Abstract
It is generally thought that dendritic cells (DCs) loaded with full-length tumor antigen could improve immunotherapy by stimulating broad T-cell responses and by allowing treatment irrespective of the patient's human leukocyte antigen (HLA) type. To investigate this, we determined the specificity of T cells from melanoma patients treated with DCs loaded with mRNA encoding a full-length tumor antigen fused to a signal peptide and an HLA class II sorting signal, allowing presentation in HLA class I and II. In delayed-type hypersensitive (DTH)-biopsies and blood, we found functional CD8+ and CD4+ T cells recognizing novel treatment-antigen-derived epitopes, presented by several HLA types. Additionally, we identified a CD8+ response specific for the signal peptide incorporated to elicit presentation by HLA class II and a CD4+ response specific for the fusion region of the signal peptide and one of the antigens. This demonstrates that the fusion proteins contain newly created immunogenic sequences and provides evidence that ex vivo-generated mRNA-modified DCs can induce effector CD8+ and CD4+ T cells from the naive T-cell repertoire of melanoma patients. Thus, this work provides definitive proof that DCs presenting the full antigenic spectrum of tumor antigens can induce T cells specific for novel epitopes and can be administered to patients irrespective of their HLA type.
Introduction
Although dendritic cells (DCs) are the most proficient of all of the professional antigen-presenting cells of the immune system, clinical results from cancer immunotherapy trials with DCs are relatively disappointing.1 Consequently, DC treatment has been scrutinized.2,3
In most clinical trials, DCs are loaded with tumor-associated antigens (TAAs) in the form of defined peptides.4 As a result, the treatment is suitable only for a select group of patients with a matching human leukocyte antigen (HLA) type, often exclusively HLA-A2+ patients. Loading the DCs with full-length TAA would lead to presentation of many epitopes contained within the TAA by the patient's unique set of HLA molecules. Theoretically, a broader T-cell repertoire will be stimulated than if the DCs were loaded with peptide antigen; this repertoire will include T cells specific for both known and unknown epitopes in the TAA that are presented by several HLA types. If this is the case, a larger group of patients could benefit from DC therapy. However, there are few data on the broadness of the T-cell response after the treatment of patients with full-length tumor antigen-loaded DCs.
The loading of DCs with full-length TAA can be achieved via different methods, including mRNA electroporation.5 The use of mRNA electroporation has several advantages. First, mRNA is transiently expressed and lacks the potential to integrate into the host genome, making it a very safe tool for clinical use.6,7 Second, the technique results in a high transfection efficiency of DCs and is applicable on a large scale, and there is a GMP-approved system available for this technique, which is necessary for clinical administration.8 Third, the use of in vitro transcribed mRNA allows the TAA to be linked to an amino-terminal signal peptide and a carboxy-terminal HLA class II sorting signal to obtain presentation of the TAA by both HLA class I and II molecules and, subsequently, stimulation of CD8+ and CD4+ T cells, respectively.9 This is important as CD4+ T cells play a crucial role in cancer immunotherapy.10,11 It has been shown that mRNA encoding a TAA fused to an HLA class II trafficking signal, e.g., derived from the transmembrane and cytoplasmic domains of the invariant chain, MHC class I, lysosome-associated membrane protein (LAMP)-1 (CD107a), or DC-LAMP (CD208), indeed leads to the stimulation of CD4+ T cells, as well as an increased stimulation of CD8+ T cells.12,13,14 Moreover, this strategy allows simultaneous polyepitopic expansion of CD8+ and CD4+ T cells, resulting in a broad T-cell response in vitro.12 However, this has not yet been investigated in vivo.
At our institution, clinical trials are performed with melanoma patients, irrespective of their HLA type. These patients are treated with DCs loaded with mRNA encoding one of four TAAs (gp100, tyrosinase, MAGE-C2, or MAGE-A3).15,16 All TAAs are fused to the sorting signal of the human DC-LAMP protein for presentation by HLA class II. In addition, MAGE-A3 and MAGE-C2 include, at their amino terminus, the signal peptide of the murine LAMP-1 protein (sig) to allow translocation toward the endoplasmic reticulum.
In this report, we performed an in-depth evaluation of the T-cell repertoire of some of the patients included in these clinical trials and demonstrated that the treatment of metastatic melanoma patients with DCs loaded with full-length TAA fused to an HLA class II sorting signal leads to the stimulation of both CD8+ and CD4+ T cells. These T cells are specific for unknown antigenic epitopes, presented by both common and rare HLA types of the Caucasian population. This finding underscores the potential of DC therapy. Moreover, we provide evidence that mRNA-loaded DCs that are generated ex vivo can induce de novo T-cell responses directed against the antigens encoded by the mRNA. In addition, we show that genetically modifying the TAA can create fusion proteins containing new immunogenic epitopes.
Results
Treatment with mRNA-loaded DCs stimulates TAA-specific CD8+ T cells recognizing a previously unknown epitope
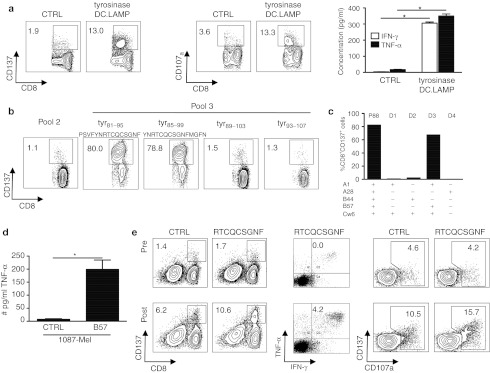
Patients were treated with autologous DCs matured with TriMix mRNA3 and co-electroporated with the above-mentioned TAAs linked to an HLA class II targeting sequence. One week after the fourth biweekly injection, an additional intradermal injection of DCs was performed to elicit a delayed-type hypersensitive (DTH) reaction. A biopsy was taken, and skin-infiltrating lymphocytes (SKILs) were analyzed. Patient 88 had a marked CD8+ T-cell response against tyrosinase, as characterized by the upregulation of CD137, CD107a and the secretion of interferon (IFN)–γ and tumor necrosis factor (TNF)–α by the SKILs (Figure 1a). No CD4+ T-cell response was observed in this patient (Table 1).
Figure 1.
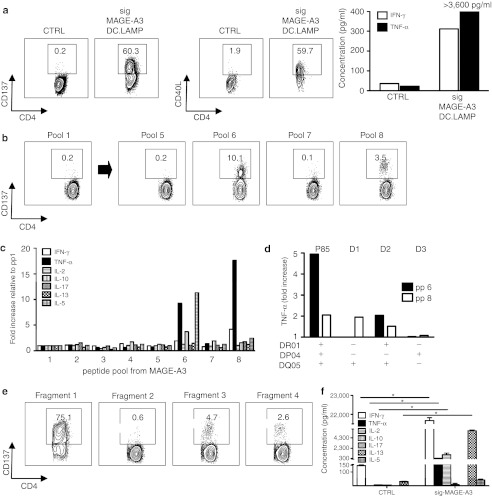
CD8+ T cells specific for a previously unidentified tyrosinase epitope are stimulated by treating a melanoma patient with DCs that had been electroporated with tyrosinase encoding mRNA. (a) During the initial screening, the SKILs of patient 88 were cocultured with sig-Nef-DC.LAMP as a control (CTRL) or tyrosinase-DC.LAMP presenting aEBV-B cells, followed by a CD137 and CD107a assay and measurement of specific IFN-γ/TNF-α secretion. The concentration of IFN-γ/TNF-α is presented as the mean + SD of duplicate cocultures. * indicates statistically significant increased cytokine secretion compared to control. (b) CD137 assay of the SKILs after stimulation with aEBV-B-cells loaded for 2 hours with overlapping 15-mer peptides (individual or pools of 10) spanning the complete tyrosinase protein, to determine the recognized region of tyrosinase. The recognized peptides and the adjacent peptides are shown. (c) Identification of the presenting HLA molecule. Percentage of CD137+CD8+ SKILs after stimulation with aEBV-B cells from patient 88 (P88) and allogeneic EBV-B-cells from four selected donors (D1-4). All EBV-B cells were electroporated with tyrosinase-DC.LAMP mRNA. (d) Presentation of the tyrosinase epitope by tumor cells. SKILs were cocultured with the tyrosinase+ HLA-B57− melanoma cell line 1087 Mel, and then electroporated with control mRNA (CTRL) or HLA-B57 mRNA (B57). After 24 hours, the TNF-α secretion by the SKILs was quantified. One representative experiment of two is shown; the values are the mean + SD of triplicate cocultures. * indicates statistically significant increased cytokine secretion compared to control. (e) SKILs were stimulated in vivo by the DC treatment. Purified CD8+ T cells from pre- and post-therapy PBMCs were stimulated three times in vitro with autologous TriMix-DC loaded with 10 µg/ml of the newly identified tyrosinase peptide. CD137 and CD107a assays and intracellular IFN-γ and TNF-α staining were performed after coculture with aEBV-B-cells loaded with the same peptide or a MelanA peptide as control (CTRL). All plots are displayed after gating on the CD8+ T cells except for the CD137 assay on stimulated CD8+ T cells isolated from the blood. The percentages shown are within the total CD8+ population. For the intracellular cytokine staining, the depicted percentages are as determined after subtraction of the background. CTRL, control; DC, dendritic cell; DC.LAMP, HLA class II sorting signal of the human DC-LAMP protein; HLA, human leukocyte antigen; IFN, interferon; LAMP, lysosome-associated membrane protein; PBMC, peripheral blood mononuclear cell; SKIL, skin-infiltrating lymphocyte; TNF, tumor necrosis factor.
Table 1. Summary of the antigenic regions of the treatment TAA recognized by post-therapy CD8+ and CD4+ SKILs and their HLA restriction.
Further characterization of the antigenic region recognized by the CD8+ SKILs of patient 88 was performed using overlapping peptides spanning the entire tyrosinase protein (pooled in groups of 10). Because pool 3 was recognized (Supplementary Figure S1a), we tested the recognition of the individual peptides from pool 3, revealing the recognition of peptides tyr81–95 (PSVFYNRTCQCSGNF) and tyr85–99 (YNRTCQCSGNFMGFN) (Figure 1b). Subsequently, we demonstrated that the minimal epitope was the 9-mer tyr87–95 (RTCQCSGNF) (Supplementary Figure S2a). To our knowledge, this epitope has not previously been described.17 Moreover, using EBV-B cells from four selected donors with one or more HLA types in common with patient 88, we determined that this epitope is presented by HLA-B57 (Figure 1c). This was confirmed by the activation of the SKILs by tyrosinase− HLA-B57+ MOLT-4 cells loaded with the 9-mer peptide (data not shown). Next, we established that the epitope RTCQCSGNF can be endogenously processed and presented by tyrosinase-expressing melanoma cells. Indeed, a coculture of the SKILs with tyrosinase+ HLA-B57− 1087-mel cells, electroporated with HLA-B57 encoding mRNA, stimulated TNF–α secretion by the SKILs (Figure 1d). All results were confirmed by the CD137 assay and IFN–γ/TNF–α secretion (data not shown).
Importantly, analysis of peripheral blood T cells collected pre- and post-treatment demonstrated that a higher percentage of CD8+ T cells recognized the RTCQCSGNF peptide post-treatment than pretreatment (Figure 1e).
In conclusion, these results show that treatment with DCs loaded with full-length TAA-encoding mRNA stimulates cytolytic CD8+ T cells, recognizing a previously unknown TAA-derived epitope. These cells are capable of infiltrating the skin and recognizing melanoma tumor cells.
Treatment with mRNA-loaded DCs stimulates TAA-specific CD4+ T cells recognizing a previously unknown epitope
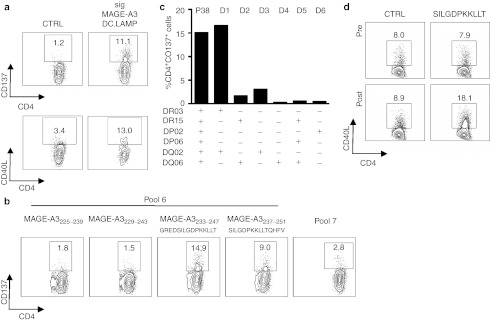
As shown by the upregulation of 4-1BB (CD137) and CD40L (CD154), patient 38 had CD4+ SKILs that were reactive against MAGE-A3 after DC treatment (Figure 2a). The same approach used for the CD8+ SKILs from patient 88 was applied in order to characterize the recognized epitope and the presenting HLA molecule. We demonstrated that within pool 6 (Supplementary Figure S1b), the peptides MAGE-A3233–247 (GREDSILGDPKKLLT) and MAGE-A3237–251 (SILGDPKKLLTQHFV) and their common sequence MAGE-A3237–247 (SILGDPKKLLT) were recognized (Figure 2b). The latter is the shortest well-recognized peptide (Supplementary Figure S2b). These data were confirmed in TNF–α and CD137 assays, respectively (data not shown). Presentation of this epitope was HLA-DRB1*03 restricted (Figure 2c). Furthermore, characterization of the CD4+ SKILs showed enhanced secretion of IFN–γ, TNF–α, and IL-2, but not IL-5 or IL-13, in response to antigenic stimulation (data not shown), suggesting a Th1 response.18 Importantly, similar to the observation made for patient 88, analysis of peripheral blood T cells of patient 38 collected pre- and post-treatment demonstrated that more SILGDPKKLLT-specific CD4+ T cells were present post-treatment than pretreatment (Figure 2d), showing that mRNA-loaded DCs stimulated these CD4+ T cells in vivo.
Figure 2.
Treatment of melanoma patient 38 with mRNA-loaded DCs stimulates CD4+ T cells specific for a novel MAGE-A3 epitope. (a) CD137 and CD40L assay of the SKILs after stimulation with aEBV-B-cells electroporated with control (CTRL) or MAGE-A3 mRNA; sig-Nef-DC.LAMP served as a control (n = 2). (b) CD137 assay of the SKILs after stimulation with aEBV-B-cells pulsed overnight with 15-mer peptides (individual or pooled in groups of 10) (n = 2). The recognized peptides from MAGE-A3 and the adjacent peptides are shown. (c) The presenting HLA molecule was determined with a CD137 assay using autologous EBV-B cells (P38) and allogeneic EBV-B cells from six selected donors (D1-6), pulsed overnight with peptide pool 6 of MAGE-A3. (d) SKILs were stimulated in vivo by the treatment DCs. Enriched CD4+ T cells from pre- and post-treatment PBMCs were stimulated once in vitro with TriMix-DCs loaded with 10 µg/ml of the 11-mer peptide SILGDPKKLLT. Screening was performed by CD40L staining after coculture of the CD4+ T cells with aEBV-B-cells pulsed overnight with either a control (CTRL) or the recognized 11-mer peptide from MAGE-A3. All plots are displayed after gating on the CD4+ T cells, and the indicated percentages are within the total CD4+ T-cell population. CTRL, control; DC, dendritic cell; DC.LAMP, HLA class II sorting signal of the human DC-LAMP protein; HLA, human leukocyte antigen; LAMP, lysosome-associated membrane protein; PBMC, peripheral blood mononuclear cell; SKIL, skin infiltrating lymphocyte.
In conclusion, these data confirm that treatment of melanoma patients with DCs loaded with mRNA encoding a TAA linked to an HLA class II sorting signal leads to the in vivo stimulation of TAA-specific CD4+ T cells that recognize a previously unknown epitope and display skin-infiltrating capacity.
Treatment with mRNA-loaded DCs induces CD8+ T cells recognizing the signal peptide fused to the TAA
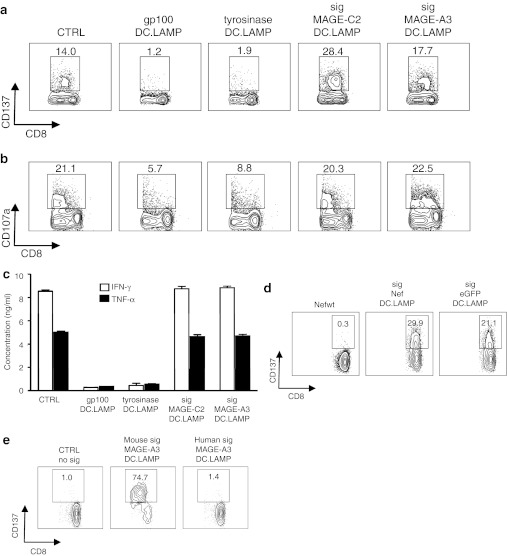
In addition to MAGE-A3-specific CD4+ SKILs, patient 38 had CD8+ SKILs showing upregulation of CD137 (Figure 3a) and CD107a expression (Figure 3b), as well as IFN–γ and TNF–α secretion (Figure 3c), in response to MAGE-C2, MAGE-A3, and the control antigen Nef (and not in response to gp100 or tyrosinase).
Figure 3.
Treatment of melanoma patient 38 with mRNA-loaded DCs also induces CD8+ T cells to recognize the exogenous signal sequence attached to some of the treatment TAAs. (a) CD137 assay, (b) CD107a assay, and (c) specific IFN-γ/TNF-α secretion during the initial screening of the SKILs of patient 38 after coculture with aEBV-B-cells presenting the control antigen sig-Nef-DC.LAMP (CTRL) and all four vaccine antigens (gp100-DC.LAMP, tyrosinase-DC.LAMP, sig-MAGE-C2-DC.LAMP, and sig-MAGE-A3-DC.LAMP). The concentration of IFN-γ/TNF-α is presented as the mean + SD of duplicate cocultures. (d) CD137 assay of the SKILs after stimulation with aEBV-B-cells electroporated with mRNA encoding Nefwt, sig-Nef-DC.LAMP, or sig-eGFP-DC.LAMP. One representative experiment of four is shown. (e) CD137 assay of the SKILs after stimulation with aEBV-B cells electroporated with mRNA encoding MAGE-A3-DC.LAMP, preceded by the murine or the human LAMP-1 derived signal sequence. One representative experiment of two is shown. All plots are displayed after gating on the CD8+ T cells, and the indicated percentages are within the total CD8+ population. CTRL, control; DC, dendritic cell; DC.LAMP, HLA class II sorting signal of the human DC-LAMP protein; eGFP, enhanced green fluorescent protein; Nefwt, mRNA encoding the unmodified sequence of Nef without sig nor DC.LAMP; HLA, human leukocyte antigen; LAMP, lysosome-associated membrane protein; sig, signal peptide of the murine LAMP-1 protein, unless indicated otherwise; SKIL, skin infiltrating lymphocyte; TAA, tumor-associated antigens.
Because the control antigen is recognized, either the Nef antigen itself, the signal sequence (sig) or the sorting sequence (DC.LAMP) must be recognized (Figure 6a). The latter can be excluded, as CD8+ SKILs are not activated by the gp100 and tyrosinase protein fused to the same DC.LAMP sequence (Figure 3a–c). Specific recognition of Nef is unlikely because the absence of HIV infection was an eligibility criterion for the study. To exclude the occurrence of an HIV infection during the treatment period, we showed that the SKILs did not recognize Nef that was not fused to sig.DC.LAMP (wild-type Nef (Nefwt)), which is readily presented by HLA class I. In contrast, all proteins containing sig were recognized, including the sig-eGFP-DC.LAMP protein containing the same sig as the TAAs and encoding a protein for which specific T cells are expected to be absent in humans (Figure 3d). These data were confirmed by IFN–γ and TNF–α secretion assays (data not shown). Therefore, we concluded that the CD8+ SKILs were specific for the signal peptide of the murine LAMP-1 protein. Furthermore, the sig-derived peptide was presented by HLA-B7 (data not shown). Three other patients (77, 80, and 123) treated in our clinical study were also HLA-B7 positive. All of these patients mounted an immune response against the signal peptide (Supplementary Figure S3).
Figure 6.
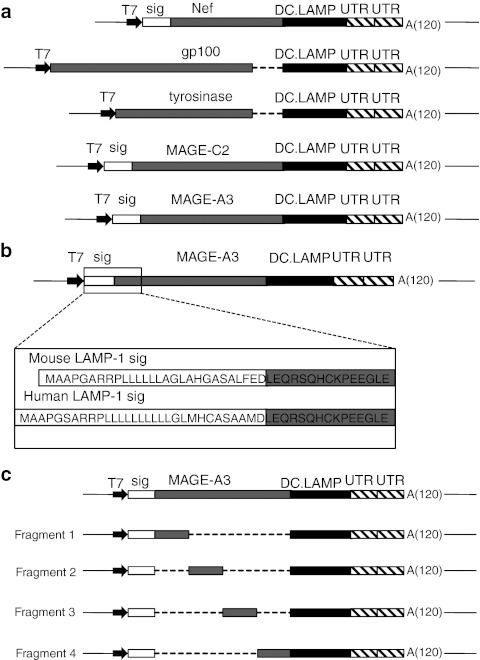
Schematic representation of the TAA-encoding vectors used for in vitro transcription of the mRNA. (a) pST1 vectors encoding the different TAAs (or control antigen Nef) flanked by the signal peptide of the mouse LAMP-1 protein (sig) and/or the HLA class II sorting signal of the human DC-LAMP protein (DC.LAMP). The deleted sequences of gp100 and tyrosinase, encoding their original transmembrane and intracytoplasmic domains, are indicated (---). (b) pST1 vectors encoding the MAGE-A3 antigen flanked by the murine or human LAMP-1 signal peptides. The protein sequences of the signal peptides and of the adjacent MAGE-A3 region are shown. (c) pST1 vectors encoding the adjacent fragments of MAGE-A3. The deleted portions (---), mouse LAMP-1 signal peptide (sig), and HLA class II sorting signal (DC-LAMP) are shown. A(120), poly-A-tail of 120 consecutive adenines; DC, dendritic cell; HLA, human leukocyte antigen; LAMP, lysosome-associated membrane protein; T7, bacteriophage T7 promoter; TAA, tumor-associated antigen; UTR, untranslated region of the human β-globin gene.
In addition to the sig-specific CD8+ response, patient 38 did not mount a TAA-specific CD8+ T-cell response to the DC treatment, as 15-mer peptides spanning MAGE-A3 and MAGE-C2 did not stimulate the CD8+ SKILs (data not shown).
Although a Basic Local Alignment Search Tool (BLAST) search indicated that the signal peptide derived from the murine LAMP-1 protein shows a high homology with its human counterpart (Figure 6b), the CD8+ SKILs could not be activated by the chimaeric construct consisting of MAGE-A3-DC.LAMP preceded by the signal peptide of the human LAMP-1 protein (Figure 3e).
In conclusion, treatment with modified DCs expressing exogenous sequences (in this case to allow presentation by HLA class II molecules) stimulates antigen-specific CD8+ T cells. The exogenous nature of the T-cell target proves the de novo induction of these cytotoxic CD8+ T cells from the naive CD8+ T-cell pool.
Treatment with mRNA-loaded DCs stimulates CD4+ T cells that recognize a peptide created by the fusion of the signal peptide to a TAA, as well as CD4+ T cells that recognize novel TAA-specific epitopes in the same patient
We observed a CD4+ T-cell response against MAGE-A3 after a single in vitro restimulation of the SKILs of patient 85 (Figure 4a). Autologous EBV-B cells loaded with either peptide pool 6 or peptide pool 8 induced upregulation of CD137 (Figure 4b). Peptides MAGE-A3217–231 (PEEKIWEELSVLEVF) and MAGE-A3221–235 (IWEELSVLEVFEGRE) and peptides MAGE-A3297–311 (PHISYPPLHEWVLRE) and MAGE-A3301–314 (YPPLHEWVLREGEE) from pools 6 and 8, respectively, were recognized by the CD4+ SKILs (Supplementary Figure S4). The SKILs recognizing peptide pool 6 showed increased TNF–α, IL-10, and IL-5 secretion upon antigen recognition, a profile in accordance with a mixed Th1/Th2 response. In contrast, the SKILs recognizing peptide pool 8 showed upregulation of IFN–γ and TNF–α, in accordance with a Th1 cytokine profile (Figure 4c).18
Figure 4.
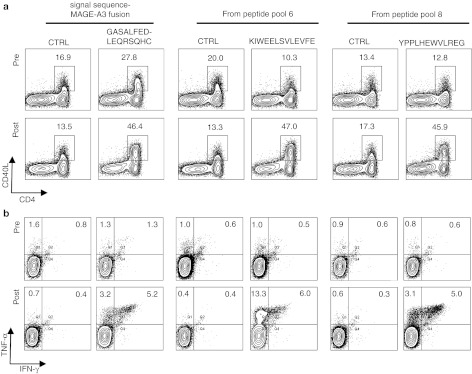
CD4+ T cells recognizing multiple MAGE-A3 epitopes and an epitope created by fusing an exogenous signal sequence to the TAA for presentation by HLA class II are induced through DC therapy of a melanoma patient using mRNA-loaded DCs. (a) After one in vitro restimulation, the CD4+ SKILs of patient 85 specifically upregulated CD137 and CD40L expression, as well as IFN-γ and TNF-α secretion, in response to aEBV-B-cells presenting MAGE-A3 in the context of HLA class I and II (sig-MAGE-A3-DC.LAMP). Sig-Nef-DC.LAMP-electroporated aEBV-B cells were used as a control (CTRL). (b) Identification of the recognized MAGE-A3 region. CD137 assay with the SKILs of patient 85 after stimulation with aEBV-B-cells pulsed overnight with overlapping 15-mer peptides (pooled per 10) spanning the entire MAGE-A3 protein (n = 4). (c) A 7-plex cytokine bead array was performed using the supernatant of stimulated SKILs. Results are representative of two independent experiments. (d) Identification of the presenting HLA molecules. Fold increases in TNF-α secretion by the CD4+ SKILs after stimulation with autologous EBV-B-cells (P85) and allogeneic EBV-B-cells of three selected donors (D1-3), loaded with peptide pool 6 and peptide pool 8, in comparison to MAGE-A3 peptide pool 1 loaded EBV-B cells. Results are representative of two independent experiments. (e) SKILs from patient 85 recognize more than two regions of the MAGE-A3 protein encoded by the mRNA loaded into the DCs. Upregulation of CD137 by the CD4+ SKILs after stimulation with aEBV-B-cells electroporated with mRNA encoding adjacent MAGE-A3 fragments. (f) The 7-plex cytokine bead array in response to the best recognized 16-mer peptide covering the fusion region of sig and MAGE-A3. Peptide pool 1 from MAGE-A3 served as a control (CTRL). The values shown are the mean + SD of duplicate cocultures. * indicates statistically significant increased cytokine secretion compared to control. All plots are displayed after gating on the CD4+ T cells. The percentages indicated are within the total CD4+ T-cell population. CTRL, control; DC, dendritic cell; DC.LAMP, HLA class II sorting signal of the human DC-LAMP protein; HLA, human leukocyte antigen; IFN, interferon; LAMP, lysosome-associated membrane protein; SKIL, skin infiltrating lymphocyte; TAA, tumor-associated antigens; TNF, tumor necrosis factor.
We deduced that the epitope in pool 6 is presented by HLA-DRB1*01, and the epitope in pool 8 is presented by HLA-DQB1*05 (Figure 4d). The 13-mer MAGE-A3220–232 (KIWEELSVLEVFE) and the 12-mer MAGE-A3301–312 (YPPLHEWVLREG) were shown to be the shortest peptides that were recognized with a similar affinity to the 15-mers (Supplementary Figure S2c,d). All results obtained from the IFN–γ/TNF–α secretion assays were confirmed by the CD137 upregulation assay (data not shown).
Although we identified two new MAGE-A3 epitopes recognized by the SKILs of patient 85, we observed a discrepancy between the percentage of SKILs recognizing the chimaeric protein derived from sig-MAGE-A3-DC.LAMP mRNA (60%) and the percentage of SKILs recognizing the newly identified MAGE-A3 peptides (10% and 3.5% of the SKILs recognized pool 6 and 8, respectively) (Figure 4a,b). This finding suggested the recognition of an additional epitope by the CD4+ SKILs of this patient. Because the control antigen did not activate the CD4+ SKILs (Figure 4a), we excluded the recognition of sig or DC-LAMP.
To identify the additionally recognized epitope in the protein derived from sig-MAGE-A3-DC.LAMP mRNA, four MAGE-A3 mRNA constructs were cloned, each encompassing two pools of peptides (Figure 6c). Each of these constructs was linked to the sig- and DC.LAMP-sequence. Fragments 3 and 4 were recognized by 4.7% and 2.6% of the SKILs (Figure 4e), corresponding to the recognition of the epitopes in pool 6 and pool 8, respectively. Fragment 1 was recognized by 75% of the SKILs (Figure 4e). The only region shared by mRNA fragment 1 and the sig-MAGE-A3-DC.LAMP mRNA that was not present in the peptides or the control mRNA is the fusion region between sig and the amino-terminal end of MAGE-A3 (Figure 6a–c). Three synthetic peptides spanning this fusion region were tested, of which peptide GASALFEDLEQRSQHC was most optimally recognized (Supplementary Figure S2e). The SKILs showed a mixed Th1/Th2 cytokine profile in response to this peptide (Figure 4f). A BLAST comparison of this sequence with the human genome uncovered no homologous human proteins.
Finally, the upregulation of CD40L (Figure 5a) and the secretion of TNF–α and TNF–α/IFN–γ (Figure 5b) by peripheral blood CD4+ T cells were analyzed in cells collected pre- and post-treatment. A higher percentage of CD4+ T cells specific for these three MAGE-A3 epitopes was found post-treatment than pretreatment. It is notable that we observed a discrepancy between the percentage of CD40L-expressing CD4+ T cells and cytokine-producing T cells. This could reflect a heterogeneous functionality of the CD4+ T cells or a partially impaired functionality.
Figure 5.
The CD4+ T-cell responses in patient 85 were all stimulated by the TriMix-DC treatment in vivo. Enriched CD4+ T cells obtained from pre- and post-treatment PBMCs were stimulated in vitro with autologous TriMix-DCs loaded with 10 µg/ml of the recognized 16-, 13-, or 12-mer peptides. To evaluate the presence of specific CD4+ T cells, aEBV-B-cells were used as target cells; the aEBV-B-cells were loaded with either the recognized peptides or a control MAGE-A3 peptide with a corresponding length (CTRL). (a) Upregulation of CD40L expression in the CD4+ T cells was quantified after two in vitro stimulations. (b) Intracellular staining of the CD4+ T cells for IFN-γ and TNF-α is shown after three in vitro stimulations. The indicated numbers are the percentages of positive cells within the total CD4+ T-cell population. CTRL, control; DC, dendritic cell; PBMC, peripheral blood mononuclear cell.
In conclusion, our results demonstrate that therapy with mRNA-loaded DCs stimulates a broad TAA-specific CD4+ T-cell response and that fusion constructs of the TAA contain newly created immunogenic epitopes. These findings also show that mRNA-loaded DCs can induce de novo CD4+ T-cell responses from the naive CD4+ T-cell pool.
Discussion
The patients in this report participated in clinical trials to assess the feasibility, toxicity, and immunogenicity of treatment with mature DCs loaded with full-length TAA-encoding mRNA.16,19 Here, we looked closer at the specificity of the stimulated T cells to estimate the diversity of the T-cell response that was stimulated in vivo by autologous DCs. We show that DC treatment of advanced melanoma patients with mRNA-loaded autologous DCs stimulates both CD8+ and CD4+ T cells specific for epitopes that have not been described elsewhere (Table 1).17 These T cells are functional, as they can migrate toward the skin, upregulate the activation marker CD137, and secrete the inflammatory cytokines IFN–γ and TNF–α.20,21 Moreover, the CD8+ SKILs can degranulate after stimulation with target cells (as shown by CD107a surface expression),22 and they recognize a melanoma cell line that endogenously presents the TAA-derived epitope. The CD4+ SKILs upregulate CD40L after stimulation, and both Th1 T-cell populations and mixed Th1/Th2 T-cell populations were found. The latter cytokine profile is typically observed after DC therapy of cancer patients.23 Although Th1 type responses are generally assumed to be favorable, both Th1 and Th2 types have been shown to play a role in tumor clearance.10,24,25 Importantly, these data provide evidence that the addition of an HLA class II targeting sequence to a TAA does indeed lead to the stimulation of TAA-specific CD4+ T cells in vivo. As such, proteins that naturally reside in the cytoplasm can be targeted by the CD4+ T cells. Moreover, we describe the coordinate evolution of CD4+ and CD8+ T-cell responses in one patient. To date, we have observed this in 8/17 patients mounting a CD8+ T-cell response against one or more of the treatment antigens.26
The diversity of the T-cell repertoire stimulated by antigen mRNA-loaded DCs is also reflected by the recognition of several epitopes within the same antigen, which has also been described by others.27 In addition, our study demonstrates that multiple epitopes are presented by different HLA types. Loss or downregulation of HLA class I expression is one of the escape mechanisms of melanoma tumors.25,28,29 It can also be speculated that melanomas escape immune surveillance through a similar mechanism for HLA class II. Indeed, a considerable percentage of melanoma cells present HLA class II on their surface,25,30,31 and direct cytotoxic effects on tumor cells by specific CD4+ T cells have been described in vitro.24 These data indicate that therapy with mRNA-loaded DCs might reduce the chance of tumor escape, as this would require the loss of expression of more than one HLA allele or an HLA haplotype in the tumor cells.
The epitopes to which the CD8+ and CD4+ T cells responded were presented by both frequently and rarely expressed HLA types in the Caucasian population (CD8+ SKILs: 3% for HLA-B57; CD4+ SKILs: 11% for HLA-DRB1*03, 9% for HLA-DRB1*01, and 39% for HLA-DQB1*05).32 To date, no HLA-B57-restricted tyrosinase epitopes or HLA-DRB1*03/HLA-DQB1*05-restricted MAGE-A3 epitopes have been described,17 whereas a single MAGE-A3 epitope presented by HLA-DRB1*01 is known.33 Furthermore, of all of the TAA-derived epitopes described thus far,17 no epitope has been found to be restricted to HLA-DQB1*05, and only one epitope (a MAGE-A1 peptide) has been described to be presented by HLA-B57.34 This underscores the potential for treatment with mRNA that encodes full-length TAAs, as more patients might benefit from DC-based immunotherapy, and clinical trials should therefore no longer be restricted to patients with a predefined HLA type.
As TAA-specific T cells can arise spontaneously in melanoma patients,35,36,37 we verified whether the T cells isolated from the post-treatment DTH skin biopsies were stimulated by our DC treatment. Compared to pretreatment T cells, higher frequencies of the antigen-specific CD8+ and CD4+ T cells were found in the peripheral blood after therapy, demonstrating that this was indeed the case. This finding is in agreement with our previous report showing that treatment-antigen-specific CD8+ T cells, recognizing epitopes other than the known HLA-A2-restricted epitopes, were increased in number after DC treatment.15 Other groups have observed that DC-treatment-specific T cells found at a DTH site cannot always be detected in the peripheral blood by direct tetramer staining, possibly because of their low frequency.38,39 Accordingly, in patient 88, the percentage of the CD8+ T cells observed post-treatment after three additional in vitro stimulations was relatively low, although the DCs modified with TriMix and TAA-encoding mRNA have proven to be potent CD8+ T-cell stimulators in in vitro cultures.40
Importantly, this work also shows that treatment with DCs that present full-length TAA bears implications for immune monitoring. Indeed, because T cells recognizing unknown epitopes are stimulated, the patients' responses should be screened with a technique independent of the presented epitope or its presenting HLA type to avoid misclassifying patients (such as the ones described here) as nonresponders.
As an immunologic side effect, CD8+ and CD4+ T cells recognizing the signal sequence preceding the TAA or its fusion region with the MAGE-A3 antigen were detected. The HLA-restriction element responsible for the presentation of the signal peptide in patient 38 appeared to be HLA-B7 (data not shown), which is expressed by 9% of the Caucasian population.32 According to the SYFPEITHI database,41 the signal peptide can engage several HLA types with sufficient strength for presentation, but we did not find sig-specific CD8+ T cells in HLA-B7− patients. In contrast, analysis of other HLA-B7+ patients indicates that they also responded to the sig-antigen.
Because the murine LAMP-1 signal sequence is not part of the native TAA sequences, the sig-specific CD4+ and CD8+ T cells will not directly contribute to tumor rejection. The influence of sig-specific T cells on the immune response to TAAs is unclear. These T cells may lead to bystander memory T-cell stimulation and contribute to a favorable environment for the development of TAA-specific T cells, and/or they could narrow the immune response by competing with TAA-specific T cells.42,43,44 Additionally, sig-specific T cells could lead to the induction of autoimmunity if they recognized a human protein. This does not appear to be an issue, as the functional CD8+ sig-specific T cells detected in patient 38 did not show cross-reactivity with the amino-terminal part of the human LAMP-1 protein, the most homologous human protein counterpart of the mouse LAMP-1 signal sequence. Also, no human analogue peptide was found for the overlapping region of the signal peptide and MAGE-A3, recognized by the CD4+ T cells of patient 85.
The presence of these CD8+ and CD4+ T cells ultimately shows that ex vivo-generated and manipulated DCs can induce de novo effector T-cell responses against mRNA encoded antigens in vivo from the naive T-cell repertoire of melanoma patients. In contrast to TAA-specific T cells, these T cells could not have been present before DC therapy, even in frequencies too low to be detected, because the signal sequence used in this study is derived from the mouse LAMP-1 protein. Such an observation could not be made beyond doubt with an endogenous TAA.
As expected owing to the xenogeneic or chimaeric origin of the recognized region, the results from the blood analysis demonstrated stimulation of these CD8+ and CD4+ T cells by the DC treatment. Nevertheless, the CD8+ and CD4+ T cells were also detected in the pretreatment assay (Figure 5 and data not shown). We assume that because of the nature of these epitopes, the sequences recognized by these T cells are sufficiently antigenic for in vitro stimulation by the DCs. Because patient 38 also participated in an earlier clinical trial that involved the administration of ampligen/CD40L-matured mRNA-loaded DCs, it is also possible that the sig-specific CD8+ T cells were induced during that therapy. In that case, the increased presence of sig-specific CD8+ T cells after the current trial demonstrates that they were reactivated by TriMix-DCs.
To our knowledge, only one other clinical trial has involved DCs loaded with mRNA encoding a chimaeric construct of a TAA flanked by a signal peptide and an endosomal/lysosomal targeting sequence.14 In this trial, the prostate carcinoma patients showed no intrinsic immune response to the sequences fused to the TAA. However, the signal sequence of the human gp96 protein was used in this study, which is probably protected by central and peripheral tolerance mechanisms.45
In conclusion, we show the potential of mRNA-loaded DCs for inducing T-cell responses in vivo from the naive T-cell repertoire. These DCs stimulate a broad spectrum of TAA-specific CD8+ and CD4+ T cells targeting previously unknown epitopes presented by common and rare HLA types in the Caucasian population. Therefore, this work forms the basis for extending DC therapy to all patients, irrespective of their HLA type, as they all might benefit from such an autologous cellular therapy.
Materials and Methods
Cell lines, media, and reagents. Autologous EBV-B (aEBV-B) cells and 1087 Mel cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Lonza, Verviers, Belgium) supplemented with 10% foetal bovine serum (BioChrom AG, Berlin, Germany), 100 U/ml penicillin, 100 µg/ml streptomycin, and 2 mmol/l L–glutamine (penicilin, streptomycin (PS)/ L-Glu) (all obtained from Lonza).
SKILs were cultured in “SKIL medium” consisting of RPMI 1640 medium supplemented with 7% heat-inactivated human AB serum (huAB serum; PAA Laboratories, Linz, Austria), 0.24 mmol/l L-asparagine, 0.55 mmol/l L-arginine, and PS/L-Glu.
Peripheral blood T cells and DCs were cultured in Iscove's modified Dulbecco's medium (IMDM; Gibco, Paisley, UK) supplemented with 1% huAB serum, PS/L-Glu, 1 mmol/l sodium pyruvate (Lonza), and nonessential amino acids (Lonza), herein referred to as “stimulation medium”.
Recombinant IL-2 and IL-7 were purchased from Chiron (Emeryville, CA) and PeproTech (Rocky Hill, NJ), respectively, and used at the indicated concentrations. Amphotericin B was purchased from Invitrogen (Paisley, UK) and used where indicated at a concentration of 1 µg/ml.
Patients, DC preparation, and treatment schedule. Melanoma patients 88, 38, 77, 80, and 85 participated in a single institution (UZ Brussel) clinical trial programme of therapy with autologous mRNA electroporated DCs.16 Patient 123 and 77 participated in a follow-up trial in which DCs were administered both intradermally and intravenously (EudraCT2009- 015748-40).19 Patients were recruited after written informed consent was obtained for the study protocol, which had been approved by the institutional ethical commission and national competent authorities. The declaration of Helsinki protocol was followed. The patients' baseline characteristics and clinical outcome are summarized in Supplementary Table S1.
All patients were treated with autologous DCs co-electroporated with TriMix mRNA and TAA-encoding mRNA linked to an HLA class II sorting signal;40 TAAs MAGE-A3, MAGE-C2, tyrosinase, and gp100 were used, and the corresponding genetic constructs are described below. DC preparation and the treatment schedule have been described previously.3
Patient 88 was serologically typed HLA-A1, A28, B44, B57, and Cw6 for HLA class I and started on TriMix-DC treatment in August 2008. Patient 38 was typed HLA-A3, A24, B7, B18, Cw7 for HLA class I and HLA-DRB1*03, DRB1*15, DPB1*0201, DPB1*0601, DQB1*02, DQB1*06 for HLA class II. He started TriMix-DC treatment in December 2009. From May 2006 until April 2007, patient 38 was included in an earlier institutional pilot trial using DCs co-electroporated with polyI:C12U (Ampligen; Bioclones, Sandton, South Africa), CD40 ligand (CD40L) mRNA, and the same TAA-encoding mRNA.46,47
Patient 85 was typed HLA-DRB1*01, DPB1*0402, DQB1*05 and started TriMix-DC treatment in June 2008.
Genetic constructs and in vitro transcription of capped mRNA. The pST1-SIINFEKL vector was kindly provided by U. Sahin (Johannes Gutenberg University, Mainz, Germany) and served as the backbone for all genetic constructs. This vector contains a bacteriophage T7 promoter to allow in vitro transcription and a tandem repeat of two 3′ untranslated regions of the human β-globin gene followed by a poly-A tail of 120 adenines and a SapI restriction site.48 From this vector, the original SIINFEKL insert was excised and replaced with the genetic sequence of interest: the TAAs MAGE-A3, MAGE-C2, gp100, and tyrosinase or the dysfunctional form of the HIV antigen Nef as a control.49 To achieve translocation of the MAGE-A3, MAGE-C2, and Nef proteins to the endoplasmic reticulum, these sequences were fused at their amino terminus to the signal sequence of murine LAMP-1 (sig). The gp100 and tyrosinase mRNAs encode their own signal peptide. To obtain translocation of the translated proteins to the HLA class II processing compartments, all antigens included, at their carboxy terminus, the HLA class II targeting sequence of the human DC-LAMP protein, consisting of its transmembrane and cytoplasmic domains (DC.LAMP).13 To avoid multiple targeting sequences within one protein, the cytoplasmic and transmembrane domains of the gp100 and tyrosinase antigens were deleted. A schematic representation of the different TAA-encoding mRNA constructs is shown in Figure 6a.
An alternative MAGE-A3 construct was made in which the murine LAMP-1 signal peptide was replaced by its human counterpart. For this purpose, the human LAMP-1 signal peptide was amplified from a synthetic template (GENEART, Regensburg, Germany) by PCR and cloned 5′ to the MAGE-A3-DC.LAMP in the pST1 vector as a SpeI-BamHII fragment. The amino acid sequences of the murine and human LAMP-1 signal peptides are shown in Figure 6b.
A series of plasmids was constructed that encoded truncated fragments of MAGE-A3. Four adjacent MAGE-A3 fragments were amplified by PCR and cloned as a BglII-BglII fragment between the signal peptide of the murine LAMP-1 protein and the sorting signal of the human DC-LAMP protein in the pST1 plasmid. A schematic representation of the different MAGE-A3 mRNA constructs is shown in Figure 6c.
Prior to in vitro transcription, all pST1 plasmids were linearized with SapI. The in vitro transcription and mRNA capping were performed using a commercially available kit (Ambion mMessage mMachine Ultra T7 Kit, Austin, TX).
The vectors encoding sig-eGFP-DC.LAMP, Nef wild type (Nefwt), and the components of TriMix have been described elsewhere.13,40,49
HLA-B57 mRNA was transcribed from the pcDNA3-HLA-B57 plasmid. After in vitro transcription and 5′ capping, a poly-A tail was added enzymatically in vitro, following the manufacturer's instructions (mMessage mMachine Ultra T7 Kit).
The mRNA concentration was measured spectrophotometrically, and the mRNA quality was evaluated by agarose gel electrophoresis. The mRNA was stored in aliquots of 20 µg at −20 °C.
Electroporation of aEBV-B cells and 1087-mel. The generation and storage of EBV-B cells is described elsewhere.26 Electroporation of EBV-B cells was performed with the same TAA-encoding mRNA as used for the DC vaccine. Prior to electroporation, cells were first washed with DPBS (Lonza) and then with Optimem without phenol red (Invitrogen, Paisley, UK). Four million cells were electroporated in a 4 mm gap cuvette using the EQUIBIO EasyjecT Plus apparatus (EQUIBIO, Ashford, UK) with 20 µg TAA-encoding mRNA (for EBV-B cells) or HLA-B57 mRNA (for 1087-mel) in a total volume of 200 µl Optimem. An exponential decay pulse of 450 V (EBV-B cells) or 300 V (1087-mel) was used with a capacity of 150 µF and a resistance of 99 Ω, resulting in a pulse time of ~6 ms (EBV-B cells) or 5 ms (1087-mel). Immediately after electroporation, EBV-B cells and 1087-mel cells were diluted to an estimated density of 1 × 106 cells/ml, in SKIL medium or 1087-mel medium, respectively, and incubated for at least 30 minutes at 37 °C/5% CO2 before further use.
Peptides and peptide pulsing of EBV-B cells. As described previously, 15-mer peptides covering the complete TAA, each with an 11 amino acid overlap, were used to characterize the peptide recognized by TAA-specific T cells.50 The peptides covering the tyrosinase or MAGE-A3 protein sequence were purchased from EMC microcollections GmbH (Tuebingen, Germany). Pools of 10 overlapping peptides were prepared, resulting in eight pools for MAGE-A3 (76 individual peptides) and 13 pools for tyrosinase (130 individual peptides). All other peptides were purchased from Eurogentec (Seraing, Belgium). Peptides were dissolved in 10% DMSO/10 mmol/l acetic acid to a final concentration of 5 mg/ml and stored in aliquots at −20 °C. Peptides are indicated by the name of the protein from which they are derived and the number of their first and last amino acid.
To load EBV-B cells with peptides for CD8+ SKIL stimulation, EBV-B cells were diluted to a final density of 1 × 106 cells/ml SKIL medium and were pulsed for 2 hours at 37 °C/5% CO2 with 5 µg/ml of the peptide pools or the individual peptides, unless indicated otherwise. To stimulate CD4+ SKILs with individual peptides and peptide pools and to titrate the minimal recognized epitope, EBV-B cells were pulsed with 5 µg/ml peptide (unless indicated otherwise) at a final density of 1 × 106 EBV-B cells/ml. Pulsing was performed overnight or for 2 hours at 37 °C in SKIL medium with or without 7% huAB serum.
DTH. One week after a treatment cycle consisting of four biweekly TriMix-DC injections, a DTH skin test was performed. A mean of 1.8 × 106 treatment DCs were injected intradermally in the backs of the patients at two sites. After 48 hours (patient 38) or 72 hours (patients 77, 80, 88, 85, and 123), 6-mm punch biopsies were taken from the injection site and cut into pieces. Leukocytes emigrating from these tissue fragments were cultured in SKIL medium. From day 1, 100 IU/ml IL-2 and 1 µg/ml amphotericin B were added to the SKIL culture. Twice a week, half of the medium was replaced with fresh IL-2 containing SKIL medium. Once a week, amphotericin B was readministered. After 2.5 weeks of culture, the SKILs were separated from the remaining tissue clumps using a MACS pre-separation filter (Miltenyi Biotec, Bergisch Gladbach, Germany) and screened for treatment antigen specificity.
Cytokine bead array and CD137 assay. Fifty-thousand SKILs were cocultured with 5 × 104 stimulator cells (electroporated or peptide pulsed aEBV-B cells) in 100 µl SKIL medium in a round bottom 96-well and incubated for 24 hours at 37 °C/5% CO2. EBV-B cells were electroporated with treatment TAA-encoding mRNA or a dysfunctional Nef-encoding mRNA as a control antigen.49 After coculture of SKILs with EBV-B cells, the supernatant was collected, and the supernatant's IFN-γ and TNF-α content was determined by a cytokine bead array (R&D Systems, Abingdon, UK). To evaluate the CD4+ T-cell responses, IFN-γ, TNF-α, IL-2, IL-10, IL-17, IL-13, and IL-5 were quantified in some experiments using a 7-plex cytokine bead array from BioRad (Hercules, CA). All procedures were performed following the manufacturer's instructions. Samples were analyzed with the Bio-Plex Manager 4.1.1 software, using the Bio-Plex 200 System (BioRad, Nazareth, Belgium).
At the same time that the supernatant was collected, cells were harvested, stained for CD4, CD8, and CD137 expression and analyzed by flow cytometry using the FACSCanto I flow cytometer and FACSDiva software (Becton Dickinson) as described elsewhere (A.M.T. Van Nuffel, unpublished data).
CD107a and CD40L assay. For CD107a and CD40L assays, 5 × 104 SKILs were restimulated with 1 × 104 tyrosinase- or MAGE-A3-electroporated EBV-B cells in 100 µl SKIL medium. For the CD107a assay, GolgiStop (1/2000; monensin; BD Biosciences Pharmingen, Erembodegem, Belgium) and anti-CD107a mAb (eBioscience, Frankfurt, Germany) were added during an overnight coculture.22 Subsequently, the cells were harvested and analyzed by flow cytometry. For the CD40L staining, 1 µg/ml anti-CD28 antibody (NA/LE purified, clone CD28.2, BD Biosciences Pharmingen) and 5 µg/ml anti-CD40 antibody (clone MAB89, Abcam, Cambridge, UK) were added.51 After a 24-hour incubation at 37 °C, cells were harvested and analyzed by FACS as described elsewhere (A.M.T. Van Nuffel, unpublished data).
In vitro expansion and cryopreservation of treatment-specific SKILs. To expand treatment-specific T cells, SKILs were restimulated weekly (for CD8+ T cells) or biweekly (for CD4+ T cells) with irradiated (90 Gy) aEBV-B-cells electroporated with tyrosinase or MAGE-A3 mRNA, respectively. If fewer than 5 × 105 SKILs were available, restimulation was induced by adding irradiated aEBV-B-cells expressing the appropriate tumor antigen until a total of 1 × 106 cells per well was obtained. If more than 5 × 105 SKILs were available, restimulation occurred by coculture of irradiated aEBV-B cells expressing the appropriate tumor antigen with SKILs and irradiated allogeneic EBV-B cells as feeder cells in a 0.4:1:3 ratio. The culture contained SKIL medium enriched with 50 IU/ml IL-2 and 5 ng/ml IL-7. When the proliferation capacity decreased, increasing amounts of IL-2 were added (up to 1,000 IU/ml). If more SKILs were obtained than necessary, SKILs were cryopreserved in 90% huAB serum with 10% DMSO using a cryofreezer container to slowly freeze the cells to −80 °C (Cryo 1 °C freezing container, rate of cooling −1 °C/minute; Nalgene, Hereford, UK). Thereafter, the cryotubes were transferred to liquid nitrogen until further use. In case no further expansion of SKILs could be achieved or the cytokine secretion capacity of the SKILs was lost, SKILs that had been frozen at an earlier time point were thawed and cultured in SKIL medium supplemented with 25 IU/ml IL-2 at a density of ~4 × 106 cells per ml. Cells were incubated for at least 2 hours at 37 °C/5% CO2 before further use.
Tracing the antigen-specific T cells in peripheral blood. Autologous DCs were generated from leukapheresis material collected after the fourth DC administration. DCs were matured by TriMix mRNA electroporation and cryopreserved until further use as described before.40 CD8+ T cells were purified from the nonadherent fraction of peripheral blood mononuclear cells (PBMCs) by immunomagnetic selection with CD8 microbeads (Miltenyi, Bergisch Gladbach, Germany). CD8+ T cells were consistently >95% pure (data not shown). The CD8− flow-through was used as a source of CD4+ T cells. The CD4+ T cell purity of the CD8− flow-through was ~60%, except in the pretreatment cell population of patient 38, where the purity was ~45% (data not shown).
Of note, post-therapy material was collected at the same time as the DTH skin biopsy was taken.
For stimulation, 20 × 106 enriched CD8+ or CD4+ T cells were cocultured with 2 × 106 TriMix-DCs loaded with 10 µg/ml of the indicated peptide. Coculture was performed in 7.5 ml of stimulation medium in a 6-well dish in the presence of 50 IU/ml IL-2 and 5 ng/ml IL-7. The medium was replaced when necessary. Cells were stimulated three times at weekly intervals for CD8+ T cells or biweekly for the CD4+ T cells. The presence of antigen-specific CD8+ and CD4+ T cells was evaluated by challenging the cells with peptide-loaded aEBV-B cells and was determined by CD137/CD107a or CD40L assay, respectively. For intracellular cytokine staining, stimulated T cells were cocultured overnight with peptide-loaded EBV-B cells (ratio 10:1) in 250 µl in a round bottom 96-well. After 2 hours, GolgiPlug (1/1,000; Brefeldin A, BD biosciences Pharmingen) was added. The cells were fixed and permeabilized using the BD Cytofix/Cytoperm Fixation/Permeabilization Kit (BD Biosciences Pharmingen) following the manufacturer's instructions, using anti-IFN-γ –PE and anti-TNF-α –antigen-presenting cells antibodies (both from eBioscience). Cells were analyzed by using a FACSCanto I flow cytometer using BD FACSDiva software.
Statistics. Significant increases in cytokine secretion were evaluated by an unpaired Student's t-test using Graphpad Prism software version 5.0c. Data were considered significant when P < 0.05.
SUPPLEMENTARY MATERIAL Figure S1. Tracing the location of the antigenic regions recognized by the CD8+ SKILs of patient 88 and the CD4+ SKILs of patient 38. Figure S2. Identification of the peptide most optimally recognized by the CD8+ SKILs of patient 88 and of the minimal peptides recognized by the CD4+ SKILs of patient 38 and 85. Figure S3. Responses to the signal peptide by the other HLA-B7+ patients included in the clinical TriMix-DC studies. Figure S4. Identification of the regions of MAGE-A3 recognized by the CD4+ SKILs of patient 85. Table S1. Patients' baseline characteristics and clinical outcomes.
Acknowledgments
The authors wish to thank Elsy Vaeremans, Xavier Debaere, Gwenny De Metter, Abderahim Hbeddou, Inge Betz, Carine Wartel, and Chiraz Mahmoud for their excellent technical assistance and the Department of radiotherapy of the UZ Brussels for the irradiation of the EBV-B cells. The authors also thank Karine Breckpot (Laboratory of Molecular and Cellular Therapy, VUB) for careful and critical reading of the manuscript. TriMix-DCs are the topic of a current patent application (WO2009/034172). A.B. and K.T. are mentioned as inventors of this application. None of the authors involved in this study receives any form of support or remuneration related to this platform. This work was supported by grants from the Interuniversity Attraction Poles Program—Belgian State—Belgian Science Policy, the National Cancer Plan of the Federal Ministry of Health, the Stichting tegen Kanker, the Vlaamse Liga tegen Kanker, an Integrated Project and a Network of Excellence sponsored by the EU FP-6, an IWT-TBM program, the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO-Vlaanderen) and the Willy Gepts Wetenschappelijk Fonds of the UZ Brussel. S.W. is a PhD fellow, and A.B. is a postdoctoral fellow of the FWO-Vlaanderen. All work was performed at the Laboratory of Molecular and Cellular Therapy, Department of Immunology-Physiology, Medical School of the Vrije Universiteit Brussel (VUB), Brussels, Belgium.
Supplementary Material
Tracing the location of the antigenic regions recognized by the CD8+ SKILs of patient 88 and the CD4+ SKILs of patient 38.
Identification of the peptide most optimally recognized by the CD8+ SKILs of patient 88 and of the minimal peptides recognized by the CD4+ SKILs of patient 38 and 85.
Responses to the signal peptide by the other HLA-B7+ patients included in the clinical TriMix-DC studies.
Identification of the regions of MAGE-A3 recognized by the CD4+ SKILs of patient 85.
Patients' baseline characteristics and clinical outcomes.
REFERENCES
- Eggermont AM. Immunotherapy: Vaccine trials in melanoma – time for reflection. Nat Rev Clin Oncol. 2009;6:256–258. doi: 10.1038/nrclinonc.2009.42. [DOI] [PubMed] [Google Scholar]
- Figdor CG, de Vries IJ, Lesterhuis WJ., and, Melief CJ. Dendritic cell immunotherapy: mapping the way. Nat Med. 2004;10:475–480. doi: 10.1038/nm1039. [DOI] [PubMed] [Google Scholar]
- Van Nuffel AM, Corthals J, Neyns B, Heirman C, Thielemans K., and, Bonehill A. Immunotherapy of cancer with dendritic cells loaded with tumor antigens and activated through mRNA electroporation. Methods Mol Biol. 2010;629:405–452. doi: 10.1007/978-1-60761-657-3_27. [DOI] [PubMed] [Google Scholar]
- Neller MA, López JA., and, Schmidt CW. Antigens for cancer immunotherapy. Semin Immunol. 2008;20:286–295. doi: 10.1016/j.smim.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Van Tendeloo VF, Ponsaerts P, Lardon F, Nijs G, Lenjou M, Van Broeckhoven C.et al. (2001Highly efficient gene delivery by mRNA electroporation in human hematopoietic cells: superiority to lipofection and passive pulsing of mRNA and to electroporation of plasmid cDNA for tumor antigen loading of dendritic cells Blood 9849–56. [DOI] [PubMed] [Google Scholar]
- Mitchell DA., and, Nair SK. RNA-transfected dendritic cells in cancer immunotherapy. J Clin Invest. 2000;106:1065–1069. doi: 10.1172/JCI11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA., and, Nair SK. RNA transfected dendritic cells as cancer vaccines. Curr Opin Mol Ther. 2000;2:176–181. [PubMed] [Google Scholar]
- Tuyaerts S, Noppe SM, Corthals J, Breckpot K, Heirman C, De Greef C.et al. (2002Generation of large numbers of dendritic cells in a closed system using Cell Factories J Immunol Methods 264135–151. [DOI] [PubMed] [Google Scholar]
- Bonehill A, Heirman C., and, Thielemans K. Genetic approaches for the induction of a CD4+ T cell response in cancer immunotherapy. J Gene Med. 2005;7:686–695. doi: 10.1002/jgm.713. [DOI] [PubMed] [Google Scholar]
- Knutson KL., and, Disis ML. Tumor antigen-specific T helper cells in cancer immunity and immunotherapy. Cancer Immunol Immunother. 2005;54:721–728. doi: 10.1007/s00262-004-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranski P., and, Restifo NP. Adoptive immunotherapy of cancer using CD4(+) T cells. Curr Opin Immunol. 2009;21:200–208. doi: 10.1016/j.coi.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiter S, Selmi A, Diken M, Sebastian M, Osterloh P, Schild H.et al. (2008Increased antigen presentation efficiency by coupling antigens to MHC class I trafficking signals J Immunol 180309–318. [DOI] [PubMed] [Google Scholar]
- Bonehill A, Heirman C, Tuyaerts S, Michiels A, Breckpot K, Brasseur F.et al. (2004Messenger RNA-electroporated dendritic cells presenting MAGE-A3 simultaneously in HLA class I and class II molecules J Immunol 1726649–6657. [DOI] [PubMed] [Google Scholar]
- Su Z, Dannull J, Yang BK, Dahm P, Coleman D, Yancey D.et al. (2005Telomerase mRNA-transfected dendritic cells stimulate antigen-specific CD8+ and CD4+ T cell responses in patients with metastatic prostate cancer J Immunol 1743798–3807. [DOI] [PubMed] [Google Scholar]
- Bonehill A, Van Nuffel AM, Corthals J, Tuyaerts S, Heirman C, François V.et al. (2009Single-step antigen loading and activation of dendritic cells by mRNA electroporation for the purpose of therapeutic vaccination in melanoma patients Clin Cancer Res 153366–3375. [DOI] [PubMed] [Google Scholar]
- Wilgenhof S, Van Nuffel AM, Corthals J, Heirman C, Tuyaerts S, Benteyn D.et al. (2011Therapeutic vaccination with an autologous mRNA electroporated dendritic cell vaccine in patients with advanced melanoma J Immunother 34448–456. [DOI] [PubMed] [Google Scholar]
- van der Bruggen, P, Stroobant, V, Vigneron, N., and, Van den Eynde, B.T-cell defined tumor antigen . < http://www.cancerimmunity.org/peptidedatabase/Tcellepitopes.htm >. Accessed 28 June 2011.
- Annunziato F., and, Romagnani S. Heterogeneity of human effector CD4+ T cells. Arthritis Res Ther. 2009;11:257. doi: 10.1186/ar2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyns, B, Wilgenhof, S, Van Nuffel, AMT, Benteyn D, Heirman C, Van Riet I.et al. (2011A phase I clinical trial on the combined intravenous (IV) and intradermal (ID) administration of autologous TriMix-DC cellular therapy in patients with pretreated melanoma (TriMixIDIV) ASCO meeting abstractsJun 9: 2519.
- Wolfl M, Kuball J, Ho WY, Nguyen H, Manley TJ, Bleakley M.et al. (2007Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities Blood 110201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder RA, Darrah PA., and, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M.et al. (2003Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation J Immunol Methods 28165–78. [DOI] [PubMed] [Google Scholar]
- Kyte JA, Trachsel S, Risberg B, thor Straten P, Lislerud K., and, Gaudernack G. Unconventional cytokine profiles and development of T cell memory in long-term survivors after cancer vaccination. Cancer Immunol Immunother. 2009;58:1609–1626. doi: 10.1007/s00262-009-0670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennadi R, Abdel-Wahab Z, Seigler HF., and, Darrow TL. Generation of melanoma-specific, cytotoxic CD4(+) T helper 2 cells: requirement of both HLA-DR15 and Fas antigens on melanomas for their lysis by Th2 cells. Cell Immunol. 2001;210:96–105. doi: 10.1006/cimm.2001.1809. [DOI] [PubMed] [Google Scholar]
- Campoli M., and, Ferrone S. HLA antigen changes in malignant cells: epigenetic mechanisms and biologic significance. Oncogene. 2008;27:5869–5885. doi: 10.1038/onc.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nuffel AMT, Tuyaerts S, Benteyn D, Wilgenhof S, Corthals J, Heirman C.et al. (2012Epitope and HLA-type independent monitoring of antigen-specific T cells after treatment with dendritic cells presenting full-length tumor antigens J Immunol Methods e-pub ahead of print 16 January 2012; [DOI] [PubMed] [Google Scholar]
- Van Tendeloo VF, Van de Velde A, Van Driessche A, Cools N, Anguille S, Ladell K.et al. (2010Induction of complete and molecular remissions in acute myeloid leukemia by Wilms' tumor 1 antigen-targeted dendritic cell vaccination Proc Natl Acad Sci USA 10713824–13829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lora A, Algarra I., and, Garrido F. MHC class I antigens, immune surveillance, and tumor immune escape. J Cell Physiol. 2003;195:346–355. doi: 10.1002/jcp.10290. [DOI] [PubMed] [Google Scholar]
- Seliger B, Maeurer MJ., and, Ferrone S. Antigen-processing machinery breakdown and tumor growth. Immunol Today. 2000;21:455–464. doi: 10.1016/s0167-5699(00)01692-3. [DOI] [PubMed] [Google Scholar]
- Bernsen MR, Håkansson L, Gustafsson B, Krysander L, Rettrup B, Ruiter D.et al. (2003On the biological relevance of MHC class II and B7 expression by tumour cells in melanoma metastases Br J Cancer 88424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramelli D, Fossati G, Mazzocchi A, Delia D, Ferrone S., and, Parmiani G. Classes I and II HLA and melanoma-associated antigen expression and modulation on melanoma cells isolated from primary and metastatic lesions. Cancer Res. 1986;46:433–439. [PubMed] [Google Scholar]
- Marsh, SGE, Parham P., and, Barber LD. Academic Press: Waltham, MA; 2000. The HLA FactsBook. [Google Scholar]
- Zhang Y, Chaux P, Stroobant V, Eggermont AM, Corthals J, Maillère B.et al. (2003A MAGE-3 peptide presented by HLA-DR1 to CD4+ T cells that were isolated from a melanoma patient vaccinated with a MAGE-3 protein J Immunol 171219–225. [DOI] [PubMed] [Google Scholar]
- Corbière V, Nicolay H, Russo V, Stroobant V, Brichard V, Boon T.et al. (2004Identification of a MAGE-1 peptide recognized by cytolytic T lymphocytes on HLA-B*5701 tumors Tissue Antigens 63453–457. [DOI] [PubMed] [Google Scholar]
- Germeau C, Ma W, Schiavetti F, Lurquin C, Henry E, Vigneron N.et al. (2005High frequency of antitumor T cells in the blood of melanoma patients before and after vaccination with tumor antigens J Exp Med 201241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanagiri T, van Baren N, Neyns B, Boon T., and, Coulie PG. Analysis of a rare melanoma patient with a spontaneous CTL response to a MAGE-A3 peptide presented by HLA-A1. Cancer Immunol Immunother. 2006;55:178–184. doi: 10.1007/s00262-005-0063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurquin C, Lethé B, De Plaen E, Corbière V, Théate I, van Baren N.et al. (2005Contrasting frequencies of antitumor and anti-vaccine T cells in metastases of a melanoma patient vaccinated with a MAGE tumor antigen J Exp Med 201249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries IJ, Bernsen MR, Lesterhuis WJ, Scharenborg NM, Strijk SP, Gerritsen MJ.et al. (2005Immunomonitoring tumor-specific T cells in delayed-type hypersensitivity skin biopsies after dendritic cell vaccination correlates with clinical outcome J Clin Oncol 235779–5787. [DOI] [PubMed] [Google Scholar]
- Lesterhuis WJ, Schreibelt G, Scharenborg NM, Brouwer HM, Gerritsen MJ, Croockewit S.et al. (2011Wild-type and modified gp100 peptide-pulsed dendritic cell vaccination of advanced melanoma patients can lead to long-term clinical responses independent of the peptide used Cancer Immunol Immunother 60249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonehill A, Tuyaerts S, Van Nuffel AM, Heirman C, Bos TJ, Fostier K.et al. (2008Enhancing the T-cell stimulatory capacity of human dendritic cells by co-electroporation with CD40L, CD70 and constitutively active TLR4 encoding mRNA Mol Ther 161170–1180. [DOI] [PubMed] [Google Scholar]
- Rammensee HG, Bachmann J, Emmerich NN, Bachor OA., and, Stevanovic S.1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics50213–219. < http://www.syfpeithi.de >. Accessed 5 June 2011. [DOI] [PubMed]
- Di Genova G, Roddick J, McNicholl F., and, Stevenson FK. Vaccination of human subjects expands both specific and bystander memory T cells but antibody production remains vaccine specific. Blood. 2006;107:2806–2813. doi: 10.1182/blood-2005-08-3255. [DOI] [PubMed] [Google Scholar]
- Gnjatic S, Jäger E, Chen W, Altorki NK, Matsuo M, Lee SY.et al. (2002CD8(+) T cell responses against a dominant cryptic HLA-A2 epitope after NY-ESO-1 peptide immunization of cancer patients Proc Natl Acad Sci USA 9911813–11818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmowski MJ, Choi EM, Hermans IF, Gilbert SC, Chen JL, Gileadi U.et al. (2002Competition between CTL narrows the immune response induced by prime-boost vaccination protocols J Immunol 1684391–4398. [DOI] [PubMed] [Google Scholar]
- Worbs T., and, Förster R. A key role for CCR7 in establishing central and peripheral tolerance. Trends Immunol. 2007;28:274–280. doi: 10.1016/j.it.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Wilgenhof, S, Van Nuffel AMT, Bentyen D, Corthals J, Heirman C, Pierret L.et al. (2010Clinical outcome following therapeutic vaccination with autologous mRNA electroporated dendritic cell (DC) vaccines in patients with advanced melanoma ASCO meeting abstractsJun 14: 8547.
- Michiels A, Breckpot K, Corthals J, Tuyaerts S, Bonehill A, Heirman C.et al. (2006Induction of antigen-specific CD8+ cytotoxic T cells by dendritic cells co-electroporated with a dsRNA analogue and tumor antigen mRNA Gene Ther 131027–1036. [DOI] [PubMed] [Google Scholar]
- Holtkamp S, Kreiter S, Selmi A, Simon P, Koslowski M, Huber C.et al. (2006Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells Blood 1084009–4017. [DOI] [PubMed] [Google Scholar]
- Allard SD, Pletinckx K, Breckpot K, Heirman C, Bonehill A, Michiels A.et al. (2008Functional T-cell responses generated by dendritic cells expressing the early HIV-1 proteins Tat, Rev and Nef Vaccine 263735–3741. [DOI] [PubMed] [Google Scholar]
- Breckpot K, Heirman C, De Greef C, van der Bruggen P., and, Thielemans K. Identification of new antigenic peptide presented by HLA-Cw7 and encoded by several MAGE genes using dendritic cells transduced with lentiviruses. J Immunol. 2004;172:2232–2237. doi: 10.4049/jimmunol.172.4.2232. [DOI] [PubMed] [Google Scholar]
- Frentsch M, Arbach O, Kirchhoff D, Moewes B, Worm M, Rothe M.et al. (2005Direct access to CD4+ T cells specific for defined antigens according to CD154 expression Nat Med 111118–1124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tracing the location of the antigenic regions recognized by the CD8+ SKILs of patient 88 and the CD4+ SKILs of patient 38.
Identification of the peptide most optimally recognized by the CD8+ SKILs of patient 88 and of the minimal peptides recognized by the CD4+ SKILs of patient 38 and 85.
Responses to the signal peptide by the other HLA-B7+ patients included in the clinical TriMix-DC studies.
Identification of the regions of MAGE-A3 recognized by the CD4+ SKILs of patient 85.
Patients' baseline characteristics and clinical outcomes.