Abstract
UU and UA dinucleotides are rare in mammalian genes and may offer natural selection against endoribonuclease-mediated mRNA decay. This study hypothesized that reducing UU and UA (UW) dinucleotides in the mRNA-coding sequence, including the codons and the dicodon boundaries, may promote resistance to mRNA decay, thereby increasing protein production. Indeed, protein expression from UW-reduced coding regions of enhanced green fluorescent protein (EGFP), luciferase, interferon-α, and hepatitis B surface antigen (HBsAg) was higher when compared to the wild-type protein expression. The steady-state level of UW-reduced EGFP mRNA was higher and the mRNA half-life was also longer. Ectopic expression of the endoribonuclease, RNase L, did not reduce the wild type or UW-reduced mRNA. A mutant form of the mRNA decay-promoting protein, tristetraprolin (TTP/ZFP36), which has a point mutation in the zinc-finger domain (C124R), was used. The wild-type EGFP mRNA but not the UW-reduced mRNA responded to the dominant negative action of the C124R ZFP36/TTP mutant. The results indicate the efficacy of the described rational approach to formulate a general scheme for boosting recombinant protein production in mammalian cells.
Introduction
Mammalian mRNA decay is a complex process comprising an essential component of transient responses. It is usually controlled by cis-acting factors, such as AU-rich elements (AREs) in the 3′UTR, and transacting factors, such as RNA-binding proteins, miRNA, and ribonucleases (reviewed in refs. 1,2). Traditionally, the 3′UTR has been viewed as the region most recognized by RNA-binding proteins such as tristetraprolin (TTP/zfp36) and HuR, and that exoribonucleases largely mediate mRNA decay.3 Conversely, very few mammalian endoribonucleases are known and they were shown to cleave at UA di-ribonucleotides.4,5 One endoribonuclease—RNase L—has a substrate cleavage preference at UU and UA in single-stranded viral mRNAs during acute viral infections and possibly in cellular mRNAs under certain conditions.6,7,8,9 Another endoribonuclease has been described that preferentially cleaves single-stranded mRNA-coding regions inside UA and UG in several mRNAs, including c-myc, MDR1, and β-globin.10 Since UA and UU dinucleotides are common targets for these known endoribonucleases, we sought to eliminate, or at least reduce, UW (W = A or U) dinucleotides in the coding region of several genes, without affecting the protein code.
The UA dinucleotides are generally under-represented in human and other vertebrate genomes and are further deficient in the mRNA when compared to nontranscribed DNA.5 The UA dinucleotides occur less frequently in the coding region than in the 3′UTR.11 Deficiency of UW frequency in coding regions may impose a selection against endoribonuclease-mediated mRNA decay inside UA dinucleotides.5,11 In this report, we showed that mRNA stability increased as a result of reduction in UW frequency, leading to increased protein production. This observation suggests a general approach for rational design of synthetic genes to yield robust protein expression.
Results
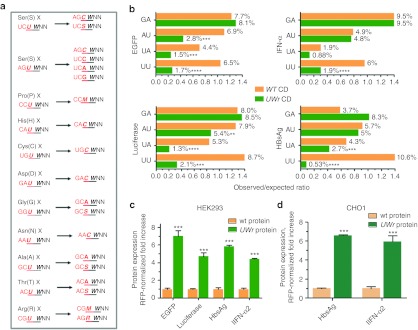
The UW frequency was reduced in synthetic genes by choosing alternative codons that do not contain UW, as well as by replacing UW in the dicodon boundary by changing the first codon (Figure 1a). This dicodon change completely destroys UU and UA at boundary sites. Several coding regions were utilized to assess the approach, namely the reporter genes enhanced green fluorescent protein (EGFP) and luciferase and the biotechnology related genes interferon-α and hepatitis B surface antigen (HBsAg; Figure 1b). The modification resulted in a significant reduction of UU dinucleotides (>68%, P < 0.001, χ2-test). The UA and, to a lesser extent, AU dinucleotides were reduced as a result of the sequence modification, whereas GA dinucleotide frequency (used as a control) did not decrease (Figure 1b). The mammalian expression vectors harboring these sequences were cotransfected with red fluorescent protein plasmid into the HEK293 cell line, and then the protein expression levels were assessed in comparison to unmodified sequences (Figure 1c). There was increased protein expression (five- to eight-fold) from expression constructs with UW-reduced coding regions. When inserted, with red fluorescent protein plasmid also, into the CHO1 cell line, which is a biotechnology standard cell line, the modified expression cassettes also gave higher protein expression (Figure 1d). Thus, the data demonstrate that UW reduction in coding regions leads to higher protein expression.
Figure 1.
Impact of UW dinucleotide frequency on protein expression. (a) The dicodon change algorithm eliminates UU or UA at the boundary by replacing the first amino acid codon that ends with U with a synonymous codon that ends with an S nucleotide. X is any amino acid. S (G or C), M (A or C), R (A or G) nucleotides are IUPAC symbols. (b) Dinucleotide frequency analysis in WT and UW reduced mRNA-coding region constructs. The Compseq program, an EMBOSS algorithm29 that calculates nucleotide composition in a sequence, was used http://mobyle.pasteur.fr/cgi-bin/portal.py#forms::compseq. The expected frequency of any dinucleotide is 1/16 (6.25%). Thus, the observed to expected ratio = % observed dinucleotide/6.25%. The observed dinucleotide (%) is shown on the columns. Several coding regions were designed based on the algorithm shown in figure. These were custom synthesized, and then subcloned into expression vectors. Two cell lines (c) HEK293 or (d) CHO1 were cotransfected with expression vectors containing wild-type (WT) coding region or UW-reduced coding region (UWr) of different genes and red fluorescent protein (RFP) plasmid at a ratio of 1/4. Protein expression at 18 hours after transfection was quantified by fluorescence (EGFP), chemiluminescence (luciferase), or enzyme-linked immunosorbent assay (ELISA) [interferon-α (IFN-α) and hepatitis B surface antigen (HBsAg)]. Data are presented as RFP-normalized fold increase which = (Expression values of UWr plasmid/WT plasmid) × (RFP normalization ratio). Data are mean ±SEM of three independent experiments.
In order to evaluate the effect of sequence modification on DNA uptake as a factor in transfection efficiency, we cotransfect the different expression vectors with a rhodamine-conjugated linear polyethylenimine derivative. We found no statistically significant difference in DNA uptake between the wild type and the sequence modified UW-reduced constructs (Supplementary Figure S2). A marginal significance (P = 0.06) was observed for the UW-reduced HBsAg construct, but this small increase (19% increase) played only a little role in the substantial increase in protein expression (400%, Figure 1c). The polyethylenimine-mediated transfection of WT and UW-reduced EGFP gave comparable results to liposomal-mediated transfection in that UW-reduced EGFP showed increased expression (Supplementary Figure S1).
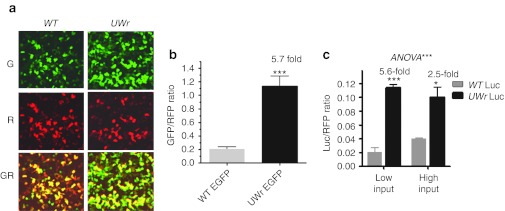
Next, we focused on EGFP so that we could study both mRNA and protein expression (by fluorescence). A representative image of WT EGFP and UW-reduced EGFP shows a dramatically enhanced fluorescence, whereas no increase is shown for the cotransfected red fluorescent protein,plasmid (Figure 2a and b). Expression of the UW-reduced luciferase-coding region under conditions of low amounts of transfected DNA (10 ng/well) was higher when compared to 75 ng/well (Figure 2c). The data in Figure 2 and Supplementary Figure S2, taken together, indicate that the enhancing effects of UW reduction on protein expression are not related to preferential DNA uptake or to variations in transfection efficiency.
Figure 2.
Effect of sequence modification and normalization on DNA uptake and expression. (a) Representative image showing green (G), red (R; red fluorescent protein (RFP) plasmid normalization plasmid), and superimposed GR channels. (b) HEK293 cells were cotransfected with green fluorescent protein (GFP) (100 ng) and RFP (25 ng) expression plasmids. The fold ratio changes between the expression levels as a result of WT and UWr-coding region modification are shown (mean ± SEM) for three independent experiments. (c) HEK293 cells (3 × 104 cells/well) in 96-well microplates were cotransfected with RFP expression plasmid and expression vectors containing either the WT-coding region or the UWr-coding region using low DNA input (10 ng/well) or high DNA input (75 ng/well).
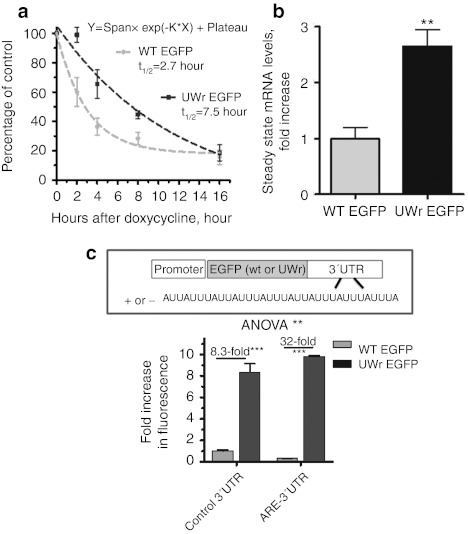
Furthermore, we have examined the mRNA half-life changes due to the UW sequence reduction. We employed the one-phase exponential decay model which describes kinetics in which the levels of a substance never decay to zero but reach a plateau. This is the case of our mRNA decay curves here, which fit the model with superior goodness of fit (R2 ≥ 0.95). Also, others' work applied the one-phase exponential decay model on mRNA kinetics that behaves in a manner where decaying mRNA levels do not reach zero but a plateau.12,13,14,15,16 We found that the mRNA for the UW-reduced EGFP had a longer half-life (2.5–3-fold increase, P = 0.01, F-test) compared to the WT mRNA, indicating that UW frequency reduction led to mRNA stabilization (Figure 3a). It is generally known that such a modest degree of mRNA half-life increase can lead to a substantial enhancement of protein expression. Steady-state mRNA levels were also higher (2.75-fold, P < 0.05) when expressed from the UWr-EGFP construct compared to the wild-type unmodified EGFP DNA (Figure 3b). When EGFP and UWr-EGFP-coding regions were fused with 3′UTR that contained ARE from the unstable tumor necrosis factor -α mRNA, higher protein expression was observed from the UWr-EGFP construct compared to the WT EGFP (32-fold versus 8-fold, P < 0.001; Figure 3c). In general, under conditions of low mRNA expression, transgene expression usually correlated positively with protein expression.17
Figure 3.
mRNA stability and UW reduction. (a) Half-life mRNA determinations of the two enhanced green fluorescent protein (EGFP) mRNA sequence variants. HeLa Tet-off cell line was transfected with the indicated expression vectors, and transcriptional activity was blocked by doxycycline (1 µg/ml). RNA was extracted at the indicated points and subjected to reverse transcription (RT)-QPCR. The one-phase decay model7 was used to quantify the half-life for WT and UW-reduced (UWr) EGFP mRNAs; R2 = 0.96 and 0.95, respectively. Data are average of two experiments (mean ± SEM); each with three replicates. (b) The steady state mRNA levels of WT and UW-reduced EGFP-coding region expression. (c) The EGFP variants were expressed from coding regions fused with a control growth hormone stable 3′UTR that lacks ARE or 3′ UTR that contains strong AU-rich elements (AREs) [from tumor necrosis factor (TNF)-α mRNA]. Data represent the expression ratio in comparison to WT EGFP (normalized to a ratio of 1). Data above are mean ± SEM of three replicates from at least two independent experiments.
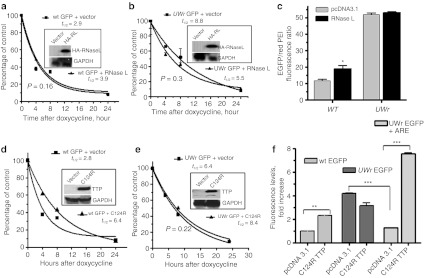
In order to understand the mechanism of increased mRNA stability due to UW reduction in coding regions, we first investigated the involvement of RNase L by overexpression and small interfering RNA experiments. We found that overexpression of HA-tagged RNase L did not reduce mRNA stability of either EGFP or UWr-EGFP (Figure 4a,b). Western blotting demonstrate expression of transfected RNase L by probing with antibody to the HA tag (Figure 4a,b; insets). At the protein level, we also noted that RNase L did not reduce the levels of WT or UWr EGFP, although there was a slight increase in WT EGFP expression (Figure 4c). Hence, in general, we found either no effect or an inconsistent effect of RNase L overexpression or of treatment with a small interfering RNA to RNase L (data not shown). We therefore resorted to another strategy that utilized the C124R zinc finger mutant form of TTP (TTP/ZFP36).
Figure 4.
Response of the enhanced green fluorescent protein (EGFP)-coding region variants to RNase L and dominant negative zinc-finger mutant, C124R tristetraprolin (TTP). HeLa Tet-off cell cells (3 × 104 cells/well) in 96-well microplates were cotransfected with expression vectors containing (a) wild-type (WT) coding region or (b) UW-reduced coding region (UWr) for EGFP mRNA and each of RNase L or pcDNA3.1 control expression vector. Subsequently, the transcription from the cotransfected EGFP plasmid was blocked by doxycycline (1 µg/ml) for the indicated periods of time. Total RNA was extracted and subjected in reverse transcription (RT)-QPCR using TaqMan primers specific to WT and UWr EGFP mRNA (details in Materials and Methods). The one-phase exponential mRNA decay model7 was used to quantify the mRNA half-life for WT (R2 = 0.96 and 0.99 for vector and RNase L curves) and UW-reduced EGFP mRNAs (R2 = 0.9 and 0.85, vector and RNase L, respectively). The data are mean ± SEM of replicate from one representative experiment of two. Insets show Western blotting for transfected HA-tagged RNase L (HA-RL) expression using a specific antibody to HA tag. (c) Protein expression changes as a result of RNase L expression using fluorescence level ratio of green fluorescence to red fluorescence. Details of experiment are similar to those described in legend A and B. (d,e) HeLa Tet-off cells (3 × 104 cells/well) in 96-well microplates were transfected for 18 hours with expression vectors containing (d) wild-type (WT) coding region or (e) UWr-coding region for EGFP in the presence of C124R expression vector or pcDNA3.1 control vector. Subsequently, the transcription was blocked by doxycycline (1 µg/ml) for the indicated periods of time. The one-phase exponential mRNA decay model7 was used to quantify the mRNA half life for WT (R2 = 0.97 and 0.99 for vector and C124R, respectively) and UWr EGFP mRNAs (R2 = 0.96 and 0.9.5 for vector and C124R, respectively). The data are mean ± SEM of three replicates from one representative experiment of two independent experiments performed. Insets show Western blotting for C124R expression. (f) Protein expression from WT EGFP-coding region, UWr EGFP-coding region, and UWr EGFP-coding region fused with tumor necrosis factor (TNF)-α ARE 3′UTR. Data is presented as fluorescence levels (mean ± SEM) of three independent experiments, each with three to four replicates. WT EGFP + pcDNA3.1 vector control was normalized to 1.0-fold ratio. The UWr EGFP and UWr EGFP+ARE data were normalized to UWr EGFP + pcDNA 3.1 control (fold ratio = 1.0).
The TTP binds to AREs as small as UAUUUAU18,19 and the C124R mutant fails to bind to AREs.20 However, C124R can still interact with a number of general mRNA decay enzymes such as those involved in deadenylation (e.g., Caf1), decapping, and exonuclease activity.17 Hence, the nonbinding mutant TTP can sequester general mRNA decay factors.17,18 This dominant negative activity appears to be specific to the 3′UTR-ARE at lower concentrations, but we hypothesized that it may have broader effects at higher concentrations in a manner independent of the 3′UTR. Thus, we overexpressed C124R TTP in Hela Tet-off cell line and studied the effects on UW-reduced EGFP mRNA and wild-type mRNA. Western blotting confirmed expression of the transfected C124R TTP (Figure 4d,e insets). The mRNA half-life was assessed for both WT EGFP and UWr EGFP transcripts released from constructs under regulation of Tet-off cytomegalovirus promoter. The dominant negative effect of C124R TTP, as demonstrated by mRNA stabilization, was clearly observed in the case of the response of WT EGFP mRNA turnover (Figure 4d). Further increases in UWr EGFP mRNA stabilization were not observed as a consequence of the dominant negative effect of C124R TTP (Figure 4e). Subsequently, no increase in protein production (fluorescence) from UWr EGFP mRNA was observed in response to the dominant negative effect of C124R (Figure 4f, middle columns). In contrast, wt EGFP protein levels were affected by the stabilizing effect of C124R on WT EGFP mRNA (Figure 4f, left columns). In addition to tumor necrosis factor-α mRNA,20,21 C124R TTP (but not wild-type TTP) has been shown recently to increase an array of AU-rich mRNAs such as uPA, uPA receptor, NF-kappa B inhibitor, matrix metalloproteinase-1, and interleukin-10.22 The C124R has the ability to increase protein expression from UWr EGFP transcripts only when the UWr EGFP-coding region is fused with ARE-3′UTRs, such as those of tumor necrosis factor-α (Figure 4f, right columns). This aspect may lead to an improved use of UW-reduced reporter genes such as EGFP and luciferase, which is post-transcriptional effect of gene expression specific to the 3′UTR.
Discussion
The present approach differs from the classical codon optimization that depends on species-specific bias of codon usage, and it is thought to be a natural selection for translational optimization. This served as the basis for improving overexpression of proteins in host cells of a different species.23,24,25 Our approach is different: first, unlike classical codon bias optimization, both codon and dicodon boundaries are considered here. Second, this rationalized approach is based on reduction of UW dinucleotides; these dinucleotides are rare in mammalian coding regions and have been shown to correlate with accelerated mRNA decay.4,5 Our approach has led to increased mRNA stabilization and, subsequently, increased protein expression levels. However, other possible post-transcriptional effects, such as translation, are likely to take part as well. Validation of this role of modification on translation efficiency would require a different set of experiments. The very modest increase (for example, a 14% increase) in mRNA stability observed with classical codon optimization26 may possibly be due to an inherited partial UW reduction. For example, the preferred human codon for isoleucine is AUC rather than AUU or AUA.
Overall, by resisting mRNA decay using reduced UW frequency and elimination of UW in dicodon boundaries, this rationalized approach may find utility as a general method to boost recombinant protein production in mammalian cells. The UW reduction approach may find broader applications if confirmed in other expression systems, such as those based on bacterial and insect cells. Also, the approach remains to be tested with stable cell lines expressing the UW-reduced coding regions, as unlike transient transfection, potential regulatory mechanisms such as DNA methylation may adversely affect stable transgene expression. The approach can enhance reporter genes for difficult to transfect cells or can be used to study 3′ UTR-mediated post-transcriptional effects due to instability sequences, such as AREs, or miRNA action.
Materials and Methods
Synthetic biology. Coding regions of EGFP, luciferase, interferon α2a, and HBsAg were modified according the following algorithm: (i) by exchanging UW-containing codons with synonymous codons that lack or reduce UW (Supplementary Table S1) and (ii) by destroying the UW dicodon boundary by exchanging the first codon with synonymous codons (Supplementary Table S2). The modified coding region sequences (Supplementary Table S3) were synthesized by GeneArt (Regensburg, Germany) and all constructs, including wild-type coding regions, weresubcloned using SalI and BamH1 sites in the same vector (Gwiz; Genlantis, San Diego, CA). The vector is expressed under control of the cytomegalovirus/intron A promoter and with the stable bovine growth hormone 3′UTR. Cloning of the ARE derived from the tumor necrosis factor-α mRNA 3′UTR was previously described.27 Dinucleotide frequency analysis was executed by the Compseq program, an EMBOSS algorithm29 that calculates nucleotide composition in a given sequence (http://mobyle.pasteur.fr/cgi-bin/portal.py#forms::compseq). The formula of observed (0) over expected (e) frequency ratio is as follows: (XY o/e = dXY/nX·nY, where nX denotes the frequency of nucleotide X, and dXY is the frequency of the dinucleotide (XY).
Cell lines and transfection. The HEK293 and CHO1-K cell lines were obtained from American Type Culture Collection (Rockville, MD) and cultured in DMEM medium (Invitrogen, Carlsbad, CA) or DMEM/F12 (in case of CHO1-K) supplemented with 10% FBS and antibiotics. The Tet-off Hela cell line, obtained from Clontech (Mountain View, CA), was maintained in DMEM with selection drug G418 (Sigma, St Louis, MO). Cells were transfected with vectors using lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. All transfections were performed in several replications, and our intrawell transfection efficiency was <6% variation. The red fluorescent protein expression vector (Evrogen, San Diego, CA) was used at ¼ ratio of the test plasmid concentration for the purpose of normalization. For RNA experiments, EGFP (50 ng per 1 × 105 cells) was used for monitoring transfection efficiency.
Quantification of protein expression and western blotting. EGFP expression was assessed and quantified by fluorescence imaging (described below). Luciferase expression was assessed using the luciferase assay system from Promega (Madison, WI) and a luminometer. Interferon-α and HBsAg was quantified by enzyme-linked immunosorbent assay (ELISA) obtained from PBL (Piscataway, NJ) and Abzyme (Piscataway, NJ).
For transfected cells with C124R TTP expression vector (a gift from Dr Perry J. Blackshear, NIH, US) and HA-tagged RNase L expression vector (synthesized by GeneArt), western blottings were performed to confirm the protein expression from the vectors. Equal amounts of protein samples were subjected to electrophoresis on 10% polyacrylamide–SDS gels (Protogel; National Diagnostics, Atlanta, GA) followed by transfer to nitrocellulose membranes (Hybond ECL; Amersham Biosciences, Piscataway, NJ). Membranes were hybridized with primary antibody to goat anti-TTP (1/500) (Santa Cruz Biotech, Santa Cruz, CA), rat anti-HA A (1/50) (Roche, Basel, Switzerland), or mouse anti-GAPDH (1/500; Abcam, Cambridge, UK) followed by the appropriate secondary horseradish peroxidase -conjugated antibody. Signal detection was performed with ECL Western blotting detection reagents (Amersham Biosciences, Amersham, UK). Protein molecular weight markers were used to verify the protein size.
Reporter fluorescence, normalization, imaging, and quantitation. The fluorescence intensity was assessed using high-resolution images obtained from an automated laser-focus high-throughput BD Pathway 435 imager (BD Biosciences, San Jose, CA). The variance in GFP fluorescence among replicate microwells was <6%. The experiments were normalized with red fluorescent protein plasmid at ¼ ratio of the test plasmid concentration. Image processing, segmentation, and fluorescence quantification were facilitated by the ProXcell program previously described.28 Fluorescence of EGFP (excitation, 488 nm; emission, 507 nm) and red fluorescent protein (excitation, 553 nm; 574 nm) was processed with appropriate filters of the BD imager. Data are presented as mean ± SEM of total fluorescence intensity in each well, with replicate readings ranging from three to four, as indicated in the text. The Student t-test was used when comparing two data groups, while analysis of variance was performed for each data set having three or more data groups.
DNA uptake. The rhodamine-conjugated linear polyethylenimine derivative (excitation at 550 nm; emission at 575 nm) was used to assess the uptake of DNA by the wild type and modified plasmids. Cells were transfected with a complex of 0.75 µl of jetPEI-FluoR (Polyplus, Paris, France) and 200 ng of the plasmid DNA. After 18-hour incubation, the cells were visualized by automated fluorescence microscopy and the fluorescence quantified as described above. This protocol was also repeated in the presence of the lipofectamine 2000 to assess its compatibility during liposomal transfection and was found compatible.
RNA, reverse transcription, and real-time PCR. Total RNA was extracted with Trireagent (Sigma). Reverse transcription was performed using 200 ng total RNA, 500 ng oligo dT(18–23), 500 mmol/l dNTP mixture, 20 U RNAsin (Pharmacia, Milton Keyries, UK), and 200 U of SuperScript II (Invitrogen). Real-time PCR was performed using custom-made primer sets (Applied Biosystems, Foster City, CA) and TaqMan master mix according to the manufacturer's instructions. The 6-carboxyfluorescein-labeled TaqMan hybridization probes were designed to target the junction of Exons 1 and 2 of EGFP or UWr EGFP to minimize DNA contamination. The primer set for EGFP: forward CTCCATCTTCGCGGTAGCT, reverse TTCTTCTCCTTTGCTAGCCATTTCT, probe: CCGCCGTTCAGTCG CCGT. The primer set for UWr EGFP: forward CTCCATCTTCGCGG TAGCT, reverse CCATGGTGGCAAGCTTGTC, Probe: CCGCCGTT CAGTCGCCGT. As internal control, we used a VIC-labeled GAPDH mRNA probe (Applied Biosystems). The primer sets failed to amplify vector DNA (negative control) and were specific for the complementary DNA. PCR protocol was as follows: Initiation steps: 2 minutes at 50 °C followed by 10 minutes at 95 °C, denaturing step: 15 seconds at 95 °C, and annealing/extension 1 minute at 60 °C. The amounts of molecules of complementary DNA were calculated using Chroma 4 DNA Engine cycler (Bio Rad, Hercules, CA) software and an eight-point standard curve using serial dilutions of purified and quantified amplicon DNA. The use of standard curve method ensures that all amplifications proceed with a comparable and high efficiency. The variations in GAPDH levels in one experiment were typically <10%. The lowest internal control measurement was set as 1 fold and the copy number of the WT EGFP and UWr EGFP were normalized.
The mRNA half-life determinations. HeLa Tet-off cells were cotransfected with TetO-regulated WT or UWr EGFP constructs, along with vector, C124R TTP, or RNase L expression vector overnight, and then treated with doxycycline (1 µg/ml) to shutoff transcription. The RNA was extracted by the RNAzol method at multiple points and then subjected to reverse transcription-QPCR as described above. To ensure equal proportions of transfected cells for each time point, cotransfections were performed in 10-cm culture plates using 4 µg of the vector DNA, and 24 hours later the cells were split into 6-well plates. The mRNA half-life was determined using a one-phase exponential decay model. The equation Y = Span × exp(–K × X) + Plateau describes the kinetics of mRNA decay. X is time, and Y depicts the response. Y starts out equal to SPAN + PLATEAU and decreases to PLATEAU with a rate constant K. The half-life of the decay is 0.6932/K. SPAN and PLATEAU are expressed in the same units as the Y axis. K is expressed in the inverse of the units used by the X axis. Data fit to the model was performed using Least Squares fit method. Comparison between mRNA curves was performed using an F-test.
SUPPLEMENTARY MATERIAL Figure S1. Expression of WT and UWr EGFP after PEI-mediated transfection. Figure S2. Effect of sequence modification on DNA uptake. Table S1. UU/UA reduction in codons. Table S2. UU/UA elimination in di-codon boundaries. Table S3. UW-reduced sequences.
Acknowledgments
This study was supported by the intramural funding, including open access charges, from King Faisal Specialist Hospital and Research Center. We thank Dr Perry J. Blackshear and Dr W. Lai (NIH) for providing the C124R mutant TTP expression vector. The technical assistance and advices of Ms Lina Mahmoud, Wijdan Al-Ahmadi, Latifa Al-Haj, and Dr Edwared Hitti are highly appreciated (Bimolecular Research Program). The assistance of Dr Sahar Al-Thawadi at Pathology Department for the HBsAg quantitation is also acknowledged.
Supplementary Material
Expression of WT and UWr EGFP after PEI-mediated transfection.
Effect of sequence modification on DNA uptake.
UU/UA reduction in codons.
UU/UA elimination in di-codon boundaries.
UW-reduced sequences.
REFERENCES
- Khabar KS. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell Mol Life Sci. 2010;67:2937–2955. doi: 10.1007/s00018-010-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M., and, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Fritz DT, Bergman N, Kilpatrick WJ, Wilusz CJ., and, Wilusz J. Messenger RNA decay in mammalian cells: the exonuclease perspective. Cell Biochem Biophys. 2004;41:265–278. doi: 10.1385/CBB:41:2:265. [DOI] [PubMed] [Google Scholar]
- Duan J., and, Antezana MA. Mammalian mutation pressure, synonymous codon choice, and mRNA degradation. J Mol Evol. 2003;57:694–701. doi: 10.1007/s00239-003-2519-1. [DOI] [PubMed] [Google Scholar]
- Beutler E, Gelbart T, Han JH, Koziol JA., and, Beutler B. Evolution of the genome and the genetic code: selection at the dinucleotide level by methylation and polyribonucleotide cleavage. Proc Natl Acad Sci USA. 1989;86:192–196. doi: 10.1073/pnas.86.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisbal C, Silhol M, Laubenthal H, Kaluza T, Carnac G, Milligan L.et al. (2000The 2′-5′ oligoadenylate/RNase L/RNase L inhibitor pathway regulates both MyoD mRNA stability and muscle cell differentiation Mol Cell Biol 204959–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabar KS, Siddiqui YM, al-Zoghaibi F, al-Haj L, Dhalla M, Zhou A.et al. (2003RNase L mediates transient control of the interferon response through modulation of the double-stranded RNA-dependent protein kinase PKR J Biol Chem 27820124–20132. [DOI] [PubMed] [Google Scholar]
- Andersen JB, Mazan-Mamczarz K, Zhan M, Gorospe M., and, Hassel BA. Ribosomal protein mRNAs are primary targets of regulation in RNase-L-induced senescence. RNA Biol. 2009;6:305–315. doi: 10.4161/rna.6.3.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ahmadi W, Al-Haj L, Al-Mohanna FA, Silverman RH., and, Khabar KS. RNase L downmodulation of the RNA-binding protein, HuR, and cellular growth. Oncogene. 2009;28:1782–1791. doi: 10.1038/onc.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom K, Urquhart JC, Tafech A, Doyle E., and, Lee CH. Purification and characterization of a novel mammalian endoribonuclease. J Cell Biochem. 2006;98:519–537. doi: 10.1002/jcb.20726. [DOI] [PubMed] [Google Scholar]
- Duret L., and, Galtier N. The covariation between TpA deficiency, CpG deficiency, and G+C content of human isochores is due to a mathematical artifact. Mol Biol Evol. 2000;17:1620–1625. doi: 10.1093/oxfordjournals.molbev.a026261. [DOI] [PubMed] [Google Scholar]
- Maurer JA., and, Wray S. Luteinizing hormone-releasing hormone (LHRH) neurons maintained in hypothalamic slice explant cultures exhibit a rapid LHRH mRNA turnover rate. J Neurosci. 1997;17:9481–9491. doi: 10.1523/JNEUROSCI.17-24-09481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams B, Obertone TS, Wang X., and, Murphy TJ. Relationship between internalization and mRNA decay in down-regulation of recombinant type 1 angiotensin II receptor (AT1) expression in smooth muscle cells. Mol Pharmacol. 1999;55:1028–1036. doi: 10.1124/mol.55.6.1028. [DOI] [PubMed] [Google Scholar]
- Guo F., and, Cech TR. In vivo selection of better self-splicing introns in Escherichia coli: the role of the P1 extension helix of the Tetrahymena intron. RNA. 2002;8:647–658. doi: 10.1017/s1355838202029011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR.et al. (2008Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1 Mol Cell 29263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak W, Parameswaran N, Hall CS, Aiyar N, Sparks HV., and, Spielman WS. Novel regulation of adrenomedullin receptor by PDGF: role of receptor activity modifying protein-3. Am J Physiol, Cell Physiol. 2002;282:C1322–C1331. doi: 10.1152/ajpcell.00561.2001. [DOI] [PubMed] [Google Scholar]
- Barnes LM., and, Dickson AJ. Mammalian cell factories for efficient and stable protein expression. Curr Opin Biotechnol. 2006;17:381–386. doi: 10.1016/j.copbio.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Brewer BY, Malicka J, Blackshear PJ., and, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J Biol Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- Worthington MT, Pelo JW, Sachedina MA, Applegate JL, Arseneau KO., and, Pizarro TT. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. J Biol Chem. 2002;277:48558–48564. doi: 10.1074/jbc.M206505200. [DOI] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS., and, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schichl YM, Resch U, Hofer-Warbinek R., and, de Martin R. Tristetraprolin impairs NF-kappaB/p65 nuclear translocation. J Biol Chem. 2009;284:29571–29581. doi: 10.1074/jbc.M109.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Souhibani N, Al-Ahmadi W, Hesketh JE, Blackshear PJ., and, Khabar KS. The RNA-binding zinc-finger protein tristetraprolin regulates AU-rich mRNAs involved in breast cancer-related processes. Oncogene. 2010;29:4205–4215. doi: 10.1038/onc.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess-Brown NA, Sharma S, Sobott F, Loenarz C, Oppermann U., and, Gileadi O. Codon optimization can improve expression of human genes in Escherichia coli: A multi-gene study. Protein Expr Purif. 2008;59:94–102. doi: 10.1016/j.pep.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Kim CH, Oh Y., and, Lee TH. Codon optimization for high-level expression of human erythropoietin (EPO) in mammalian cells. Gene. 1997;199:293–301. doi: 10.1016/s0378-1119(97)00384-3. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Mousa A., and, Ribeiro P. Codon optimization improves heterologous expression of a Schistosoma mansoni cDNA in HEK293 cells. Parasitol Res. 2002;88:583–586. doi: 10.1007/s00436-001-0585-0. [DOI] [PubMed] [Google Scholar]
- Fath S, Bauer AP, Liss M, Spriestersbach A, Maertens B, Hahn P.et al. (2011Multiparameter RNA and codon optimization: a standardized tool to assess and enhance autologous mammalian gene expression PLoS ONE 6e17596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitti E, Al-Yahya S, Al-Saif M, Mohideen P, Mahmoud L, Polyak SJ.et al. (2010A versatile ribosomal protein promoter-based reporter system for selective assessment of RNA stability and post-transcriptional control RNA 161245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al-Haj L, Al-Ahmadi W, Al-Saif M, Demirkaya O., and, Khabar KS. Cloning-free regulated monitoring of reporter and gene expression. BMC Mol Biol. 2009;10:20. doi: 10.1186/1471-2199-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I., and, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of WT and UWr EGFP after PEI-mediated transfection.
Effect of sequence modification on DNA uptake.
UU/UA reduction in codons.
UU/UA elimination in di-codon boundaries.
UW-reduced sequences.