, 1999 [DOI] [PubMed] [Google Scholar]
- 32. Evenepoel P, Meijers BK, Bammens BR, Verbeke K. Uremic toxins originating from colonic microbial metabolism. Kidney Int Suppl S12–S19, 2009 [DOI] [PubMed] [Google Scholar]
- 34. Freeland KR, Wilson C, Wolever TM. Adaptation of colonic fermentation and glucagon-like peptide-1 secretion with increased wheat fibre intake for 1 year in hyperinsulinaemic human subjects. Br J Nutr 103: 82–90, 2010 [DOI] [PubMed] [Google Scholar]
- 35. Garner CE, Smith S, de Lacy Costello B, White P, Spencer R, Probert CS, Ratcliffe NM. Volatile organic compounds from feces and their potential for diagnosis of gastrointestinal disease. FASEB J 21: 1675–1688, 2007 [DOI] [PubMed] [Google Scholar]
- 36. Geboes KP, De Preter V, Luypaerts A, Bammens B, Evenepoel P, Ghoos Y, Rutgeerts P, Verbeke K. Validation of lactose[15N,15N]ureide as a tool to study colonic nitrogen metabolism. Am J Physiol Gastrointest Liver Physiol 288: G994–G999, 2005 [DOI] [PubMed] [Google Scholar]
- 37. Geypens B, Claus D, Evenepoel P, Hiele M, Maes B, Peeters M, Rutgeerts P, Ghoos Y. Influence of dietary protein supplements on the formation of bacterial metabolites in the colon. Gut 41: 70–76, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 125: 1401–1412, 1995 [DOI] [PubMed] [Google Scholar]
- 39. Goldin BR. Intestinal microflora: metabolism of drugs and carcinogens. Ann Med 22: 43–48, 1990 [DOI] [PubMed] [Google Scholar]
- 40. Goldin BR, Swenson L, Dwyer J, Sexton M, Gorbach SL. Effect of diet and Lactobacillus acidophilus supplements on human fecal bacterial enzymes. J Natl Cancer Inst 64: 255–261, 1980 [DOI] [PubMed] [Google Scholar]
- 41. Goossens D, Jonkers D, Russel M, Stobberingh E, Van Den Bogaard A, Stockbrugger R. The effect of Lactobacillus plantarum 299v on the bacterial composition and metabolic activity in faeces of healthy volunteers: a placebo-controlled study on the onset and duration of effects. Aliment Pharmacol Ther 18: 495–505, 2003 [DOI] [PubMed] [Google Scholar]
- 42. Gostner A, Blaut M, Schaffer V, Kozianowski G, Theis S, Klingeberg M, Dombrowski Y, Martin D, Ehrhardt S, Taras D, Schwiertz A, Kleessen B, Luhrs H, Schauber J, Dorbath D, Menzel T, Scheppach W. Effect of isomalt consumption on faecal microflora and colonic metabolism in healthy volunteers. Br J Nutr 95: 40–50, 2006 [DOI] [PubMed] [Google Scholar]
- 43. Grasten SM, Juntunen KS, Poutanen KS, Gylling HK, Miettinen TA, Mykkanen HM. Rye bread improves bowel function and decreases the concentrations of some compounds that are putative colon cancer risk markers in middle-aged women and men. J Nutr 130: 2215–2221, 2000 [DOI] [PubMed] [Google Scholar]
- 44. Grun CH, van Dorsten FA, Jacobs DM, Le Belleguic M, van Velzen EJ, Bingham MO, Janssen HG, van Duynhoven JP. GC-MS methods for metabolic profiling of microbial fermentation products of dietary polyphenols in human and in vitro intervention studies. J Chromatogr B Analyt Technol Biomed Life Sci 871: 212–219, 2008 [DOI] [PubMed] [Google Scholar]
- 45. Haberer P, du Toit M, Dicks LM, Ahrens F, Holzapfel WH. Effect of potentially probiotic lactobacilli on faecal enzyme activity in minipigs on a high-fat, high-cholesterol diet-a preliminary in vivo trial. Int J Food Microbiol 87: 287–291, 2003 [DOI] [PubMed] [Google Scholar]
- 46. Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. The role of butyrate on colonic function. Aliment Pharmacol Ther 27: 104–119, 2008 [DOI] [PubMed] [Google Scholar]
- 47. Hou JK, Abraham B, El-Serag H. Dietary intake and risk of developing inflammatory bowel disease: a systematic review of the literature. Am J Gastroenterol 106: 563–573, 2011 [DOI] [PubMed] [Google Scholar]
- 48. Hughes R, Magee EA, Bingham S. Protein degradation in the large intestine: relevance to colorectal cancer. Curr Issues Intest Microbiol 1: 51–58, 2000 [PubMed] [Google Scholar]
- 49. Imray CH, Radley S, Davis A, Barker G, Hendrickse CW, Donovan IA, Lawson AM, Baker PR, Neoptolemos JP. Faecal unconjugated bile acids in patients with colorectal cancer or polyps. Gut 33: 1239–1245, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jacobs DM, Gaudier E, van Duynhoven J, Vaughan EE. Non-digestible food ingredients, colonic microbiota and the impact on gut health and immunity: a role for metabolomics. Curr Drug Metab 10: 41–54, 2009 [DOI] [PubMed] [Google Scholar]
- 51. Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, Tysk C, Schmitt-Kopplin P. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One 4: e6386, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jenkins DJ, Vuksan V, Kendall CW, Wursch P, Jeffcoat R, Waring S, Mehling CC, Vidgen E, Augustin LS, Wong E. Physiological effects of resistant starches on fecal bulk, short chain fatty acids, blood lipids and glycemic index. J Am Coll Nutr 17: 609–616, 1998 [DOI] [PubMed] [Google Scholar]
- 53. Kovatcheva-Datchary P, Egert M, Maathuis A, Rajilic-Stojanovic M, de Graaf AA, Smidt H, de Vos WM, Venema K. Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ Microbiol 11: 914–926, 2009 [DOI] [PubMed] [Google Scholar]
- 54. Krogius-Kurikka L, Lyra A, Malinen E, Aarnikunnas J, Tuimala J, Paulin L, Makivuokko H, Kajander K, Palva A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol 9: 95, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sorensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5: e9085, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lauridsen MB, Bliddal H, Christensen R, Danneskiold-Samsoe B, Bennett R, Keun H, Lindon JC, Nicholson JK, Dorff MH, Jaroszewski JW, Hansen SH, Cornett C. 1H NMR spectroscopy-based interventional metabolic phenotyping: a cohort study of rheumatoid arthritis patients. J Proteome Res 9: 4545–4553, 2010 [DOI] [PubMed] [Google Scholar]
- 57. Legido-Quigley C, Stella C, Perez-Jimenez F, Lopez-Miranda J, Ordovas J, Powell J, van-der-Ouderaa F, Ware L, Lindon JC, Nicholson JK, Holmes E. Liquid chromatography-mass spectrometry methods for urinary biomarker detection in metabonomic studies with application to nutritional studies. Biomed Chromatogr 24: 737–743, 2010 [DOI] [PubMed] [Google Scholar]
- 58. Leschelle X, Robert V, Delpal S, Mouille B, Mayeur C, Martel P, Blachier F. Isolation of pig colonic crypts for cytotoxic assay of luminal compounds: effects of hydrogen sulfide, ammonia, and deoxycholic acid. Cell Biol Toxicol 18: 193–203, 2002 [DOI] [PubMed] [Google Scholar]
- 59. Lewis S, Cochrane S. Alteration of sulfate and hydrogen metabolism in the human colon by changing intestinal transit rate. Am J Gastroenterol 102: 624–633, 2007 [DOI] [PubMed] [Google Scholar]
- 60. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124: 837–848, 2006 [DOI] [PubMed] [Google Scholar]
- 61. Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Wei H, Chen Y, Lu H, Zuo J, Su M, Qiu Y, Jia W, Xiao C, Smith LM, Yang S, Holmes E, Tang H, Zhao G, Nicholson JK, Li L, Zhao L. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA 105: 2117–2122, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ling WH, Korpela R, Mykkanen H, Salminen S, Hanninen O. Lactobacillus strain GG supplementation decreases colonic hydrolytic and reductive enzyme activities in healthy female adults. J Nutr 124: 18–23, 1994 [DOI] [PubMed] [Google Scholar]
- 63. Luhrs H, Gerke T, Muller JG, Melcher R, Schauber J, Boxberge F, Scheppach W, Menzel T. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol 37: 458–466, 2002 [DOI] [PubMed] [Google Scholar]
- 64. Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA 106: 5859–5864, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Marteau P, Pochart P, Flourie B, Pellier P, Santos L, Desjeux JF, Rambaud JC. Effect of chronic ingestion of a fermented dairy product containing Lactobacillus acidophilus and Bifidobacterium bifidum on metabolic activities of the colonic flora in humans. Am J Clin Nutr 52: 685–688, 1990 [DOI] [PubMed] [Google Scholar]
- 66. McBain AJ, MacFarlane GT. Modulation of genotoxic enzyme activities by non-digestible oligosaccharide metabolism in in-vitro human gut bacterial ecosystems. J Med Microbiol 50: 833–842, 2001 [DOI] [PubMed] [Google Scholar]
- 67. Medani M, Collins D, Docherty NG, Baird AW, O'Connell PR, Winter DC. Emerging role of hydrogen sulfide in colonic physiology and pathophysiology. Inflamm Bowel Dis 17: 1620–1625, 2011 [DOI] [PubMed] [Google Scholar]
- 68. Meijer K, de Vos P, Priebe MG. Butyrate and other short-chain fatty acids as modulators of immunity: what relevance for health? Curr Opin Clin Nutr Metab Care 13: 715–721, 2010 [DOI] [PubMed] [Google Scholar]
- 69. Meijers BK, De Preter V, Verbeke K, Vanrenterghem Y, Evenepoel P. p-Cresyl sulfate serum concentrations in haemodialysis patients are reduced by the prebiotic oligofructose-enriched inulin. Nephrol Dial Transplant 25: 219–224, 2010 [DOI] [PubMed] [Google Scholar]
- 70. Meijers BK, Van Kerckhoven S, Verbeke K, Dehaen W, Vanrenterghem Y, Hoylaerts MF, Evenepoel P. The uremic retention solute p-cresyl sulfate and markers of endothelial damage. Am J Kidney Dis 54: 891–901, 2009 [DOI] [PubMed] [Google Scholar]
- 71. Nakamura J, Kubota Y, Miyaoka M, Saitoh T, Mizuno F, Benno Y. Comparison of four microbial enzymes in Clostridia and Bacteroides isolated from human feces. Microbiol Immunol 46: 487–490, 2002 [DOI] [PubMed] [Google Scholar]
- 72. Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29: 1181–1189, 1999 [DOI] [PubMed] [Google Scholar]
- 73. Nilsson AC, Ostman EM, Knudsen KE, Holst JJ, Bjorck IM. A cereal-based evening meal rich in indigestible carbohydrates increases plasma butyrate the next morning. J Nutr 140: 1932–1936, 2010 [DOI] [PubMed] [Google Scholar]
- 74. O'Keefe SJ. Nutrition and colonic health: the critical role of the microbiota. Curr Opin Gastroenterol 24: 51–58, 2008 [DOI] [PubMed] [Google Scholar]
- 75. Ouwehand AC, Derrien M, de Vos W, Tiihonen K, Rautonen N. Prebiotics and other microbial substrates for gut functionality. Curr Opin Biotechnol 16: 212–217, 2005 [DOI] [PubMed] [Google Scholar]
- 76. Penders J, Stobberingh EE, van den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy 62: 1223–1236, 2007 [DOI] [PubMed] [Google Scholar]
- 77. Pouteau E, Vahedi K, Messing B, Flourie B, Nguyen P, Darmaun D, Krempf M. Production rate of acetate during colonic fermentation of lactulose: a stable-isotope study in humans. Am J Clin Nutr 68: 1276–1283, 1998 [DOI] [PubMed] [Google Scholar]
- 78. Probert CS, Ahmed I, Khalid T, Johnson E, Smith S, Ratcliffe N. Volatile organic compounds as diagnostic biomarkers in gastrointestinal and liver diseases. J Gastrointestin Liver Dis 18: 337–343, 2009 [PubMed] [Google Scholar]
- 79. Rajilic-Stojanovic M, Heilig HG, Molenaar D, Kajander K, Surakka A, Smidt H, de Vos WM. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol 11: 1736–1751, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Robroeks CM, van Berkel JJ, Dallinga JW, Jobsis Q, Zimmermann LJ, Hendriks HJ, Wouters MF, van der Grinten CP, van de Kant KD, van Schooten FJ, Dompeling E. Metabolomics of volatile organic compounds in cystic fibrosis patients and controls. Pediatr Res 68: 75–80, 2010 [DOI] [PubMed] [Google Scholar]
- 81. Roediger WE, Duncan A, Kapaniris O, Millard S. Sulphide impairment of substrate oxidation in rat colonocytes: a biochemical basis for ulcerative colitis? Clin Sci (Lond) 85: 623–627, 1993 [DOI] [PubMed] [Google Scholar]
- 82. Roediger WE, Moore J, Babidge W. Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig Dis Sci 42: 1571–1579, 1997 [DOI] [PubMed] [Google Scholar]
- 83. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9: 313–323, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rowan FE, Docherty NG, Coffey JC, O'Connell PR. Sulphate-reducing bacteria and hydrogen sulphide in the aetiology of ulcerative colitis. Br J Surg 96: 151–158, 2009 [DOI] [PubMed] [Google Scholar]
- 85. Rowland IR. The role of the gastrointestinal microbiota in colorectal cancer. Curr Pharm Des 15: 1524–1527, 2009 [DOI] [PubMed] [Google Scholar]
- 86. Rowland IR, Mallett AK, Wise A. The effect of diet on the mammalian gut flora and its metabolic activities. Crit Rev Toxicol 16: 31–103, 1985 [DOI] [PubMed] [Google Scholar]
- 87. Rowland IR, Rumney CJ, Coutts JT, Lievense LC. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis 19: 281–285, 1998 [DOI] [PubMed] [Google Scholar]
- 88. Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, Gordon JI. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA 105: 16767–16772, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Schwiertz A, Taras D, Schafer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 18: 190–195, 2009 [DOI] [PubMed] [Google Scholar]
- 90. Sharma BC, Sharma P, Agrawal A, Sarin SK. Secondary prophylaxis of hepatic encephalopathy: an open-label randomized controlled trial of lactulose versus placebo. Gastroenterology 137: 885–891, 2009 [DOI] [PubMed] [Google Scholar]
- 91. Sleeth ML, Thompson EL, Ford HE, Zac-Varghese SE, Frost G. Free fatty acid receptor 2 and nutrient sensing: a proposed role for fibre, fermentable carbohydrates and short-chain fatty acids in appetite regulation. Nutr Res Rev 23: 135–145, 2010 [DOI] [PubMed] [Google Scholar]
- 92. Spanhaak S, Havenaar R, Schaafsma G. The effect of consumption of milk fermented by Lactobacillus casei strain Shirota on the intestinal microflora and immune parameters in humans. Eur J Clin Nutr 52: 899–907, 1998 [DOI] [PubMed] [Google Scholar]
- 93. Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis in inflammatory bowel disease. Gut 53: 1–4, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tang Y, Chen Y, Jiang H, Robbins GT, Nie D. G-protein-coupled receptor for short-chain fatty acids suppresses colon cancer. Int J Cancer 128: 847–856, 2011 [DOI] [PubMed] [Google Scholar]
- 95. Tannock GW. Molecular assessment of intestinal microflora. Am J Clin Nutr 73: 410S–414S, 2001 [DOI] [PubMed] [Google Scholar]
- 96. Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, Mellinger JD, Smith SB, Digby GJ, Lambert NA, Prasad PD, Ganapathy V. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res 69: 2826–2832, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Toden S, Bird AR, Topping DL, Conlon MA. Resistant starch attenuates colonic DNA damage induced by higher dietary protein in rats. Nutr Cancer 51: 45–51, 2005 [DOI] [PubMed] [Google Scholar]
- 98. Tong JL, Ran ZH, Shen J, Fan GQ, Xiao SD. Association between fecal bile acids and colorectal cancer: a meta-analysis of observational studies. Yonsei Med J 49: 792–803, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Van Berkel JJ, Dallinga JW, Moller GM, Godschalk RW, Moonen EJ, Wouters EF, Van Schooten FJ. A profile of volatile organic compounds in breath discriminates COPD patients from controls. Respir Med 104: 557–563, 2010 [DOI] [PubMed] [Google Scholar]
- 100. van Munster IP, Tangerman A, Nagengast FM. Effect of resistant starch on colonic fermentation, bile acid metabolism, and mucosal proliferation. Dig Dis Sci 39: 834–842, 1994 [DOI] [PubMed] [Google Scholar]
- 101. Ventura M, Turroni F, Canchaya C, Vaughan EE, O'Toole PW, van Sinderen D. Microbial diversity in the human intestine and novel insights from metagenomics. Front Biosci 14: 3214–3221, 2009 [DOI] [PubMed] [Google Scholar]
- 102. Verbeke K, Ferchaud-Roucher V, Preston T, Small AC, Henckaerts L, Krempf M, Wang H, Vonk RJ, Priebe MG. Influence of the type of indigestible carbohydrate on plasma and urine short-chain fatty acid profiles in healthy human volunteers. Eur J Clin Nutr 64: 678–684, 2010 [DOI] [PubMed] [Google Scholar]
- 103. Vitali B, Ndagijimana M, Cruciani F, Carnevali P, Candela M, Guerzoni ME, Brigidi P. Impact of a synbiotic food on the gut microbial ecology and metabolic profiles. BMC Microbiol 10: 4, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Waldram A, Holmes E, Wang Y, Rantalainen M, Wilson ID, Tuohy KM, McCartney AL, Gibson GR, Nicholson JK. Top-down systems biology modeling of host metabotype-microbiome associations in obese rodents. J Proteome Res 8: 2361–2375, 2009 [DOI] [PubMed] [Google Scholar]
- 105. Walker AW, Sanderson JD, Churcher C, Parkes GC, Hudspith BN, Rayment N, Brostoff J, Parkhill J, Dougan G, Petrovska L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol 11: 7, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Watanabe A, Sakai T, Sato S, Imai F, Ohto M, Arakawa Y, Toda G, Kobayashi K, Muto Y, Tsujii T, Kawasaki H, Okita K, Tanikawa K, Fujiyama S, Shimada S. Clinical efficacy of lactulose in cirrhotic patients with and without subclinical hepatic encephalopathy. Hepatology 26: 1410–1414, 1997 [DOI] [PubMed] [Google Scholar]
- 107. Weaver GA, Krause JA, Miller TL, Wolin MJ. Short chain fatty acid distributions of enema samples from a sigmoidoscopy population: an association of high acetate and low butyrate ratios with adenomatous polyps and colon cancer. Gut 29: 1539–1543, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Weber FL, Jr, Banwell JG, Fresard KM, Cummings JH. Nitrogen in fecal bacterial, fiber, and soluble fractions of patients with cirrhosis: effects of lactulose and lactulose plus neomycin. J Lab Clin Med 110: 259–263, 1987 [PubMed] [Google Scholar]
- 109. Williams HR, Cox IJ, Walker DG, North BV, Patel VM, Marshall SE, Jewell DP, Ghosh S, Thomas HJ, Teare JP, Jakobovits S, Zeki S, Welsh KI, Taylor-Robinson SD, Orchard TR. Characterization of inflammatory bowel disease with urinary metabolic profiling. Am J Gastroenterol 104: 1435–1444, 2009 [DOI] [PubMed] [Google Scholar]
- 110. Wolever TM, Fernandes J, Rao AV. Serum acetate:propionate ratio is related to serum cholesterol in men but not women. J Nutr 126: 2790–2797, 1996 [DOI] [PubMed] [Google Scholar]
- 111. Xue R, Dong L, Zhang S, Deng C, Liu T, Wang J, Shen X. Investigation of volatile biomarkers in liver cancer blood using solid-phase microextraction and gas chromatography/mass spectrometry. Rapid Commun Mass Spectrom 22: 1181–1186, 2008 [DOI] [PubMed] [Google Scholar]
- 112. Zimber A, Gespach C. Bile acids and derivatives, their nuclear receptors FXR, PXR and ligands: role in health and disease and their therapeutic potential. Anticancer Agents Med Chem 8: 540–563, 2008 [DOI] [PubMed] [Google Scholar]
- 113. Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57: 1605–1615, 2008 [DOI] [PubMed] [Google Scholar]