Abstract
Adeno-associated virus (AAV) vectors have shown remarkable efficiency for gene delivery to cultured cells and in animal models of human disease. However, limitations to AAV vectored gene transfer exist after intravenous transfer, including off-target gene delivery (e.g., liver) and low transduction of target tissue. Here, we show that during production, a fraction of AAV vectors are associated with microvesicles/exosomes, termed vexosomes (vector-exosomes). AAV capsids associated with the surface and in the interior of microvesicles were visualized using electron microscopy. In cultured cells, vexosomes outperformed conventionally purified AAV vectors in transduction efficiency. We found that purified vexosomes were more resistant to a neutralizing anti-AAV antibody compared to conventionally purified AAV. Finally, we show that vexosomes bound to magnetic beads can be attracted to a magnetized area in cultured cells. Vexosomes represent a unique entity which offers a promising strategy to improve gene delivery.
Introduction
Adeno-associated virus (AAV) vectors are one of the frontrunners for gene therapy in humans due to the vector's good safety profile in clinical trials. They have shown excellent gene expression ability in a variety of tissues in rodents, nonhuman primates1 and large animal models,2 as well as in humans.3 The robust and stable gene transfer in nondividing cells conferred by AAV is owed both to the virus's protein capsid and the episomal configuration of its DNA in the cell nucleus. The tropisms of AAV are very different between different serotypes4 and advances using self-complementary DNA and modified capsids have expanded the efficacy of these vectors even further.5,6 AAV vectors are showing promising results in phase I clinical trials in organs such as the eye,7 muscle,8 and brain.9 However, as with other viral vectors, intravenous delivery of AAV mainly results in liver sequestration, a major issue when other organs are the target of gene transfer.
Most cells shed a variety of membrane-bound vesicles varying in size from 20 nm to 1 µm in diameter, which have been termed exosomes, microparticles, and microvesicles, referred to here collectively as microvesicles.10,11 These microvesicles have been found to contain a selective set of lipids, proteins,12,13 and nucleic acids.14,15,16,17 Microvesicles have been shown to effectively shuttle between different cell types and deliver proteins and nucleic acids to the recipient cells.14 AAV capsids in association with host cell-derived microvesicles may alter cell binding and transduction properties. We hypothesized that during standard AAV vector production, a fraction of capsids may be released from the cell inside or in association with microvesicles which may bestow unique properties onto the vector.
Here, we show that AAV capsids are indeed associated with microvesicles (termed vexosomes) that can be isolated from conditioned medium of the packaging cell line. Furthermore, vexosomes mediate enhanced transduction of cells in culture compared to conventionally purified AAV vector and their specificity of transduction can be modified by changing surface molecules on the membrane.
Results
AAV vectors are associated with microvesicles released from AAV producer cells
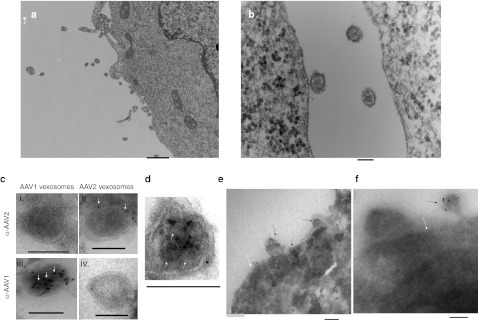
To test the idea that AAV capsids may be incorporated into microvesicles during virus vector production, a single-stranded AAV2 encoding GFP (AAV-GFP) was produced by standard triple-transfection of 293T cells with AAV plasmids. Forty-eight hours post-transfection, cells were harvested and examined by transmission electron microscopy. We observed many microvesicles near the surface of cells as well as budding from the plasma membrane (Figure 1a,b and Supplementary Figure S1). Occasionally, clusters of AAV capsids were clearly visible near the cell surface (Supplementary Figure S1). Next, we sought to detect AAV vectors associated with microvesicles shed into the media of 293T producer cells. First, we developed a differential centrifugation protocol that would preferentially pellet the relatively large microvesicles rather than the free AAV capsids that may be present. A 25-minute centrifugation at 20,000g effectively pelleted the AAV-associated microvesicles (data not shown). As the sedimentation coefficient of AAV is reported to be 110S,18 the pelleting of free AAV is very inefficient at this speed and time. Different AAV vector types may have different abilities to associate with microvesicles in the producer cell line. We therefore generated AAV with two vector serotypes, AAV1 and AAV2, and characterized their microvesicle packaging efficiency. The microvesicle pellets from both AAV1 and AAV2 producer cell media was analyzed by transmission electron microscopy and immunogold labeling to indirectly observe the relationship of AAV capsids and microvesicles. The microvesicle pellet was fixed, cryosectioned, and stained with anti-AAV antibodies (which recognize intact capsids) followed by a secondary antibody conjugated to 10-nm gold particles. We observed AAV capsids associated with microvesicles for both AAV1 and AAV2 serotypes (Figure 1c and Supplementary Figure S2). Some microvesicles contained AAV capsids in their interior as the double membrane of microvesicles is observed (white ring around the vesicle) indicating that the microvesicle has been cut open (Figure 1c and Supplementary Figure S2). As a negative staining control, AAV2 vexosomes stained with anti-AAV1 or AAV1 vexosomes stained with anti-AAV2 followed by gold-conjugated secondary antibody yielded no specific signal (Figure 1c). Under high magnification of the immunogold labeled AAV1 microvesicles, we directly observed capsids (size ~25 nm) of which some were stained with 5 nm gold (Figure 1d). We termed these AAV/microvesicle associations vector-exosomes, abbreviated as vexosomes. Capsids were associated with vesicles ranging in diameter from ~50 to 200 nm (data not shown). The average number of AAV capsids/microvesicle was quantitated for AAV1 and AAV2. For AAV1 there was an average of 8.2 capsids/microvesicle while for AAV2 there was only 1.2 capsids/microvesicle. This difference was found to be statistically significant (P = 0.025, Table 1). These cryosections were also analyzed for the percentage of microvesicles/field which contained AAV vectors. This analysis revealed that 12.2% ± 2.9% and 9% ± 7.6% of microvesicles contained AAV capsids from AAV1 and AAV2 preparations, respectively. We also examined cryosections of AAV1 producing 293T cells using immunogold labeling of the capsid. Labeling of clusters of AAV capsids in the nucleus was observed (data not shown). We observed specific labeling of capsids at the cell surface in association with microvesicle (Figure 1e). Figure 1f shows a budding microvesicle which is labeled with anti-AAV1/10 nm gold. Lower magnification images showed immunogold labeling of AAV1 particles in the nucleus, cytoplasm, and at the cell surface (Supplementary Figure S3). Next, we analyzed whether AAV production in 293T cells stimulated microvesicle release. Microvesicles were isolated from cell cultures transfected with a control plasmid encoding firefly luciferase (Fluc), or from cells transfected with AAV1-Fluc plasmids or AAV2-Fluc plasmids. Forty-eight hours post-transfection, we counted microvesicles using nanoparticle tracking analysis (NTA). A slight, yet statistically significant reduction (1.23–1.28-fold) in microvesicle count (NTA detectable size range 50–1,000 nm) for AAV transfected cells compared to control plasmid transfected cells was observed, suggesting that AAV vector production does not enhance microvesicle release (Supplementary Figure S4).
Figure 1.
293T cells producing adeno-associated virus (AAV) vectors secrete microvesicles associated with AAV particles. (a,b) Transmission electron micrograph (TEM) of 293T cells shedding microvesicles. Bar in (a) = 500 nm. Bar in (b) = 100 nm. (c) Cryosectioned microvesicle fractions from the media of AAV1 and AAV2-producing 293T cells. Sections were stained with an anti-AAV1 capsid antibody (panels iii, iv) or an anti-AAV2 antibody (panels i, ii) followed by a 10-nm gold-labeled secondary antibody. White arrows indicate AAV capsids apparently within microvesicles; black arrows indicate AAV capsids on surface of microvesicles; Bars =100 nm. Note: Staining controls for specific detection of AAV1 and AAV2 capsids are displayed in panels (i) (AAV1 vexosomes stained with anti-AAV2 antibody followed by the gold-labeled secondary (antibody) and (iv) (AAV2 vexosomes stained with anti-AAV1 antibody followed by the gold-labeled secondary antibody). (d) Direct visualization of capsid inside microvesicle along with 5-nm gold labeling of capsids. The microvesicle membrane is indicated by a black arrow, while capsids are indicated by white arrows. Note the punctuate dots indicating 5-nm gold labeling on one or more capsids inside the vesicle. Bar = 100 nm. (e,f) Immunogold labeling of cryosections of AAV1-producing 293T cells. Microvesicles containing gold-labeled AAV capsids are indicated with black arrows. The cell membrane is indicated with a white arrow. Bar = 100 nm.
Table 1. Average number of AAV capsids-associated per microvesicle.
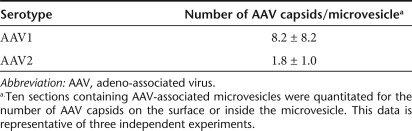
To compare the sedimentation efficiencies of AAV2 vexosomes to a conventionally purified AAV2 vector through a velocity gradient, we harvested media from 293T cells producing an AAV2 vector encoding Fluc. The microvesicles in media were pelleted using the 20,000g centrifugation described above, the pellet was resuspended and treated with Benzonase to remove any plasmid DNA bound to the microvesicle surface, and layered onto a 6–18% iodixanol velocity gradient, as described.19 We also concentrated the 20,000g microvesicle pellet-depleted media using centrifugal concentrators with a 100-kDa molecular weight cutoff and loaded it onto a separate gradient. Finally, as a control another gradient was layered with a 20,000g microvesicle pellet obtained by mixing conventionally purified AAV2-Fluc with microvesicle-containing media from nontransfected 293T cells. Fractions were collected, starting from the top of the gradient. 293T cells were infected with aliquots from different fractions of the gradient and 72 hours later, cells were harvested and luciferase activity was measured. For conventionally purified AAV2-Fluc vector mixed with purified microvesicle-containing media, 90% of the luciferase activity was contained between fractions 30–36 with only 16% being localized in the bottom fraction (fraction 36; Figure 2a). In contrast, luciferase activity in the 20,000g vexosome pellet sample was concentrated in the bottom fraction (81% of the total activity; Figure 2a). Finally, when the gradient sample containing microvesicle-depleted media was analyzed, the distribution of luciferase activity was similar to free AAV mixed with microvesicle-containing media, suggesting that a substantial amount of AAV2 in the media of producer cells is microvesicle-free (Figure 2a). The data with free AAV mixed with microvesicles also suggests that the majority of free AAV does not stick to the microvesicle surface.
Figure 2.
Characterization of vexosomes. (a) Functional vexosomes are contained in different gradient fractions than standard adeno-associated virus (AAV). The following three samples were loaded onto different 6–18% iodixanol step gradients: (i) standard AAV2-Fluc vectors mixed with microvesicles (MV), (ii) vexosomes pelleted at 20,000g (20K × g AAV2-Fluc vexosome pellet), (iii) vexosome-depleted media (20K × g vexosome pellet-depleted media). After centrifugation the gradients were fractionated and 5 µl aliquots from each fraction were used to transduce separate wells of 293T cells. Two days later, cell lysates were examined for Fluc activity to determine which fractions contained functional vector. (b) Nanoparticle tracking analysis (NTA) of iodixanol gradient fraction #36 from the standard AAV2-Fluc vector mixed with purified 293T-derived microvesicles (dashed line) and of iodixanol fraction #36 from the vexosome sample (solid line). (c) Detection of GAPDH mRNA in vexosome sample using reverse transcription (RT)-PCR. The PCR products were visualized on a 2% agarose gel. Lane 1, 108 AAV genome copies (g.c.) from fraction #36 of iodixanol gradient vexosomes pelleted at 20K× g; lane 2, 108 AAV g.c. from fraction #36 of iodixanol gradient of vexosome 20K × g-depleted media; lane 3, #36 of iodixanol gradient, pure AAV (108 g.c.) mixed with 293T microvesicles sample; lane 4, pure AAV (108 g.c.); lane 5, HeLa cell total RNA used as template for RT-PCR (positive control); lane 6, HeLa cell total RNA used as template (no RT step, negative control); lane 7, no template control; lane 8, DNA ladder. (d,e) Bioanalyzer analysis of RNA in (d) conventionally purified AAV-Fluc vector sample (108 g.c.) or (e) 108 g.c. from vexosome sample. Black bracket indicates area of small RNA species.
Characterization of vexosome size and contents
To better understand the size of microvesicles observed to colocalize with AAV vectors in media, fraction 36 from the vexosome pellet sample or fraction 36 from the conventional AAV2-Fluc mixed with microvesicles (from Figure 2a) was analyzed for size by NTA. The AAV2-Fluc vector mixed with microvesicle-containing media from nontransfected 293T gave a profile ranging from around 50–200 nm with a mean size of 104 nm (Figure 2b). For the vexosome fraction, we observed a similar microvesicle size distribution with the addition of a distinct “shoulder” at around 200 nm in size (Figure 2b).
As microvesicles are known to contain various nucleic acids, we confirmed that the vexosomes contained mRNA. Total RNA was isolated from the vexosomes isolated in fraction 36 of the iodixanol gradient. This RNA was used as a template for reverse transcription following by PCR amplification of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA using specific primers. Agarose gel electrophoresis of this PCR product confirmed the presence of GAPDH mRNA in the vexosome fraction (Figure 2c), as expected for microvesicles.
We analyzed the RNA sample from the vexosomes for size distribution of RNA using a Bioanalyzer. Conventionally purified AAV [108 genome copies (g.c.)] did not contain a detectable small RNA peak (Figure 2d). As expected, the vexosome sample contained small RNAs which ranged from ~25–200 nucleotides in length (Figure 2e).
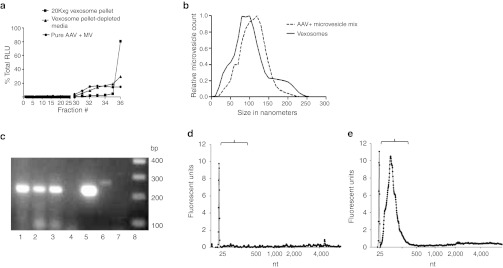
Simplified purification protocol and gene delivery properties of vexosomes in vitro. While the iodixanol step gradient was useful in characterizing vexosomes, it is an 11-step gradient, making its routine use cumbersome when dealing with multiple samples. We therefore sought to use another type of gradient with fewer density layers for a more streamlined vexosome purification approach. We chose to use a 8%, 30%, 45%, 60% sucrose density gradient as this has been successfully employed for exosome purifications.20 We layered the resuspended microvesicle pellet from AAV1-Fluc producer cell media onto the gradient and performed ultracentrifugation. Fractions were collected and microvesicles pelleted by a subsequent ultracentrifugation step at 100,000g, and resuspended in phosphate-buffered saline (PBS). We pelleted fractions 5–10 as exosomes are reported to migrate in the 30–45% density layer (fractions 5–7).20 To ascertain how microvesicle-free AAV migrated in the sucrose gradient, iodixanol purified AAV1-Fluc isolated from cell lysates was loaded onto a gradient and subjected to the same centrifugation and pelleting steps. We quantitated AAV genome copy number in fractions 5–10 for both samples. For cell lysate isolated AAV ~95% of the total g.c. were found in fraction 5. Another small peak (3% of total g.c.) was located in fraction 9. The genome copy quantitative PCR (qPCR) distribution was remarkably different for vexosomes (Figure 3a). Peaks were observed in fraction 5 (52% of total), fraction 7 (22% of total), and fraction 9 (17% of total). The peak in fraction 5 for the vexosome sample likely represents microvesicle-free AAV as the majority of cell lysate purified AAV was localized here, while the unique peak in fraction 7 most likely represents vexosomes as this fraction is located at the published density of exosomes (30–45% interface, ref. 20). The peak in fraction 9 may represent free AAV aggregates although it may also contain larger microvesicles harboring multiple AAV capsids. We also transduced 293T cells with an aliquot of fractions 5–10 for either cell lysate isolated AAV or vexosomes and performed a luciferase assay 3 days post-transduction. The transduction data showed a similar profile as the qPCR data for both samples (Figure 3b). Finally, we performed an immunoblot on fractions 5, 7, and 9 to detect the microvesicle-associated protein, Alix.11 For the vexosome sample, Alix was detected in all three fractions analyzed (#5, 7, and 9) while it was not detected for cell lysate isolated AAV (Supplementary Figure S5 and Supplementary Materials and Methods). The Alix immunoblot suggests that fraction 5 contains free AAV particles along with AAV-free microvesicles and that fraction 7 contains the vexosomes. To ascertain whether fraction 7 purified AAV1 vexosomes could transduce cells in the presence of neutralizing antibody, either conventionally purified AAV1-Fluc or AAV1-Fluc vexosomes were mixed with media containing a control anti-AAV2 antibody, or a dilution series of media containing an anti-AAV1 antibody which recognizes intact capsids. After antibody/vector incubation, the mixture was added to U87 cells for a transduction assay. Forty-eight hours post-transduction cells were harvested for a luciferase assay. At the lowest antibody dilution (1:1,000) there were only low levels of transduction (compared to transduction in the presence of the anti-AAV2 control antibody) for both AAV1 and AAV1 vexosomes. However, at higher dilutions of antibody, AAV1 vexosomes-transduced cells at a higher efficiency than did conventionally purified AAV1 (Figure 3c). For example at a 1:2,000 dilution of antibody, AAV1 vexosomes yielded a 4.6-fold (P < 0.05) higher transduction efficiency than AAV1 (Figure 3c).
Figure 3.
Streamlined vexosome purification using sucrose gradients. Vexosomes are banded by sucrose density ultracentrifugation. The resuspended microvesicle pellet from AAV1-Fluc producer cells or iodixanol purified AAV1-Fluc from cell lysates were loaded onto a sucrose gradient. After centrifugation sucrose gradient fractions were analyzed for (a) adeno-associated virus (AAV) genomes or (b) Fluc activity after transduction of 293T cells with an aliquot of each fraction. The unique peak in fraction 7 of media purified AAV is indicated by a black arrow. This likely represents the major vexosome fraction. To calculate total g.c., the number of g.c. in each fraction (titer in g.c./ml × sample volume) were added together to give the total contained in the entire gradient. The % total for each fraction was calculated by the following equation: %Total g.c. = total g.c. in fraction n/gradient total g.c. × 100. (c) Antibody neutralization assay of AAV1-Fluc vexosomes. 108 g.c. of standard purified AAV1-Fluc or AAV1-Fluc vexosomes were incubated with a control anti-AAV2 antibody or serial dilutions of a neutralizing anti-AAV1 antibody. After 1-hour incubation, the mixture was added to cells and a transduction assay performed on U87 cells. Cells were harvested at 48 hours for a luciferase assay.
To quantitate the yield of sucrose gradient purified vexosomes from small-scale preparations (one 15-cm plate), we performed qPCR for AAV g.c. on cell lysates, media (containing free and microvesicle-associated AAV), and the sucrose gradient purified vexosomes isolated from the cognate media sample. This was performed for both AAV1-Fluc and AAV2-Fluc transfected 293T cells (Table 2). For AAV1 the purified vexosome fraction constituted 0.01% of the total AAV genome yield (sum of media and cell lysate numbers), while it was 0.2% for AAV2 (Table 2).
Table 2. Vexosome yields from sucrose density gradient purification.
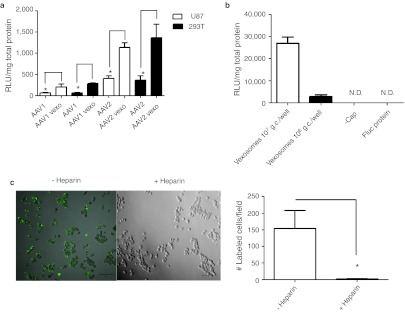
To evaluate the ability of vexosomes to deliver AAV genomes to cells in vitro, we transduced human 293T cells and human U87 glioma cells with 107 g.c./well of conventionally purified AAV1-Fluc or AAV2-Fluc vector as well as AAV1-Fluc vexosomes or AAV2-Fluc vexosomes (vexosomes isolated from fraction #7 of the sucrose gradient described in Figure 3). Forty-eight hours post-transduction cell lysates were analyzed for luciferase activity. Strikingly, both AAV1-Fluc vexosomes and AAV2-Fluc vexosomes mediated enhanced transduction (3–4.5-fold) compared to their cell lysate purified parental serotypes (Figure 4a). As previously reported, standard cell lysate purified AAV2 vector is more efficient than AAV1 vectors at transducing U87 cells6 and was also more efficient on 293T cells (Figure 4a).
Figure 4.
Gene delivery to cultured cells by vexosomes. (a) Human 293T or U87 cells were transduced with 107 genome copies (g.c.)/well of standard AAV1 or AAV2-Fluc vectors or AAV1 or AAV2-Fluc vexosomes purified from sucrose gradient fraction #7. Forty-eight hours post-transduction, cells were lysed and luciferase activity determined. Mean + SD are shown (b) Fluc transgene expression from vexosomes is not plasmid DNA, mRNA, or microvesicle-associated luciferase protein mediated. 293T cells were transduced/transfected with 107 and 106 g.c./well of AAV2-Fluc vexosomes or 0.4 µg of microvesicles from AAV-Fluc (-cap) transfected 293T cells. The microvesicles from AAV2-Fluc vexosomes were also directly lysed for analysis of Fluc protein contained in microvesicles. Mean + SD are shown. (c) Binding and uptake of 293T cell derived-microvesicles into 293T cells is blocked by coincubation with heparin. Microvesicles from 293T cells (not transfected with adeno-associated virus (AAV) plasmids) were labeled with the lipid dye, PKH67 and added to 293T cells in the presence or absence of heparin (250 µg/ml). Cells were incubated at 37 °C for 45 minutes before fixation and imaging by confocal microscopy for exosome-associated fluorescence. Bars = 100 µm. Graph depicts average quantitation of PKH+ cells in the presence or absence of heparin + SD (n = 3).
As microvesicles have been reported to deliver siRNA, mRNA, and proteins to recipient cells,14,21 we ascertained whether any of the luciferase activity observed at 48 hours in our transduction experiments using vexosomes could be due to luciferase plasmid DNA, transcribed luciferase mRNA, or translated luciferase protein packaged within microvesicles from the transfected 293T donor cells. We performed a transfection of 293T cells with the AAV plasmid containing the Fluc construct, adenovirus helper plasmid, and a plasmid which expressed the AAV rep proteins but no capsid proteins so that no AAV capsids could be produced. Omitting capsid expression (and thus formation of intact AAV vectors) would allow us to observe if any microvesicle-associated Fluc plasmid, mRNA, or protein were involved in transferring luciferase activity. Microvesicles were isolated from these cells and treated ± Benzonase nuclease to remove any DNA bound to the surface. The plasmid DNA was titered by qPCR of the 3′ end of the Fluc expression cassette. No signal above detection level was observed by qPCR in the Benzonase-treated sample, while the titer was 2.0 × 1010 g.c./ml in the absence of Benzonase, suggesting that Benzonase completely removes surface bound DNA (data not shown). This also suggests that the vexosome preparations do not contain Fluc plasmid DNA (below detection limit) on the inside (nuclease protected) of microvesicles. Next, we performed a microvesicle transfection/transduction experiment with vexosomes lacking the capsid proteins as well as the AAV2-Fluc vexosomes with capsid proteins for direct comparison. We determined microvesicle titers by NTA to allow comparison between the samples. 293T cells were incubated with cap containing AAV2-Fluc vexosomes at 107and 106 g.c./well, corresponding to 5 × 108 and 5× 109 microvesicles/well, respectively. For the vexosomes lacking capsid proteins, each well of 293T cells was incubated with 5 × 109 microvesicles. Forty-eight hours after incubation cells were harvested and assayed for luciferase activity. No luciferase activity was observed above background for the microvesicles lacking cap, indicating that the capsid is necessary for efficient gene transfer of the AAV transgene (Figure 4b). In contrast, luciferase activities were 2.7 × 104 relative light units/mg and 2.8 × 103 relative light units/mg for 107 and 106 g.c./well of AAV2 vexosomes, respectively (Figure 4b). Finally, for direct detection of luciferase activity in purified AAV2-Fluc vexosomes, we performed a luciferase assay on lysed vexosomes (1 µl) and compared this to the luciferase levels in transduced 293T cell lysates obtained after incubation of 1 µl (106 g.c./well) of the same sample with 293T cells. Luciferase activity was undetectable in lysed AAV2-Fluc vexosomes, whereas it was 2.8 × 103 relative light units/mg, as mentioned above, after transduction with AAV2-Fluc vexosomes (Figure 4b). These results indicate that the transgene expression levels observed after vexosome delivery (at 48 hours post-transduction) is not due to donor microvesicle-associated plasmid DNA, Fluc mRNA, or Fluc protein. Further supporting this conclusion is the transduction data with AAV1-Fluc and AAV2-Fluc vexosomes which package the same AAV genome and transgene cassette but yield different transduction efficiencies when delivered at the same g.c. dose (Figure 4a). If Fluc plasmid, mRNA, or protein were the cause of the observed transduction, then cell transduction efficiencies for AAV1 and AAV2 vexosomes would be expected to be the same.
We also examined cellular attachment of 293T-derived microvesicles (AAV-free) to 293T cells. It was previously shown that microvesicles (from glioma cells) are positively charged.22 Heparin sulfate is a highly negatively charged glycosaminoglycan with potential to interact with microvesicles, and heparin can also be used to block initial binding of viruses (including AAV2) that utilize heparan sulfate proteoglycan on the cell surface. We examined the ability of heparin to block uptake of 293T-derived microvesicles into recipient 293T cells. Purified microvesicles from nontransfected 293T cells were labeled with a fluorescent lipid dye, PKH67. These labeled microvesicles were incubated in the presence or absence of heparin and added to wells containing 293T cells and incubated at 37 °C for 45 minutes. Analysis of cells by confocal microscopy revealed a near complete inhibition of cellular uptake of microvesicles in the presence of heparin (Figure 4c).
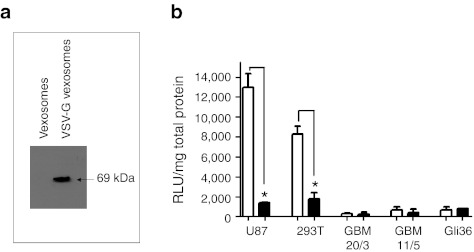
Vexosome pseudotyping with a membrane-bound receptor. Enveloped virus vectors can be retargeted/modified by insertion of a ligand into the membrane. Pseudotyping vectors with vesicular stomatitis virus glycoprotein G (VSV-G) has been used to enhance enveloped virus vector transduction in cultured cells due to the broad tropism of VSV-G.23 We employed a similar strategy for vexosomes by cotransfecting a VSV-G plasmid into 293T cells at the time of AAV2-Fluc production. Interestingly, we observed a visibly larger microvesicle pellet when including the VSV-G plasmid during vexosome production (data not shown). Isolation of nucleic acid from equal volumes of the microvesicle producer cell media from standard vexosomes or VSV-G vexosomes yielded nucleic acid concentrations of 8 and 90 ng/µl, respectively, indicating roughly a tenfold increase when pseudotyping with VSV-G. Furthermore, including the VSV-G plasmid increased the yield of AAV2 vector genomes in the microvesicle fraction by 13-fold (P = 0.0029, Supplementary Figure S6) This supports the observations reported by Mangeot et al. in which VSV-G expression stimulated microvesicle production in transfected cells.24 We confirmed VSV-G incorporation into vexosomes by immunoblotting the 20,000g microvesicle/vexosome pellets (Figure 5a and Supplementary Figure S7). Next, we transduced a panel of cell lines with vexosomes (107 g.c./well) purified from 293T cells with or without pseudotyping them with VSV-G. Remarkably, a distinct transduction profile was observed for the VSV-G vexosomes compared to the standard vexosomes (Figure 5b). For example, standard vexosomes transduced U87 cells and 293T cells nine- and five-fold more efficiently than VSV-G vexosomes, respectively (Figure 5b) whereas it was not significantly changed on the other cell lines tested.
Figure 5.
Engineering vexosomes with vesicular stomatitis virus glycoprotein G (VSV-G) glycoprotein. AAV2-Fluc vexosomes coated with VSV-G (AAV2-Fluc VSV-G vexosomes) were produced by transfecting 293T cells with adeno-associated virus (AAV) plasmids as well as a VSV-G expression vector. (a) Immunoblot detection of VSV-G on AAV-VSV-G vexosomes. (b) Transduction of a panel of cells (cell types indicated in graph) were transduced with 107 g.c./well of either AAV2-Fluc vexosomes or AAV2-Fluc VSV-G vexosomes. Forty-eight hours post-transduction cells were lysed and a luciferase assay performed. White bars, AAV2-Fluc vexosomes; black bars, AAV2-Fluc VSV-G vexosomes. Mean + SD are shown.
Magnetic targeting of vexosomes
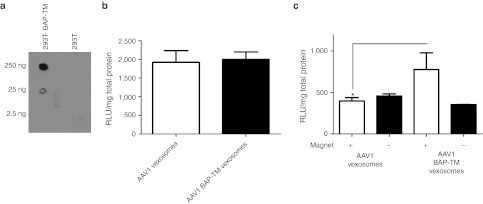
We next ascertained whether we could specifically target vexosomes to cells by expressing a transmembrane receptor on the microvesicle surface. We chose to use the previously described biotin acceptor peptide—transmembrane domain (BAP-TM) receptor 25 as this genetically engineered receptor allows binding to biotinylated ligands via a streptavidin bridge. 293T cells were transduced with two lentivirus vectors encoding BAP-TM and biotin ligase BirA, which allows efficient biotinylation in mammalian cells.26 To evaluate successful incorporation of BAP-TM into microvesicles, we spotted nitrocellulose membranes with 2.5–250 ng of microvesicle-associated protein from 293T cells or 293T cells transduced with the lentivirus vectors (called 293T-BAP-TM cells) and probed it with streptavidin-horse radish peroxidase. As expected, specific signal was detected for 293T-BAP-TM-derived microvesicles but not for 293T-derived microvesicles, indicating successful pseudotyping of the microvesicles, incorporating biotin in its membrane (Figure 6a). Next, we produced and purified AAV1-Fluc vexosomes in 293T cells and AAV1-Fluc BAP-TM vexosomes in 293T-BAP-TM cells. We first compared the transduction efficiency of untargeted vexosomes by incubating U87 glioma cells with 108 g.c./well of either AAV1-Fluc vexosomes or AAV1-Fluc BAP-TM vexosomes. No significant difference in transduction efficiency was observed at 48 hours post-transduction (Figure 6b). To target vexosomes, we incubated AAV1-Fluc vexosomes or AAV1-Fluc BAP-TM vexosomes with streptavidin-conjugated magnetic beads. A small magnet was adhered to the underside of one region in a well of a 12-well plate. We added AAV1-Fluc vexosomes or AAV1-Fluc BAP-TM vexosomes to these wells expecting vexosomes which could efficiently bind to the magnetic beads to preferentially transduce cells in the region of the magnet. At 48 hours post-transduction, cells within the magnetic zone were harvested followed by the remaining cells outside of the magnetic zone (see Supplementary Figure S8 for experimental schematic). Luciferase activity was determined and we observed a twofold enhanced transduction of BAP-TM vexosomes compared to standard vexosomes within the magnetic zone (P < 0.05), Figure 6c). The ratio of luciferase signal inside the magnetic zone to outside was 0.86 for AAV1 vexosomes while it was 2.5 for AAV1 BAP-TM vexosomes suggesting a more specific targeting by the streptavidin-conjugated magnetic nanoparticles when the biotinylated ligand (BAP-TM) was expressed on the microvesicle surface.
Figure 6.
Magnetic targeting of vexosomes. Vexosomes were engineered to express a biotinylated transmembrane receptor (BAP-TM) on their surface by transducing 293T cells with a lentivirus encoding BAP-TM. (a) Streptavidin-horseradish peroxidase (HRP) dot blot detection of biotinylation on BAP-TM microvesicles. Microvesicles (protein amount indicated on figure) from 293T cells or 293T-BAP-TM cells were spotted onto nitrocellulose. Specific binding of streptavidin-HRP was detected by chemiluminescence. (b) Transduction efficiency of 107 g.c./well of untargeted AAV1-Fluc vexosomes or AAV1-Fluc BAP-TM vexosomes on U87 cells. (c) Magnetic targeting of AAV1-Fluc BAP-TM vexosomes to U87 glioma cells. Vexosomes or BAP-TM vexosomes incubated with magnetic beads were added to a 12-well plate with a magnet attached to the underside of the well. Forty-eight hours post-transduction cells were harvested from the area inside the magnetic zone (+ magnet) as well as the cells outside the magnetic zone (- magnet). Luciferase assays were performed on each of these samples and protein levels normalized by Bradford assay. For (b–c) mean + SD is shown. AAV, adeno-associated virus.
Discussion
Our studies demonstrate that under normal AAV production conditions in 293T cells, a fraction of AAV vectors isolated from the cell culture media can be detected within microvesicles. To our knowledge, this is the first report directly visualizing virus vector capsids associated with and inside microvesicles (vexosomes) and showing that these vectors can be purified and can efficiently transduce cells. These vexosomes were capable of enhanced gene transfer in cultured cells compared to conventionally purified AAV using the same AAV genome copy number dose (Figure 4a). Finally, we were able to show that incorporation of ligands into the microvesicle membrane allowed enhanced targeting in cultured cells (Figure 6c).
The mechanism of enhanced gene transfer in culture has yet to be determined but may depend on factors such as alternate receptor usage, direct fusion of microvesicles with the recipient cell membrane or increased escape from endosomes post entry. Microvesicles are known to express a plethora of host cell-derived cell surface receptors and ligands27 so it is quite possible that one or more of these molecules are utilized in cell entry, however we showed that heparin blocked purified 293T-derived microvesicle uptake by 293T recipient cells (Figure 4c). Although it is unclear at this time if heparin is blocking an electrostatic interaction or specific heparan sulfate proteoglycan attachment, many viruses exploit this receptor for attachment in cultured cells28,29,30,31 and it is possible that 293T-derived vexosomes also exploit this receptor for attachment.
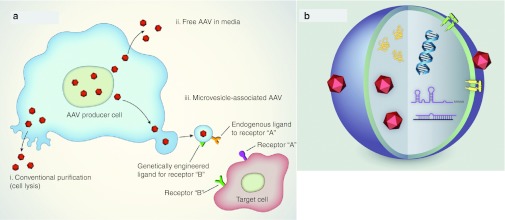
In producer cells, since 80–90% of AAV2 vector is found intracellularly, the media is typically discarded and the vector purified from cellular lysates (Figure 7a). However, two recent reports demonstrated that for some AAV serotypes (e.g., AAV1), over 80% of the total vector yield is found in the media,32,33 although the mechanism by which AAV is shed from the cell has not previously been described. As we have observed both free and microvesicle-associated AAV in the medium (Figures 2a and 3a), it is possible that more than one pathway is involved in the export of AAV vectors from cells (Figure 7a). Based on the transmission electron microscopy analysis of 293T cells we believe the majority of AAVs associate with shedding microvesicles at the plasma membrane rather than being expelled through multivesicular bodies. The yields obtained for the sucrose purified AAV1 vexosomes and AAV2 vexosomes are typically 108 g.c. from a 15-cm plate 293T cell preparation. This corresponds to ~0.01% of the total amount of AAV1 and 0.2% of AAV2 produced in these cells. This is likely to be an underestimation of the actual vexosome titer and percentage due to the loss during the multistep purification process (two pelleting steps and one density gradient). Optimization of the production and purification process may increase vexosomes yields.
Figure 7.
Overview of adeno-associated virus (AAV) vexosome vector system. (a) AAV is conventionally purified from cell lysates (i), However, AAV is also shed into the media apparently as both free particles32 (ii), as well as microvesicle-associated vectors (vexosomes) (iii). Vexosomes can be isolated from the media and used for gene delivery to target cells. (b) Illustration of vexosome components. The microvesicle may contain AAV vectors encoding a gene of interest, mRNA, miRNA or DNA as well as vector encoded and nonvector-encoded proteins (orange). The microvesicle surface may be decorated with receptors or ligands endogenously (yellow) or ectopically (green) expressed in the packaging cell.
Interestingly, we observed that expression of VSV-G on vexosomes increased microvesicle yield as well as AAV genome titers approximately tenfold (Supplementary Figure S6). In addition to increasing vexosome yields, VSV-G expression changed the cell transduction profile compared to unmodified 293T-derived vexosomes (Figure 5b). This is most likely due to an interaction of fusogenic VSV-G with its cognate receptor in lieu of the endogenous receptors and ligands expressed on the 293T cell-derived vexosome surface. Surprisingly, we observed a lower transduction efficiency on U87 and 293T using VSV-G vexosomes compared to standard vexosomes. It may be that VSV-G expression on the surface of vexosomes interferes with innate microvesicle proteins which allow efficient attachment to these cell types. Alternatively, VSV-G mediated entry of vexosomes may enable efficient endosomal escape34 before the essential endosomal acidification required by AAV for efficient transduction.35 Although it is currently not known what mechanism/pathway the vexosomes follow from the cell surface to the nucleus, it is likely that properties of both the microvesicle and the AAV capsid play a role in this process. AAV2 vectors are more efficient at gene transfer to U87 and 293T cells compared to AAV1 vectors.6,36 A recent study examining cell entry, trafficking, nuclear import and transduction of different AAV serotypes found that although AAV1 entered cells efficiently and rapidly, only low levels of transduction were observed compared to AAV2.37 This was attributed to inefficient AAV1 capsid disassembly at the nucleus.37 Thus, it is not surprising that the AAV2-Fluc vexosomes were more efficient than AAV1-Fluc vexosomes in culture (Figure 4a), as the capsid is most likely involved in vexosome transduction properties.
The discovery of AAV virus capsids associated with microvesicles may open a new window of opportunity for gene therapy applications. The AAV vector may be codelivered with microvesicles together with therapeutic proteins, mRNA, regulatory noncoding RNAs or even DNA17 by electroporation of microvesicles as performed by Alvarez-Erviti et al.21 and depicted in Figure 7b. The fact that a multitude of proteins and RNA can be codelivered to the target cell may give an opportunity to change the behavior of the vector itself. Microvesicle-transferred Fluc plasmid, mRNA, or protein did not play a significant role in the luciferase activities obtained 48 hours after vexosome incubation at the vexosome concentrations used here, indicating that such transfer is much less efficient than when a functional AAV vector is associated with the microvesicle. The reporter gene used in our study, firefly luciferase (Fluc), has a short half-life of 2 hours in mammalian cells ensuring the luciferase signal measured 48–72 hours postinfection was linked to gene transfer rather than protein transfer.38 However, the use of a reporter with a longer half-life (e.g., GFP, 26 hours;39 Gaussia princeps luciferase, 6 days40) may reveal an additional protein-mediated contribution of transgene expression after vexosome delivery. It is also important to note that while we did not detect luciferase protein in a small volume of lysed vexosome sample (Figure 4b) analysis of larger amounts of the preparation may reveal low levels of activity. As other viral particles, e.g., HIV41 and hepatitis C,42 have also been shown to be associated with microvesicles, it is possible that other viruses useful as gene therapy vectors may also be incorporated to obtain novel vexosome gene delivery vehicles. Packaging within or in association with lipid membranes offers at least one major advantage. It allows molecules incorporated into the plasma membrane to be used for targeting of the vexosomes (Figures 6c and 7b). In addition, AAV1-Fluc vexosomes were neutralized at high concentrations of an anti-AAV1 antibody, but were resistant at lower concentrations as compared to standard AAV under identical conditions (Figure 3c). This suggests that at least some of the AAV vector associated with vexosomes is protected from the neutralizing effects of the antibody. This finding warrants further investigation into the relative immunogenicity of vexosomes compared to standard AAV vectors, which may provide an additional advantage.
In summary, we have discovered a novel pathway by which AAV vectors are exported from producer cells. These microvesicle-associated vectors can be purified from the media of the vector producer cell lines, giving the vector unique properties as compared to conventionally purified vectors. Isolation and initial characterization of these enveloped AAV vectors showed that they displayed efficient gene delivery properties and may expand the uses of this vector platform both as a tool of molecular biology as well as a gene therapy vector.
Materials and Methods
Cell culture. The human glioblastoma cell line U87 and human 293T cells were obtained from American Type Culture Collection (Manassas, VA). Gli36 cells were obtained from Dr Anthony Capanogni (University of California at Los Angeles, Los Angeles, CA). Primary GBM cell isolates, GBM 20/3, and 11/5 have been previously described.6,17 All cells were cultured in high glucose Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Sigma, St Louis, MO) and 100 U/ml penicillin, 100 µg/ml streptomycin (Invitrogen) in a humidified atmosphere supplemented with 5% CO2 at 37 °C.
Vector production and microvesicle isolation
Vexosome production: AAV vectors were produced by transfecting 15 cm plates of 293T cells using the calcium phosphate method with the following plasmids: 12 µg of an ITR-flanked AAV transgene expression vector (firefly luciferase, Fluc), 25 µg of an adenovirus helper plasmid FΔ6,43 and 12 µg of the AAV2 rep/cap expression vector, pH22.44 In some experiments, pH22 was replaced with pXR1, an expression vector-encoding AAV2 rep and AAV1 capsid proteins.36 In the -capsid experiment we used a modified pH22 vector in which the DNA encoding for capsid has been digested out. Sixteen hours post-transfection, media was exchanged with fresh media supplemented with 2% microvesicle-depleted FBS.14 Forty-eight hours post-transfection media was harvested and microvesicles and associated AAV vectors were purified as described below in “gradient centrifugation.”
Conventional AAV production: Standard AAV was generated with conventional purification methods as previously described.6 Briefly, media was discarded from transfected 293T cells and cells were lysed by three freeze/thaw cycles in a dry ice/ethanol bath followed by immersing tubes in a 37 °C water bath. Cellular debris was removed by centrifugation at 1,000g for 10 minutes. The clarified lysate was treated with 50 U/ml of Benzonase for 1 hour at 37 °C. AAV vector was purified from the lysate using an iodixanol step gradient (15%, 25%, 40%, 60% layers). Vector was harvested in the 40% layer after a 1 hour centrifugation at 360,000g. Virus was concentrated and the buffer exchanged to PBS using Amicon Ultra-15 100K centrifugal filter devices (Millipore, Billerica, MA).
Vector quantitation: The vector titer (in g.c./ml) was performed using a quantitative TaqMan PCR assay as previously described.45 First, AAV genomic DNA was isolated by treating a 2 µl vexosome fraction in a 50 µl reaction with 2 U of DNaseI for 2 hours at 37 °C to remove any potential unencapsulated or extravesicular AAV genomes or plasmid DNA. Next, DNase was inactivated for 25 minutes at 75 °C. Then a PCR was prepared using TaqMan Fast Universal PCR Master Mix (Applied Biosystems, Foster City, CA), a TaqMan probe (5′-6FAM-tgccagccatctgttgtttgcc-MGB; Applied Biosystems) and primer set (forward primer, 5′-cctcgactgtgccttctag-3′ reverse primer, 5′-tgcgatgcaatttcctcat-3′) which specifically anneals to the Poly A signal sequence region in the transgene cassette. A standard curve was prepared using serial dilutions of an AAV plasmid of a known molar concentration. The quantitative PCR was performed in an Applied Biosystems 7500 Thermal cycler using the following conditions: 1 cycle, 94 °C 30 seconds; 40 cycles 94 °C 3 seconds, 60 °C 30 seconds.
Gradient centrifugation
Iodixanol gradient centrifugation: Vexosomes were purified using a 6–18% iodixanol gradient, as previously described.19 with the following modifications. The media harvested from AAV producer cells (293T) was centrifuged for 10 minutes at 300g to remove floating cells followed by a 10 minutes at 1,000g spin to remove apoptotic bodies. Media was then transferred to a fresh tube and centrifuged at 20,000g for 25 minutes, k factor, 1,279. The resulting microvesicle pellet was resuspended in PBS, pH 7.4, with 2.5 mmol/l MgCl2. The resuspended pellet was treated with 250 U of Benzonase nuclease (Sigma) for 1 hour at 37 °C. Next, a 6–18% iodixanol step gradient (1.2% increments) was prepared in 38.5-ml open-top thin-walled polyallomer tubes (Beckman, Palo Alto, CA) using Optiprep (Sigma-Aldrich, St Louis, MO). After overlaying the gradient with 1 ml of the Benzonase-treated sample, the tubes were centrifuged at 32,000 r.p.m. in a SW32 Ti rotor (Beckman-Coulter, Brea, CA) for 129 minutes with the brake off in an Optima L-90K ultracentrifuge (Beckman-Coulter). Following this centrifugation, the top 2 ml of the gradient were discarded and then 1 ml fractions from the top to the bottom of the gradient were collected.
Sucrose gradient ultracentrifugation: Vexosomes were pelleted as described in iodixanol gradient centrifugation. Resuspended vexosome pellets (500 µl in PBS with 2.5 mmol/l MgCl2) were treated with Benzonase (250 U) for 1 hour at 37 °C and then layered onto a sucrose density gradient (8, 30, 45, 60% layers) and centrifuged 38 minutes at 50,000 r.p.m. in a MLS-50 swinging bucket rotor (Beckman) in a Beckman Optima MAX-XP ultracentrifuge (Beckman) with deceleration set to 8. Fractions were collected, diluted 1:10 in PBS and microvesicles pelleted at 100,000g for 75 minutes in the MLS-50 rotor. Pellets from each fraction were resuspended in 50 µl PBS and used in Fluc transduction assays, genome copy quantitative PCR analysis, and western blot analysis for microvesicle-associated proteins.
VSV-G vexosomes. Vexosomes pseudotyped with VSV-G were produced and purified as described above with the inclusion of 4 µg of a VSV-G expression vector per plate at the time of AAV plasmid cotransfection.
Vector transduction assays. 104 cells (cell type indicated in figure) were plated the day before transduction into each well of 96 well plates. Purified AAV or vexosomes (dose indicated in figure legend) were mixed with serum-free media in a total volume of 100 µl and added to cells. After 1 hour at 37 °C, the media was changed and cells were incubated for 48–72 hours in complete media containing 10% FBS. Cells were then rinsed in PBS and lysed using Reporter Lysis Buffer (Promega, Madison, WI). A luciferase assay was performed on 20 µl lysate using a luminometer equipped with an injector that added 100 µl of luciferase substrate buffer/well. Luciferase activities (in relative light units) were normalized to protein content of the samples by performing a Bradford assay (BioRad, Hercules, CA). For the AAV1 neutralization assay, 108 g.c. of either cell lysate purified AAV1-Fluc or sucrose gradient purified AAV1-Fluc vexosomes were incubated for 1 hour at room temperature with each of the following: (i) 2% bovine serum albumin in serum-free OPTI-MEM I (Invitrogen) media with a 1:1,000 dilution of anti-AAV2 antibody (Clone A20; American Research Products, Belmont, MA), and (ii) media with a range of dilutions of anti-AAV1 antibody (Clone ADK1a; American Research Products). After incubation with antibody, the standard transduction assay described above was performed.
PKH67-labeled microvesicles and heparin-blocking assay. Purified 293T microvesicles were labeled with PKH67 green fluorescent-labeling kit (Sigma-Aldrich) as described.14 100 µl of labeled microvesicles were incubated with or without 250 µg/ml heparin (Sigma-Aldrich) in a total volume of 1 ml of Dulbecco's modified Eagle's medium supplemented with 10% FBS 30 minutes at room temperature. Next, the mixture was added to a well of 293T cells and cells incubated for 45 minutes at 37 °C before fixation in formaldehyde. Microvesicle-associated fluorescence was imaged using a Zeiss LSM 5 Pascal laser confocal microscope.
GAPDH reverse transcriptase-PCR. To detect mRNA inside microvesicles, 108 g.c. of purified AAV2-Fluc associated microvesicles were used to isolate RNA using Trizol LS reagent (Invitrogen). RNA was isolated from HeLa cells as a positive control. Next a two-step reverse transcriptase PCR was performed using Sensiscript reverse transcriptase (Qiagen, Valencia, CA) and HotStarTaq DNA polymerase (Qiagen) was performed to amplify GAPDH transcripts from microvesicle RNA or from HeLa RNA. cDNA synthesis: 50 ng of RNA was reverse transcribed using Sensiscript reverse transcriptase (Qiagen). PCR amplification of GAPDH cDNA: 1 cycle 95 °C for 3 minutes; 40 cycles, 95 °C for 30 seconds, 60 °C for 30 seconds, 70 °C for 30 seconds; 1 cycle 70 °C for 7 minutes. PCR products were analyzed on a 2% agarose gel stained with ethidium bromide.
Bioanalyzer analysis of RNA. RNA was measured using the Agilent RNA 6000 Pico Kit according to manufacturer's recommendations.
Microvesicle RNA extraction. Conditioned media was collected after 48 hours and centrifuged first at 500g for 10 minutes to pellet dead cells and debris. The supernatant was then filtered through a 0.8-µm filter and ultracentrifuged at 110,000g for 80 minutes in a 70Ti rotor. The pellet was washed in 12 ml PBS and repelleted at 100,000g for 60 minutes in a MLA-55 rotor. The pellet was resuspended in 100 µl Tris-buffered saline. The microvesicle RNA was further extracted with the miRNeasy mini kit according to manufacturer's recommendation.
NTA. The size and concentration of nanoparticles in the vexosome preparations was determined using a LM10 nanoparticle analyzer (Nanosight, Amesbury, UK) operated by NTA software version 2.0. Samples were diluted in PBS and the particle concentration and size distribution were measured.
BAP-TM vexosomes
BAP-TM vexosome production: 293T cells were transduced (10 transducing units/cell) with the previously described lentivirus vector, CSCW-BAP-TM25 as well as a lentivirus vector encoding the Escherichia coli biotin ligase, BirA, referred to as CSCW-BirA.26 The latter vector enables efficient biotinylation of BAP-TM in cultured mammalian cells.26 Vexosome production in these cells (referred to as 293T-BAP-TM) was the same as for parental 293T.
BAP-TM dot blot: Twenty milliliter of media from 293T or 293T-BAP-TM cells was harvested and microvesicles were pelleted at 20,000g for 60 minutes. The microvesicle pellet was washed with 20 ml PBS and microvesicles were pelleted again. The pellet was resuspended in 100 µl PBS, and associated protein was quantitated by Bradford assay. A nitrocellulose membrane was spotted with 2.5, 25, and 250 ng of the 293T and 293T-BAP-TM-derived intact microvesicles. The membrane was blocked overnight in 5% milk in PBS. The membrane was rinsed three times for 5 minutes each in PBS, incubated 30 minutes with a 1:10,000 dilution of streptavidin-horseradish peroxidase (GE Healthcare, Piscataway, NJ) in PBS containing 0.1% Tween 20, and then rinsed three times for 5 minutes, each in PBS. Chemiluminescence detection of horseradish peroxidase activity was performed with a Pierce Supersignal Western Pico Chemiluminescent Substrate kit (Pierce, Rockford, IL) followed by exposure of the membrane to autoradiography film (Denville Scientific, Metuchen, NJ).
Magnetic bead transduction assays: AAV1-Fluc Vexosomes or AAV1-Fluc BAP-TM vexosomes (108 g.c. of either) were incubated with 2% biotin-free bovine serum albumin in Opti-MEM (Invitrogen) for 30 minutes on ice before mixing with 10 µl of Streptavidin Microbeads (Miltenyi Biotec, Auburn, CA) (total reaction volume, 200 µl). Microbeads and the vexosome samples were incubated 30 minutes on ice. A 1.3 Tesla Neodymium rare earth disc magnet (10 mm diameter, 1 mm thickness; Indigo Instruments, Waterloo, Ontario, Canada) was taped to the bottom of a 12-well tissue culture plate containing U87 cells. Next, vexosomes or BAP-TM vexosomes incubated with magnetic beads were added to the well for 2 hours at 37 °C. The magnet was then removed, and Dulbecco's modified Eagle's medium containing 10% FBS was added and cells were incubated for 48 hours before luciferase assay analysis. For analysis, cells within the magnetic zone were carefully pipetted off and placed in a microcentrifuge tube followed before pipetting the remainder of cells outside the magnetic zone into a separate tube.
Transmission electron microscopy
Cryopreserved sections and immunogold labeling: Microvesicles and AAV in producer cell media (both AAV1 and AAV2 serotypes analyzed) were pelleted at 20,000g for 30 minutes. The pellet was resuspended in PBS and repelleted. Following this, the pellet was fixed for 2 hours in 4% formaldehyde in PBS before being cryosectioned. Sections were incubated with 1:100 dilutions of either anti-AAV1 (Clone ADK1a; American Research Products) or anti-AAV2 antibodies (Clone A20, American Research Products) (both antibodies recognize intact particles) followed by a 5 or 10 nm gold-labeled secondary anti-mouse antibody (Sigma). Controls for staining specificity were performed by incubating AAV1 vexosome samples with the AAV2 antibody followed by the secondary antibody or incubating AAV2 vexosome samples with the AAV1 antibody followed by the secondary antibody. Sample processing and images were acquired by Harvard Conventional Electron Microscopy Core. Images were taken using a Tecnai G2 Spirit Bio TWIN transmission electron microscope.
Epon-embedded sections: Monolayers of 293T cells (either control or AAV-producing cells) were fixed directly in the plate for 1 hour using 2.5% glutaraldehyde, 1.25% paraformaldehyde, and 0.03% picric acid in 0.1 mol/l sodium cacodylate buffer (pH 7.4) followed by washing in 0.1 mol/l sodium cacodylate buffer. The cells were then post-fixed for 30 minutes in 1% osmium tetroxide (OsO4)/1.5% potassiumferrocyanide (KFeCN6), washed in water three times and incubated in 1% aqueous uranyl acetate for 30 minutes followed by two washes in water and subsequent dehydration in grades of alcohol (5 minutes each; 50%, 70%, 95%, two times 100%). Cells were removed from the dish in propyleneoxide, pelleted at 3,000 r.p.m. for 3 minutes and infiltrated for 2 hours in a 1:1 mixture of propyleneoxide and TAAB Epon (Marivac Canada Inc., St Laurent, Canada). The samples were subsequently embedded in TAAB Epon and polymerized at 60° C for 48 hours. Ultrathin sections (about 60 nm) were cut on a Reichert Ultracut-S microtome, picked up on to copper grids stained with lead citrate and examined in a JEOL 1200EX Transmission electron microscope or a TecnaiG2 Spirit BioTWIN, and images were recorded with an AMT 2k CCD camera.
Acknowledgments
This work was supported by NCI P50 CA86355 (C.A.M., B.A.T., X.O.B.), NCI P01 CA069246 (X.O.B.), NCI RO1 CA141150 (X.O.B.), NIH/NINDS P30NS045776 (B.A.T.), and American Brain Tumor Association (J.S., C.A.M.). We thank the MGH Nucleic Acid Quantitation Core facility (with assistance from Luba Zagachin) supported by NINDS grant #P30NS4577 for the use of the thermal cyclers for quantitative PCR. We would also like to thank Igor Bagayev of the confocal microscopy core for assistance with fluorescence microscopy. For the electron microscopy data acquisition and analysis, we would like to thank the Harvard Conventional electron microscopy core as well as Howard Mulhern of Children's Hospital Boston Electron Microscope facility. We thank Emily Mills for generating the artwork in Figure 7. We thank Kristan van der Vos for helpful discussions and technical advice. J.S. and C.A.M. are inventors on a patent application involving vexosome technology. J.S. holds equity in, and is an employee of Exosome Diagnostics, Inc.
Supplementary Material
TEM examination of plasma membrane of 293T cells producing AAV2 vector.
Microvesicles from AAV producer cells contain AAV capsids.
AAV1 capsid localization in 293T producer cell.
AAV vector production in 293T cells does not stimulate microvesicle release.
Alix immunoblot.
VSV-G increased microvesicle-associated AAV titer.
VSV-G detection in vexosome pellets from 293T cells cotransfected with a VSV-G expression plasmid.
BAP-TM magnetic targeting schematic for Figure 6c.
REFERENCES
- Coura Rdos S., and, Nardi NB. The state of the art of adeno-associated virus-based vectors in gene therapy. Virol J. 2007;4:99. doi: 10.1186/1743-422X-4-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda VR, Stedman HH, Haurigot V, Buchlis G, Baila S, Favaro P.et al. (2010Peripheral transvenular delivery of adeno-associated viral vectors to skeletal muscle as a novel therapy for hemophilia B Blood 1154678–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High KA. Update on progress and hurdles in novel genetic therapies for hemophilia. Hematology Am Soc Hematol Educ Program. 2007. pp. 466–472. [DOI] [PubMed]
- Zincarelli C, Soltys S, Rengo G., and, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- Wu Z, Sun J, Zhang T, Yin C, Yin F, Van Dyke T.et al. (2008Optimization of self-complementary AAV vectors for liver-directed expression results in sustained correction of hemophilia B at low vector dose Mol Ther 16280–289. [DOI] [PubMed] [Google Scholar]
- Maguire CA, Gianni D, Meijer DH, Shaket LA, Wakimoto H, Rabkin SD.et al. (2010Directed evolution of adeno-associated virus for glioma cell transduction J Neurooncol 96337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K.et al. (2008Effect of gene therapy on visual function in Leber's congenital amaurosis N Engl J Med 3582231–2239. [DOI] [PubMed] [Google Scholar]
- Brantly ML, Chulay JD, Wang L, Mueller C, Humphries M, Spencer LT.et al. (2009Sustained transgene expression despite T lymphocyte responses in a clinical trial of rAAV1-AAT gene therapy Proc Natl Acad Sci USA 10616363–16368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S, Fujimoto K, Kato S, Mizukami H, Asari S, Ikeguchi K.et al. (2010A phase I study of aromatic L-amino acid decarboxylase gene therapy for Parkinson's disease Mol Ther 181731–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci E, Racchetti G., and, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Théry C, Ostrowski M., and, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- Wieckowski E., and, Whiteside TL. Human tumor-derived vs dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunol Res. 2006;36:247–254. doi: 10.1385/IR:36:1:247. [DOI] [PubMed] [Google Scholar]
- Théry C, Zitvogel L., and, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M.et al. (2008Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers Nat Cell Biol 101470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guescini M, Genedani S, Stocchi V., and, Agnati LF. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J Neural Transm. 2010;117:1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, Baran J, Urbanowicz B, Branski P.et al. (2006Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes Cancer Immunol Immunother 55808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO.et al. (2011Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences Nat Commun 2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., and, Muzyczka N. In vitro packaging of adeno-associated virus DNA. J Virol. 1998;72:3241–3247. doi: 10.1128/jvi.72.4.3241-3247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin R, Diou J, Bélanger D, Tremblay AM., and, Gilbert C. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J Immunol Methods. 2008;338:21–30. doi: 10.1016/j.jim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Sun, D, Zhuang, X, Xiang, X, Liu, Y, Zhang, S, Liu, C et al. (2010). A novel nanoparticle drug delivery system: The anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther 18: 1606–1614 [DOI] [PMC free article] [PubMed]
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S., and, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA.et al. (2009Proteomic and immunologic analyses of brain tumor exosomes FASEB J 231541–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee JK, Friedmann T., and, Burns JC. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 1994;43 Pt A:99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- Mangeot PE, Dollet S, Girard M, Ciancia C, Joly S, Peschanski M.et al. (2011Protein transfer into human cells by VSV-G-induced nanovesicles Mol Ther 191656–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous BA, Grimm J, Perry KF, Chen JW, Weissleder R., and, Breakefield XO. Metabolic biotinylation of cell surface receptors for in vivo imaging. Nat Methods. 2006;3:391–396. doi: 10.1038/nmeth875. [DOI] [PubMed] [Google Scholar]
- Niers JM, Chen JW, Weissleder R., and, Tannous BA. Enhanced in vivo imaging of metabolically biotinylated cell surface reporters. Anal Chem. 2011;83:994–999. doi: 10.1021/ac102758m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S., and, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- Summerford C., and, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JS, Byrnes AP., and, Griffin DE. Heparin-binding and patterns of virulence for two recombinant strains of Sindbis virus. Virology. 2006;347:183–190. doi: 10.1016/j.virol.2005.11.034. [DOI] [PubMed] [Google Scholar]
- O'Donnell CD, Kovacs M, Akhtar J, Valyi-Nagy T., and, Shukla D. Expanding the role of 3-O sulfated heparan sulfate in herpes simplex virus type-1 entry. Virology. 2010;397:389–398. doi: 10.1016/j.virol.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgard P., and, Stockert R. Heparan sulfate proteoglycans initiate dengue virus infection of hepatocytes. Hepatology. 2000;32:1069–1077. doi: 10.1053/jhep.2000.18713. [DOI] [PubMed] [Google Scholar]
- Lock M, Alvira M, Vandenberghe LH, Samanta A, Toelen J, Debyser Z.et al. (2010Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale Hum Gene Ther 211259–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberghe LH, Xiao R, Lock M, Lin J, Korn M., and, Wilson JM. Efficient serotype-dependent release of functional vector into the culture medium during adeno-associated virus manufacturing. Hum Gene Ther. 2010;21:1251–1257. doi: 10.1089/hum.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidelman O, Schlegel R, Tralka TS., and, Blumenthal R. pH-dependent fusion induced by vesicular stomatitis virus glycoprotein reconstituted into phospholipid vesicles. J Biol Chem. 1984;259:4622–4628. [PubMed] [Google Scholar]
- Douar AM, Poulard K, Stockholm D., and, Danos O. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J Virol. 2001;75:1824–1833. doi: 10.1128/JVI.75.4.1824-1833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JE, Rolling F, Li C, Conrath H, Xiao W, Xiao X.et al. (2002Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity J Virol 76791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser, NW, Yan Z, Zhang, Y, Lei-Butters, DC, Engelhardt, JF.Unique characteristics of AAV1, 2, and 5 viral entry, intracellular trafficking, and nuclear import define transduction efficiency in HeLa cells Hum Gene Therepub ahead of print) [DOI] [PMC free article] [PubMed]
- Ignowski JM., and, Schaffer DV. Kinetic analysis and modeling of firefly luciferase as a quantitative reporter gene in live mammalian cells. Biotechnol Bioeng. 2004;86:827–834. doi: 10.1002/bit.20059. [DOI] [PubMed] [Google Scholar]
- Corish P., and, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- Wurdinger T, Badr C, Pike L, de Kleine R, Weissleder R, Breakefield XO.et al. (2008A secreted luciferase for ex vivo monitoring of in vivo processes Nat Methods 5171–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley RD., and, Gummuluru S. Immature dendritic cell-derived exosomes can mediate HIV-1 trans infection. Proc Natl Acad Sci USA. 2006;103:738–743. doi: 10.1073/pnas.0507995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masciopinto F, Giovani C, Campagnoli S, Galli-Stampino L, Colombatto P, Brunetto M.et al. (2004Association of hepatitis C virus envelope proteins with exosomes Eur J Immunol 342834–2842. [DOI] [PubMed] [Google Scholar]
- Stone IM, Lurie DI, Kelley MW., and, Poulsen DJ. Adeno-associated virus-mediated gene transfer to hair cells and support cells of the murine cochlea. Mol Ther. 2005;11:843–848. doi: 10.1016/j.ymthe.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Hauck B., and, Xiao W. Characterization of tissue tropism determinants of adeno-associated virus type 1. J Virol. 2003;77:2768–2774. doi: 10.1128/JVI.77.4.2768-2774.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire CA, Meijer DH, LeRoy SG, Tierney LA, Broekman ML, Costa FF.et al. (2008Preventing growth of brain tumors by creating a zone of resistance Mol Ther 161695–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TEM examination of plasma membrane of 293T cells producing AAV2 vector.
Microvesicles from AAV producer cells contain AAV capsids.
AAV1 capsid localization in 293T producer cell.
AAV vector production in 293T cells does not stimulate microvesicle release.
Alix immunoblot.
VSV-G increased microvesicle-associated AAV titer.
VSV-G detection in vexosome pellets from 293T cells cotransfected with a VSV-G expression plasmid.
BAP-TM magnetic targeting schematic for Figure 6c.