Abstract
Lentiviral vectors (LV) are widely used to stably transfer genes into target cells investigating or treating gene functions. In addition, gene transfer into early murine embryos may be improved to efficiently generate transgenic mice. We applied lentiviral gene transfer to generate a mouse model transgenic for SET binding protein-1 (Setbp1) and enhanced green fluorescent protein (eGFP). Neither transgenic founders nor their vector-positive offspring transcribed or expressed the transgenes. Bisulfite sequencing of the internal spleen focus-forming virus (SFFV) promoter demonstrated extensive methylation of all analyzed CpGs in the transgenic mice. To analyze the impact of Setbp1 on epigenetic silencing, embryonic stem cells (ESC) were differentiated into cardiomyocytes (CM) in vitro. In contrast to human promoters in LV, virally derived promoter sequences were strongly methylated during differentiation, independent of the transgene. Moreover, the commonly used SFFV promoter (SFFVp) was highly methylated with remarkable strength and frequency during hematopoietic differentiation in vivo in LV but less in γ-retroviral (γ-RV) backbones. In summary, we conclude that LV using an internal SFFVp are not suitable to generate transgenic mice or perform constitutive expression studies in differentiating cells. Choosing the appropriate promoter is also crucial to allow stable transgene expression in clinical gene therapy.
Introduction
Lentiviral vectors (LV) stably express transgenes due to integration into the host cell genome. Pseudotyping with the vesicular-stomatitis virus glycoprotein1 leads to a broad target cell spectrum including stem cells or fertilized eggs.2 LV facilitate the generation of transgenic mice with a higher degree of vector-positive offspring than achieved by DNA microinjection3 while being less time consuming than homologous recombination and are therefore a promising tool for the generation of transgenic mammalians to investigate gene functions.4,5
Several studies have shown that host cells can block expression of endogenous6,7,8,9 as well as exogenous retroviral (RV) elements by complex defence mechanisms in embryonic stem cells (ESC).10,11,12,13,14,15 Silencing can be mediated through binding of transacting factors expressed by the target cell to the proviral long-terminal repeats (LTRs) or by methylation of CpG sites within the integrated provirus and flanking host cell DNA sequences. Extinction of provirus expression during long-term culture or differentiation is less well understood.13,16 CpG methylation of the viral LTR promoter leads to transcriptional repression of different retroviruses like Moloney murine leukemia virus, human T-cell leukemia virus-1, and Rous sarcoma virus. Moreover, LTR methylation has been shown to cause provirus silencing in human immunodeficiency virus type-1 LTRs and thereby defines viral latency.17,18,19
The use of self-inactivating human immunodeficiency virus type-1-based LV containing truncated LTR sequences for gene transfer into ESC in vitro allows efficient ectopic gene expression in undifferentiated and differentiated cells.20 Lentiviral gene transfer into human adult hematopoietic stem cells mediates stable long-term expression of transgenes driven by the spleen focus-forming virus (SFFV) promoter in vivo.21,22
Here, we applied lentiviral gene transfer to generate mice transgenic for the nuclear protein SET binding protein-1 (Setbp1) and the fluorescent marker enhanced green fluorescent protein (eGFP) driven by the SFFV promoter (SFFVp). We demonstrate complete transgene silencing due to promoter methylation during differentiation of embryonic and somatic stem cells. These results have significant implications for the use of lentiviral gene transfer in murine transgenesis and emphasize that long-term expression of viral promoters during differentiation has to be tested before their application in clinical stem cell gene therapy.
Results
Lentiviral gene transfer into zygotes is highly efficient resulting in transgenic mice with single vector integrations
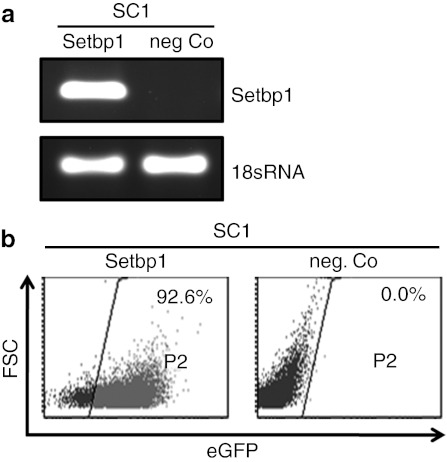
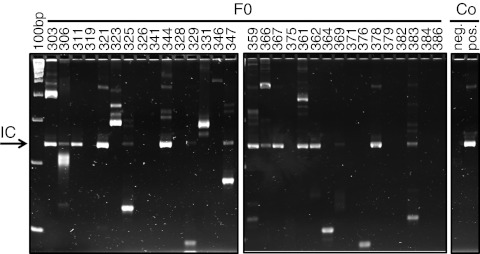
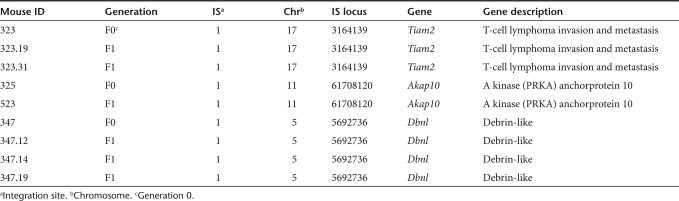
In an initial set of experiments, transcription and expression of LV encoding for Setbp1 and eGFP were tested in vitro (Figure 1). To generate mice transgenic for Setbp1 and eGFP, lentiviral particles were injected into the perivitelline space of early stage embryos (dE0.5). Infected zygotes were cultured overnight and transferred into foster mothers. To minimize insertional mutagenesis, vector particle numbers were adjusted to generate animals carrying single copies of the transgenic insert. Three weeks after birth, transgenic newborn mice (F0) were identified by PCRs specific for the Setbp1 transgene and the Wpre element within the lentiviral construct. Individual lentiviral integration loci were determined in 20 of 31 F0 mice by linear amplification-mediated (LAM)-PCR (Figure 2). A total of 11 (9 male and 2 female) mice were tested for propagation of the transgene into offspring mice. 6 out of 11 F0 mice showed germline transmission with 8–53% vector-positive offspring. Integration site analyses by LAM or nonrestrictive LAM-PCR followed by 454 deep-sequencing demonstrated single vector integrations in all analyzed transgenic mice (n = 11) (Table 1).
Figure 1.
Functionality of Setbp1 encoding self-inactivating lentiviral vector. (a) Transcription of Setbp1 mRNA in lentiviral vector (LV)-infected murine SC1-cells (Setbp1) and in uninfected control cells (neg. Co). (b) Enhanced green fluorescent protein (eGFP)-positive SC1-cells in % after infection with LV.Setbp1 and in uninfected control cells. FSC, forward scatter.
Figure 2.
Lentiviral gene transfer results in transgenic mice with single vector integrations. Lentiviral vector (LV) integration sites were detected in 65% of 31 analyzed founder (F0) mice by linear amplification-mediated (LAM)-PCR.
Table 1. Lentiviral gene transfer results in transgenic mice with single vector integrations.
Methylation of SFFVp sequences does not allow lentiviral transgene expression in vivo
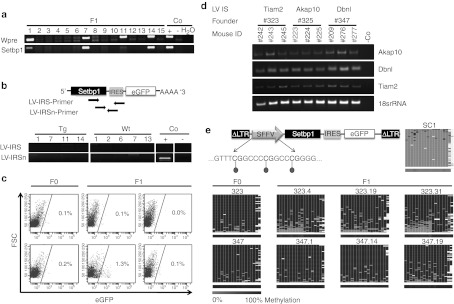
Despite genomic integration of the provirus and highly efficient germline transmission, no ectopic transcription and expression of Setbp1 or eGFP was detectable in transgenic mice (Figure 3). The presence of endogenous Setbp1 mRNA in vector-positive F1 mice (Figure 3a) (n = 14) was verified by reverse transcription-PCR, whereas no LV-specific Setbp1 mRNA (Figure 3b) or eGFP expression was detectable in whole body slices. Only in one out of 49 mice (F0, n = 20; F1, n = 24; F2, n = 5) 1.3% of all nucleated peripheral blood cells expressed eGFP. In the other 48 mice <0.3% eGFP+ blood cells were detected (Figure 3c). To investigate whether the lack of expression from the lentivirally introduced transgene is caused by epigenetic mechanisms, we first analyzed whether the chromatin allows the transcription of genes adjacent to the proviral integration sites. Akap10, Dbnl, and Tiam2 genes neighboring the lentiviral integration sites were equally expressed in peripheral blood cells of transgenic and wild-type mice (n = 9) (Figure 3d). Furthermore, we determined the methylation status of the internal SFFVp by deep bisulfite sequencing. Interestingly, 18 of 18 analyzed CpGs within the promoter region were extensively methylated (>90%) in F0 animals and all F1 progeny analyzed (n = 12) (Figure 3d). The same promoter region was completely unmethylated (<3%) in transduced control cells (SC1; Figure 3e) and lentiviral transgene expression could be detected verifying vector functionality (Figure 1).
Figure 3.
No ectopic transgene transcription and expression in transgenic mice due to extensive methylation of spleen focus-forming virus promoter (SFFVp). (a) Genotyping-PCR analyzing tail-tip DNA of F1 mice. (b) Scheme of primer localization and reverse transcription-PCR (RT-PCR) of transgenic (Tg) and wild-type (Wt) F1 mouse whole body slices; Co, controls. (c) Peripheral blood of 49 mice was analyzed for enhanced green fluorescent protein (eGFP) expression (six representative fluorescence-activated cell sorting (FACS) blots are shown here). (d) RT-PCRs verifying transcription of genes adjacent to the proviral vector integration in progeny of three different transgenic founder mice (founder mice 323, 325, and 347). Mouse numbers in black represent transgenic mice, numbers in gray the wild-type counterparts. LV IS, lentiviral integration site. (e) Scheme of transgenic LV and the analyzed CpGs within promoter region. Heat maps for 454 reads (rows) show the methylation status of 18 CpGs (columns) within the SFFVp region in transduced control (SC1) cells and in transgenic animals (n = 8 representatives). Methylated CpGs are shown in dark gray, unmethylated CpGs in light gray and gaps in white. The average methylation level for each CpG site is visualized in the lower bar using a light gray to black color scale. FSC, forward scatter.
Spontaneous differentiation of lentivirally transduced ESC in vitro leads to silencing of SFFVp sequences
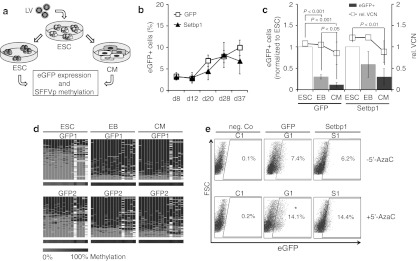
To understand the kinetics of transgene silencing, 13 embryos were transduced and cultured in vitro for additional 4 days. As early as 48 and 96 hours after injection of the lentiviral vector, no eGFP expression was detectable by fluorescence microscopy even though lentiviral vector integrations could be detected by LAM-PCR (Supplementary Figure S1). Next, we investigated whether the Setbp1 transgene may promote the observed methylation of the SFFVp sequence. To understand the influence of proliferation and differentiation on transgene silencing, murine development was mimicked in vitro by mouse ESC (ESC) differentiation into embryoid bodies (EB). EB were further differentiated into spontaneously beating cardiomyocytes (CM) by replating trypsinized single cells on cell-culture dishes. Following transduction with LV.Setbp1 or the corresponding SFFV driven control vector LV.eGFP, eGFP expression in ESC was analyzed at different time points after initiation of differentiation. The percentage of eGFP-positive cells in transduced ESC cultures increased from 3.1 ± 0.6% at day 8 to 6.8 ± 3.0% at day 37 in LV.Setbp1 transduced cells and from 3.3 ± 0.9% to 9.9 ± 1.8% in cells transduced with the control vector (Figure 4b). Interestingly, 5 days after EB formation eGFP expression decreased 1.8 ± 0.1- and 3.5 ± 0.5-fold in differentiated ESC (P < 0.05) infected with LV.Setbp1 and LV.eGFP, respectively (Figure 4c). The relative vector copy number per target cell was almost stable during differentiation. Only a slight and statistically not significant decrease of relative vector copy number from 1.1 ± 0.004 to 0.9 ± 0.3 in LV.eGFP (P >0.05) and from 1.2 to 0.9 ± 0.3 in LV.Setbp1 transduced cells was observed during differentiation (Figure 4c).
Figure 4.
Enhanced green fluorescent protein (eGFP) expression remains stable in undifferentiated mouse embryonic stem cells (ESC) transduced with spleen focus-forming virus (SFFV) driven lentiviral vectors (LV) but decreases after spontaneous differentiation in vitro caused by promoter silencing. (a) Experimental design to assess the silencing of SFFV promoter sequences in ESC before and after spontaneous differentiation. (b) eGFP expression in undifferentiated embryonic stem cells transduced with control eGFP (G) or Setbp1 (S) encoding vector for up to 37 days after transduction. (c) Left y-axis: eGFP expression of LV transduced ESC, differentiated EB and CM (normalized to eGFP expression of ESC). Right y-axis: Relative vector copy number (VCN) determined by real-time PCR using LC480 (Roche Diagnostics) in G and in S transduced cells during differentiation. (d) Heat maps for 454 reads (rows) show the methylation status of 16 CpGs (columns) within SFFV promoter (SFFVp) in LV transduced ESC during differentiation. (e) eGFP expression (representative blots) of CM at day 4 after 5′-azacytidine (5′-AzaC) treatment (0.5 µmol/l). FSC, forward scatter.
Methylation of the SFFVp after differentiation into CM was confirmed by deep bisulfite sequencing. Whereas in undifferentiated cells around 45% of analyzed CpGs within the promoter region harbored methylation marks, the degree of methylation increased up to 80% during EB formation and then remained stable after further differentiation into CM (Figure 4d). To verify silencing of SFFVp sequences by DNA methylation, spontaneously differentiated CM were treated with 5′-azacytidine and analyzed by fluorescence-activated cell sorting. eGFP expression increased from 7.9 ± 0.7% to 14.2 ± 0.1% (P < 0.05) at day 4 after 5′-AzaC treatment in LV.eGFP transduced cells and from 6.0 ± 0.4% to 12.6 ± 2.6% in LV.Setbp1 cells (Figure 4e).
Enhanced silencing of viral promoters during ESC differentiation
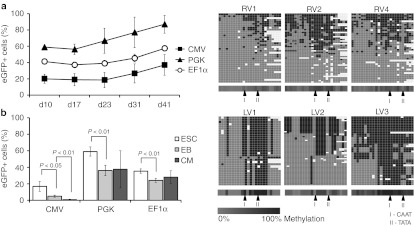
We then asked whether virally derived promoters in LV are more prone to methylation during differentiation than promoters of human origin. To address this question, undifferentiated mESC were infected with the virally derived cytomegalovirus (CMV) promoter or phosphoglycerate kinase (PGK) and elongation factor 1α (EF1α) promoter (both human derivatives) driven LV encoding for eGFP, and eGFP expression was analyzed for up to 41 days. Transduced mESC stably expressed eGFP independent of the promoter used (Figure 5a). In contrast, when ESC were induced to differentiate into CM, CMV promoter-driven eGFP expression decreased 3.6 ± 1.5-fold (P < 0.05) during EB formation whereas eGFP expression driven by the human promoters PGK and EF1α was only slightly reduced (PGK promoter 1.7 ± 0.2-fold decrease in eGFP+ cells and EF1α 1.5 ± 0.05-fold decrease in eGFP+ cells; P < 0.01). Moreover, further differentiation significantly reduced CMV promoter-driven eGFP expression in LV transduced CM 5.2 ± 3.1- fold (P < 0.01), whereas PGK and EF1α driven expression remained stable (1.2 ± 0.7-fold change in PGK cells and 0.9 ± 0.2-fold change in EF1α cells; P > 0.05) (Figure 5b). In line with this, highly purified eGFP-expressing LV.PGK transduced ESC cultures showed only a slight decrease in the percentage of eGFP+ cells after effective initiation of differentiation, whereas the proportion of sorted LV.CMV transduced eGFP+ cells was largely reduced (~147-fold; P = 0.0014) after differentiation into EB (Supplementary Figure S2).
Figure 5.
Silencing of virally derived internal promoters during differentiation in lentivirally transduced embryonic and adult stem cells. (a) Enhanced green fluorescent protein (eGFP) expression over time in undifferentiated mouse embryonic stem cells (ESC) after transduction of CMV, PGK, or EF1α driven lentiviral vectors (MOI 50). (b) Percentage of eGFP+ cells during differentiation of transduced ESC with CMV, PGK, and EF1α driven lentiviral vectors. CM, cardiomyocytes; EB, embryoid bodies. (c) Heat maps for 454 reads (rows) show the methylation status of 26 CpGs (columns) within the spleen focus-forming virus promoter (SFFVp) in lentiviral vector (LV) and γ-retroviral vector (RV) backbone, 20 weeks after bone marrow transplantation. Methylated CpGs are shown in dark gray, unmethylated CpGs in light gray and gaps in white. The average methylation level for each CpG site is visualized in the lower bar using a light gray to black color scale.
High degree of promoter methylation around CAAT- and TATA-box elements in LV in vivo
To investigate whether silencing of lentiviral promoter sequences during differentiation also occurs in adult somatic stem cells, we analyzed eGFP expression and methylation status of SFFVp sequences in LV.eGFP transduced adult bone marrow hematopoiesis. Four weeks after bone marrow transplantation (BMT), 5.4–17.6% of donor-derived mononuclear cells expressed eGFP. Subsequently, eGFP expression decreased to 0.0–0.6% eGFP+ cells at 36 weeks after BMT whereas the proportion of donor-derived CD45.1+ cells remained stable (Supplementary Table S2). In two additional experiments, murine bone marrow cells were transduced with LV.SFFV using low and very high gene transfer efficiencies and transplanted into myeloablated mice (Supplementary Table S2). Epigenetic silencing of the internal SFFVp was independent of the transduction efficiency. In the first experiment, transduction efficiency was 3.1% resulting in 3.0 ± 1.4% blood cells expressing eGFP at 4 weeks after BMT (n = 3). At 52 weeks after transplantation, eGFP expression was decreased to 0.5 ± 0.3%. In the second experiment, 65% of lineage depleted Sca1+ c-kit+ (LSK) cells expressed eGFP before transplantation (n = 2) and at 2 weeks after BMT 48.4 ± 0.9% eGFP+ donor-derived mononuclear peripheral blood cells were detected. Again, the percentage of eGFP+ donor cells decreased in vivo to 9.2 ± 3.8% at 7 weeks after BMT and to 2.7 ± 2.5% at 40 weeks after BMT.
To determine whether promoter silencing is specific to LV, we compared the methylation of CpGs within SFFVp in LV to CpGs of the same promoter in a γ-retroviral vector (γ-RV) backbone. CD45.1+ male bone marrow cells were transduced with γ-RV (transduction efficiency 29%) or LV (transduction efficiency 13%) and transplanted in congeneic CD45.2 female recipients. Peripheral blood of transplanted mice (n = 7) was collected at different time points after transplantation of γ-RV and LV transduced bone marrow. Only 1.0 and 0.1% of the LV transduced cells expressed eGFP after BMT, whereas eGFP expression in γ-RV-transduced cells remained stable ranging from 29.1% GFP+ cells (8 weeks after transplantation) to 39.3% GFP+ cells (36 weeks after transplantation) (Supplementary Table S2). Deep bisulfite sequencing revealed that SFFVp in LV (n = 3) were strongly methylated, especially around CAAT- and TATA-Box elements, resulting in lack of transgene expression (Figure 5c). In contrast, the same promoter in a γ-RV backbone (n = 3) showed a diffuse and nonrestricted methylation pattern (Figure 5c).
Lastly, to examine whether SFFVp sequences are particularly prone to silencing, eGFP expression in BM cells transduced with eGFP-expressing LV driven by CMV or PGK promoters were analyzed. Transduction efficiencies were 5.9 and 29.7% for LV.CMV and LV.PGK, respectively. Every 4 weeks after BMT, peripheral blood of the transplanted mice (n = 3 per group) was analyzed for eGFP expression in mononuclear cells. In two mice, CMV driven eGFP expression decreased from 20.2–26.2% of donor-derived cells at 4 weeks to 2.5–6.2% at 16 weeks after transplantation. In contrast, PGK-driven eGFP expression was more stable with 23.5–32.2% GFP+ donor-cells at 4 weeks after BMT and 10.9–29.2% 48 weeks later (Supplementary Table S2). To analyze the kinetics of eGFP expression ex vivo, lineage depleted (lin–) bone marrow cells were transduced with LV driven by CMV, PGK, or SFFVp. Under culture conditions that favor stem cell maintenance, stable or even increasing proportions of eGFP+ cells were detectable until d23 after transduction (Supplementary Figure S3a). In addition, we observed stable eGFP expression after γ-RV transduction of lin− bone marrow cells in vitro (Supplementary Figure S3b).
Taken together, these data indicate that SFFVp driven LV become highly methylated in close proximity to CAAT- and TATA-box elements during murine blood cell development in vivo, whereas SFFV sequences within γ-RV vectors show a scattered methylation pattern. Furthermore, transplantation of CMV regulated LV transduced bone marrow cells suggests that additional virally derived promoters are silenced during differentiation in vivo.
Discussion
Here, we demonstrate that SFFVp driven transgene expression in mice generated by lentiviral transduction is lost during differentiation. Despite highly efficient lentiviral gene transfer into early murine embryos no transcription or transgene expression was detectable in vivo. Variegated RV expression has been reported if the vector copy number is below 323 and not all single copy integrants express well.2,24 Nevertheless, it is highly unlikely that the lack of expression was vector integration site dependent for several reasons: First, in none of 15 different founder mice carrying individual LV integrations at varying sites within the genome as well as in progeny of five selected founders, transgene expression was detectable. Second, the genomic region allowed the transcription of endogenous genes as detected in the offspring of three different transgenic founder mice. And third, comparable silencing of the promoter region occurred within the lentiviral vector in differentiating ESC as well as in adult hematopoietic stem cells.
DNA methylation analysis demonstrated that the internal SFFVp was completely methylated in all analyzed founders and their progeny. It has been suggested previously that self-inactivating LV are less prone to silencing during murine development2,25 than γ-RV-derived vectors as enhancer/promoter sequences within the LTRs are deleted20,26,27 which are otherwise targeted by host defence mechanisms.12 Cellular transcriptional repressors cannot bind to these modified LTR sequences to suppress viral expression as shown for murine leukemia-based viruses. In line with these findings, it has been suggested that lentiviral extinction by means of long-term silencing of viral transgene expression in a methylation-dependent manner does not occur in lentivirally derived transgenic mice.2,25,28 However, our results clearly demonstrate complete methylation of lentiviral promoter sequences in transgenic mice.
In vitro differentiation of lentivirally transduced ESC led to a substantial decrease of eGFP expression due to CpG methylation, whereas undifferentiated ESC stably maintained eGFP expression. This differentiation driven promoter methylation was independent of the transgene Setbp1. Interestingly, it has been shown that the embryonic cancer cell line P19 partially methylates lentiviral SFFVp sequences during in vitro culture without apparent differentiation29 in a dynamic process combining DNA methylation and chromatin modification.30 Our results indicate that virally derived promoters are recognized by methylating defence mechanisms whereas endogenous promoter sequences are not silenced and stably express the transgenes.
To determine whether silencing of the SFFVp is restricted to early embryonic development we analyzed CpG methylation in the adult hematopoietic system of mice receiving LV- and γ-RV-transduced bone marrow transplants. Again, no lentiviral transgene expression was detectable in LV transduced differentiated peripheral blood cells at 8 weeks after transplantation. Interestingly, mice transplanted with γ-RV-transduced cells showed stable SFFVp mediated expression of eGFP for several months, together with scattered CpG methylation. It can be speculated that intact active enhancer sequences within the γ-RV LTR sequences might have stabilized the expression level in addition to low levels of methylation at SFFVp CpGs. We found CpG methylation of SFFVp particularly around CAAT- and TATA-box sequences in LV-infected cells as also described by Zhang et al.29 Moreover, we observed a decrease not only in SFFV but also in CMV promoter driven transgene expression in vivo, whereas the endogenous PGK promoter conferred stable long-term eGFP expression in mononuclear cells following transplantation. Even though the number of eGFP+ donor cells driven by virally derived promoters decreased in peripheral blood cells over time both after high- and low-gene transfer efficiency (3–65%), the number of mice analyzed does not allow a direct comparison of vector silencing in cells with different transduction efficiencies. However, CpG dinucleotides within the PGK promoter cluster in one CpG island (with 25% CpGs in total) whereas virally derived promoters include a lower CpG content (11–12% CpGs in total). Interestingly, also CpGs within the SFFVp sequences cluster in one CpG island (Supplementary Figure S4), usually characterized by a chromatin permissive state.31 Previously, we reported that in two patients treated by γ-RV gene therapy for chronic granulomatous disease the therapeutic transgene gp91phox was silenced due to SFFVp methylation.32,33 Nevertheless, this finding was unusual as RV vectors mediated stable long-term expression of transduced hematopoiesis in numerous successful human gene therapy studies.34,35,36 In the first clinical gene therapy study using LV, stable transgene expression without genotoxic side effects was achieved that allowed the correction of X-linked adrenoleukodystrophy.37 Interestingly, our data provide evidence that the methylation pattern of the internal SFFVp may be dependent on the viral backbone used and that the promoter within LV seems to be systematically methylated. To circumvent epigenetic silencing, it has been suggested to combine insulator elements with tissue specific endogenous promoters for long-term correction of genetic diseases like chronic granulomatous disease.38 In line with our results, it was shown that fused promoter sequences containing the A2UCOE (ubiquitous chromatin opening elements derived from the human HNRPA2B1-CBX3 locus) element and the myeloid specific MRP8 promoter in LV are methylated in P19 embryonic carcinoma cells. Careful evaluation of promoter methylation should be done during the preclinical development of therapeutic LV.
In summary, we demonstrate that the commonly used SFFVp is highly methylated with remarkable strength and frequency during development in vivo and differentiation in vitro. We conclude that self-inactivating LV are prone to DNA methylating mechanisms and that frequency as well as latency of lentiviral silencing is dependent on the internal promoter used. LV using the SFFVp are not suitable for the generation of transgenic mice or constitutive expression studies in ESC or in adult hematopoiesis. These findings have important implications for ongoing and future gene therapy trials using LV for gene correction in the hematopoietic system.
Materials and Methods
Generation of lentiviral and RV vectors. The coding sequence of murine Setbp1 gene (ncbi: NM_053099.2 GI: 197927409) was amplified from complementary DNA clone MGC:90748 (IMAGE: 30531988) with specific primers (EISI-Setbp1-N-F = 5′-GGAATTCCTGCAGGACCATGGAGCCAGAGG-′3, XIKI-Setbp1-N-R = 5′-GCTCTAGAGCGGTACCACACTTCCCAAG-′3, EIKI-Setbp1-C-F = 5′-GGAATTCCACATGGCTCGGGAGG-′3, XIAI-Setbp1-C-R = 5′-GCTCTAGAGCGATCGCCTAGGGAAGGACATCACTCTC-′3), subcloned, inserted into the 8,492 bp plasmid pCCL.SIN.cPPT.SFFV.IRES.eGFP.wPRE (LV.eGFP/LV.SFFV) using the restriction enzymes SbfI and AsiSI and verified by sequencing. VSV.G pseudotyped concentrated lentiviral vector stocks for LV.eGFP, LV.Setbp1, pCCL.SIN.cPPT.CMV.eGFP.wPRE (LV.CMV), and pCCL.SIN.cPPT.PGK.eGFP.wPRE (LV.PGK) were produced by transient cotransfection as described39 with minor modifications. In brief, cotransfection of lentiviral packaging and transfer vectors was done by complex formation with polyethylenimine (Sigma, Deisenhofen, Germany) using a DNA:PEI ratio of 1:3. Generation of γ-RV particles was done by stable transfection as described.40
Generation of transgenic mice. In brief, fertilized oocytes from superovulated B6D2F1 female mice were collected 12 hours after mating. 10–100 pl of the lentiviral solution were injected into the perivitelline space of the zygotes. The injected embryos were cultured in KSOM overnight and transferred into the oviduct of pseudopregnant foster mothers.41
Three weeks after birth, transgenic founder mice (F0) were identified by PCR analysis of tail genomic DNA (extracted using DNA Blood and Tissue Kit; Qiagen, Hilden, Germany) using Setbp1 primers (Setbp1_Mm_268-288 = 5′-CTGGGAAAAATAGCAAAGC-′3 and Setbp1_Mm_492-472 = 5′-GGCTCTGACTGCTGCTTTTT-′3) and primers matching lentiviral vector specific Wpre elements (Wpre_fwd = 5′-TCGACAATCAACCTCTGGAT-′3 and Wpre_rev = 5′-TGACAGGTGGTGGCAATGCC-′3). For generation of F1 animals, F0 mice were bred with C57Bl6/J mice. Vector-positive animals (F1) were mated with C57Bl6/J mice revealing F2. All animals were housed at the German Cancer Research Center pathogen-free animal facility according to all applicable laws and regulations.
Integration site analysis. LAM-PCR with Tsp509I and nonrestricted (nr)-LAM were performed as described.42,43,44 For high-throughput pyrosequencing (GS Flx; Roche Diagnostics, Mannheim, Germany), samples were prepared according to manufacturer's protocols. Sequences were mapped to the murine genome using UCSC blast-like alignment tool genome browser.45
Complementary DNA synthesis and semiquantitative PCR. RNA (isolated using RNeasy Kit; Qiagen) was reverse transcribed using a Superscript III First-Strand Synthesis System for reverse transcription-PCR (Invitrogen, Karlsruhe, Germany). Setbp1 primer sequences were: Setbp1_Mm_268-288 = 5′-CTGGGAAAAATAGCAAAGC-′3 and Setbp1_Mm_492-472 = 5′-GGCTCTGACTGCTGCTTTTT-′3, 18sRNA Primers: 18sRNA_fwd = 5′-GTAACCCGTTGAACCCCATT-3′ and 18sRNA_rev = 5′-CCATCCAATCGGTAGTAGCG-3′.
qPCR analysis. DNA was extracted using DNA Blood and Tissue Kit (Qiagen). Relative vector copy number (LTR-primers: LTR_fwd = 5′- AGCTTGCCTTGAGTGCTTCA-′3 and LTR_rev = 5′-GAGTCCTGCGTCGAGAGAGC-′3) was examined by quantitative real time-PCR (LC480; Roche Diagnostics). Signal intensities were normalized against murine β-actin gene (Primers: β-actin_fwd = 5′-GATATCGCTGCGCTGGTCGTC-′3 and β-actin_rev = 5′- CTTAGCACCGGCATCGATCC-′3).
Flow cytometry. Peripheral blood of transgenic and wild-type mice was collected. After lysis of erythrocytes (0.15 mol/l ammonium chloride), cells were washed with Hanks balanced salt solution (Sigma) containing 2% fetal calf serum and analyzed by fluorescence-activated cell sorting (LSRII; Becton Dickinson, Heidelburg, Germany) for eGFP expression.
mESC, EB, and differentiated ESC were dissociated to single cells by digesting with trypsin-EDTA (GIBCO; Invitrogen) for 7 minutes, washed once with Hanks balanced salt solution containing 2% fetal calf serum and analyzed for eGFP-positive cells.
Sodium bisulfite sequencing. For methylation analysis of cytosins, 500–2,000 ng DNA were deaminated using Epitect Bisulfite Kit (Qiagen). PCR on converted DNA was performed with barcoded primers (Supplementary Table S1). Amplicons were sequenced on a GS FLX 454 sequencer (Roche Diagnostics). Sequence reads that matched with the specific barcoded primers were analyzed using BISMA.46 For each amplicon, the sequences that showed a conversion >95% and a lower threshold identity >90% were visualized in heat maps, using a light gray/black color scale.
Cell culture, lentiviral transduction, spontaneous differentiation, and 5′-AzaC treatment. Murine SC1-cells were cultured in Iscove's modified Dulbecco's medium (Invitrogen) supplemented with 10% fetal calf serum (PAN-Biotech, Aidenbach, Germany), 2 mmol/l L-Glutamine (Invitrogen), and 1% penicillin/streptomycin (Invitrogen). The murine ESC line JM8.N4 (kindly provided by William C. Skarnes, Wellcome Trust Sanger Institute, Cambridge, UK) was cultured on gelatin-coated plates in ES medium consisting of knockout Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% fetal calf serum, 2 mmol/l L-glutamine, penicillin/streptomycin, 0.1 mmol/l β-mercaptoethanol (Sigma) and LIF (Millipore, Schwalbach, Germany). For passage, cells were dissociated to single cells using 0.25% trypsin (Invitrogen) and replated 1:3 every second day. Lentiviral transduction of ESC was performed in presence of protaminesulfat (8 µg/ml) 3 hours after seeding of 104 cells in a gelatin-coated 96-well plate using a multiplicity of infection of 10 or 50. 20 hours later medium was changed, cells were expanded and used for analyses. Spontaneous differentiation of LV transduced ESC was induced by LIF withdrawal. For EB formation cells were trypsinized and plated on petri dishes. Five days later EB were trypsinized and replated on tissue-culture treated dishes for additional six days. Treatment with 5′-azacytidine (5′-AzaC; Sigma) was done by adding 0.5 µmol/l daily to differentiated ESC. On day 4 cells were harvested and used for analyses.
BMT. Bone marrow cells were harvested from Boy/J males, transduced with LV or γ-RV and injected via tail vein into lethally irradiated female C57Bl6/J mice as described.40 In brief, bone marrow cells from tibias, femurs and hips of Boy/J males were harvested and washed in Hanks balanced salt solution (Sigma) supplemented with 2% FCS (PAN-Biotech). Bone marrow cells from male Boy/J mice were lineage depleted using EasySep (StemCell Technologies, Grenoble, France), transduced with LV (multiplicity of infection 100) over night or prestimulated for 2 days with cytokines and transduced 2 days with γ-RV (multiplicity of infection 100) as described40 and injected via tail vein into lethally irradiated (950 cGy) female C57Bl6/J mice (2.25 × 105 cells per mouse, n = 3 per group). Every four weeks, cell lineages were stained with fluorochrome-conjugated monoclonal antibodies (Becton Dickinson) and analyzed by fluorescence activated cell sorting (LSRII, Becton Dickinson).
Statistical analysis. Results are presented as mean ± SD. Statistical significance was determined using Student's t-test for P values <0.05.
SUPPLEMENTARY MATERIAL Figure S1. Transduced murine embryos (dE4) show lentiviral vector integrations. Figure S2. Stable eGFP expression in LV.PGK transduced ESC after induction of differentiation. Figure S3. Stable eGFP expression in lentivirally transduced bone marrow cells in vitro. Figure S4. CpG density of SFFV, CMV, and PGK promoter sequences. Table S1. Summary of methylation analysis. Table S2. Summary of bone marrow transplantation experiments.
Acknowledgments
We thank Luigi Naldini (San Raffaele Telethon Institute for Gene Therapy, Milan, Italy) for providing lentiviral packaging plasmids, the transfer vector pCCL.SIN.cPPT.SFFV.IRES.eGFP.wPRE and promoter derivatives, William C. Skarnes (Wellcome Trust Sanger Institute, Cambridge, UK) for providing the murine ESC line JM8.N4 and Annika Mengering as well as Stefanie Wenzel for technical assistance. This work was done in Heidelberg (Germany) and was supported by the Bundesministerium für Bildung und Forschung, Grant 01KV9907, by the Deutsche Forschungsgemeinschaft DFG, Grants KA 976/5-1, KA 976/5-2, Ka976/5-3, Ka976/5-4, and SFB873, and by the Deutsche Krebshilfe Grants 10-1860-GI I and 107217. The authors declared no conflict of interest.
Supplementary Material
Transduced murine embryos (dE4) show lentiviral vector integrations.
Stable eGFP expression in LV.PGK transduced ESC after induction of differentiation.
Stable eGFP expression in lentivirally transduced bone marrow cells in vitro.
CpG density of SFFV, CMV, and PGK promoter sequences.
Summary of methylation analysis.
Table S2. Summary of bone marrow transplantation experiments.
REFERENCES
- Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH.et al. (1996In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector Science 272263–267. [DOI] [PubMed] [Google Scholar]
- Pfeifer A, Ikawa M, Dayn Y., and, Verma IM. Transgenesis by lentiviral vectors: lack of gene silencing in mammalian embryonic stem cells and preimplantation embryos. Proc Natl Acad Sci USA. 2002;99:2140–2145. doi: 10.1073/pnas.251682798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA., and, Ruddle FH. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc Natl Acad Sci USA. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A, Kessler B, Ewerling S, Kabermann A, Brem G, Wolf E.et al. (2006Epigenetic regulation of lentiviral transgene vectors in a large animal model Mol Ther 1359–66. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., and, Hoogenraad CC. Lentiviral transgenesis. Methods Mol Biol. 2011;693:117–142. doi: 10.1007/978-1-60761-974-1_8. [DOI] [PubMed] [Google Scholar]
- Harbers K, Schnieke A, Stuhlmann H, Jähner D., and, Jaenisch R. DNA methylation and gene expression: endogenous retroviral genome becomes infectious after molecular cloning. Proc Natl Acad Sci USA. 1981;78:7609–7613. doi: 10.1073/pnas.78.12.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P.et al. (2005The profile of repeat-associated histone lysine methylation states in the mouse epigenome EMBO J 24800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G.et al. (2007Genome-wide maps of chromatin state in pluripotent and lineage-committed cells Nature 448553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR., and, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Jähner D, Stuhlmann H, Stewart CL, Harbers K, Löhler J, Simon I.et al. (1982De novo methylation and expression of retroviral genomes during mouse embryogenesis Nature 298623–628. [DOI] [PubMed] [Google Scholar]
- Stewart CL, Stuhlmann H, Jähner D., and, Jaenisch R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc Natl Acad Sci USA. 1982;79:4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Yokota Y, Ishida H., and, Sugahara T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983;32:1105–1113. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D., and, Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D., and, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H.et al. (2010Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET Nature 464927–931. [DOI] [PubMed] [Google Scholar]
- Laker C, Meyer J, Schopen A, Friel J, Heberlein C, Ostertag W.et al. (1998Host cis-mediated extinction of a retrovirus permissive for expression in embryonal stem cells during differentiation J Virol 72339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarik DP, Cook JA., and, Pitha PM. Inactivation of the HIV LTR by DNA CpG methylation: evidence for a role in latency. EMBO J. 1990;9:1157–1164. doi: 10.1002/j.1460-2075.1990.tb08222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A.et al. (2009CpG methylation controls reactivation of HIV from latency PLoS Pathog 5e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauder SE, Bosque A, Lindqvist A, Planelles V., and, Verdin E. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 2009;5:e1000495. doi: 10.1371/journal.ppat.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L.et al. (1998Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery J Virol 729873–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam PY, Li S, Wu J, Hu J, Zaia JA., and, Yee JK. Design of HIV vectors for efficient gene delivery into human hematopoietic cells. Mol Ther. 2002;5:479–484. doi: 10.1006/mthe.2002.0558. [DOI] [PubMed] [Google Scholar]
- Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C.et al. (2002High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter Hum Gene Ther 13803–813. [DOI] [PubMed] [Google Scholar]
- Kalberer CP, Pawliuk R, Imren S, Bachelot T, Takekoshi KJ, Fabry M.et al. (2000Preselection of retrovirally transduced bone marrow avoids subsequent stem cell gene silencing and age-dependent extinction of expression of human beta-globin in engrafted mice Proc Natl Acad Sci USA 975411–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A, Kessler B, Ewerling S, Weppert M, Vogg B, Ludwig H.et al. (2003Efficient transgenesis in farm animals by lentiviral vectors EMBO Rep 41054–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ., and, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Blömer U, Takahashi M, Gage FH., and, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamedali A, Moreau-Gaudry F, Richard E, Xia P, Nolta J., and, Malik P. Self-inactivating lentiviral vectors resist proviral methylation but do not confer position-independent expression in hematopoietic stem cells. Mol Ther. 2004;10:249–259. doi: 10.1016/j.ymthe.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Kvell K, Czömpöly T, Hiripi L, Balogh P, Kóbor J, Bodrogi L.et al. (2010Characterisation of eGFP-transgenic BALB/c mouse strain established by lentiviral transgenesis Transgenic Res 19105–112. [DOI] [PubMed] [Google Scholar]
- Zhang F, Frost AR, Blundell MP, Bales O, Antoniou MN., and, Thrasher AJ. A ubiquitous chromatin opening element (UCOE) confers resistance to DNA methylation-mediated silencing of lentiviral vectors. Mol Ther. 2010;18:1640–1649. doi: 10.1038/mt.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Yang Q., and, Chang LJ. Dynamic DNA methylation and histone modifications contribute to lentiviral transgene silencing in murine embryonic carcinoma cells. J Virol. 2005;79:13497–13508. doi: 10.1128/JVI.79.21.13497-13508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. The dinucleotide CG as a genomic signalling module. J Mol Biol. 2011;409:47–53. doi: 10.1016/j.jmb.2011.01.056. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U.et al. (2006Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1 Nat Med 12401–409. [DOI] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A.et al. (2010Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease Nat Med 16198–204. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Hauer J, Lim A, Picard C, Wang GP, Berry CC.et al. (2010Efficacy of gene therapy for X-linked severe combined immunodeficiency N Engl J Med 363355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelli B., and, Aiuti A. Gene therapy for adenosine deaminase deficiency. Immunol Allergy Clin North Am. 2010;30:249–260. doi: 10.1016/j.iac.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Boztug K, Schmidt M, Schwarzer A, Banerjee PP, Díez IA, Dewey RA.et al. (2010Stem-cell gene therapy for the Wiskott-Aldrich syndrome N Engl J Med 3631918–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I.et al. (2009Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy Science 326818–823. [DOI] [PubMed] [Google Scholar]
- Brendel C, Müller-Kuller U, Schultze-Strasser S, Stein S, Chen-Wichmann L, Krattenmacher A.et al. (2011Physiological regulation of transgene expression by a lentiviral vector containing the A2UCOE linked to a myeloid promoter Gene Therepub ahead of print). [DOI] [PubMed]
- Follenzi A, Ailles LE, Bakovic S, Geuna M., and, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- Ball CR, Pilz IH, Schmidt M, Fessler S, Williams DA, von Kalle C.et al. (2007Stable differentiation and clonality of murine long-term hematopoiesis after extended reduced-intensity selection for MGMT P140K transgene expression Blood 1101779–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilov M, Chai M, van der Hoeven F, Kloz U, Schmid W., and, Schütz G. Germ line transmission and expression of an RNAi cassette in mice generated by a lentiviral vector system. Transgenic Res. 2007;16:783–793. doi: 10.1007/s11248-007-9119-6. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Schwarzwaelder K, Bartholomae C, Zaoui K, Ball C, Pilz I.et al. (2007High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat Methods 41051–1057. [DOI] [PubMed] [Google Scholar]
- Gabriel R, Eckenberg R, Paruzynski A, Bartholomae CC, Nowrouzi A, Arens A.et al. (2009Comprehensive genomic access to vector integration in clinical gene therapy Nat Med 151431–1436. [DOI] [PubMed] [Google Scholar]
- Paruzynski A, Arens A, Gabriel R, Bartholomae CC, Scholz S, Wang W.et al. (2010Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing Nat Protoc 51379–1395. [DOI] [PubMed] [Google Scholar]
- UCSC Blast-Like Alignment Tool (BLAT) Genome Browser. (July 2007 (NCBI37/mm9)). < . < http://www.genome.ucsc.edu/cgi-bin/hgBlat >
- Rohde C, Zhang Y, Reinhardt R., and, Jeltsch A. BISMA–fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transduced murine embryos (dE4) show lentiviral vector integrations.
Stable eGFP expression in LV.PGK transduced ESC after induction of differentiation.
Stable eGFP expression in lentivirally transduced bone marrow cells in vitro.
CpG density of SFFV, CMV, and PGK promoter sequences.
Summary of methylation analysis.
Table S2. Summary of bone marrow transplantation experiments.