Abstract
Comparative integrome analyses have highlighted alpharetroviral vectors with a relatively neutral, and thus favorable, integration spectrum. However, previous studies used alpharetroviral vectors harboring viral coding sequences and intact long-terminal repeats (LTRs). We recently developed self-inactivating (SIN) alpharetroviral vectors with an advanced split-packaging design. In a murine bone marrow (BM) transplantation model we now compared alpharetroviral, gammaretroviral, and lentiviral SIN vectors and showed that all vectors transduced hematopoietic stem cells (HSCs), leading to comparable, sustained multilineage transgene expression in primary and secondary transplanted mice. Alpharetroviral integrations were decreased near transcription start sites, CpG islands, and potential cancer genes compared with gammaretroviral, and decreased in genes compared with lentiviral integrations. Analyzing the transcriptome and intragenic integrations in engrafting cells, we observed stronger correlations between in-gene integration targeting and transcriptional activity for gammaretroviral and lentiviral vectors than for alpharetroviral vectors. Importantly, the relatively “extragenic” alpharetroviral integration pattern still supported long-term transgene expression upon serial transplantation. Furthermore, sensitive genotoxicity studies revealed a decreased immortalization incidence compared with gammaretroviral and lentiviral SIN vectors. We conclude that alpharetroviral SIN vectors have a favorable integration pattern which lowers the risk of insertional mutagenesis while supporting long-term transgene expression in the progeny of transplanted HSCs.
Introduction
Retroviral vectors are important tools for human gene therapy. Compared to other gene transfer methods, retroviral transduction is versatile, efficient, and not overtly toxic. Especially in the hematopoietic system, in which the genetic modification of few hematopoietic stem cells (HSCs) can lead to the correction of cells in all descendant lineages, retroviral vectors have led to substantial successes in preclinical and clinical trials. However, in some of these trials initial success has been hampered by clonal expansion of transduced cells, potentially leading to leukemia (as reviewed in ref. 1). Such adverse events resulted from genotoxic semi-random integration events of retroviral vectors in the genome, leading to aberrant proto-oncogene expression, thus causing clonal proliferation. Genotoxicity poses one of the major challenges for human gene therapy using retroviral vectors and can be caused by several mechanisms, including promoter activation, gene transcript truncation, and erroneous splicing.
Among the most frequently used retroviral vectors for clinical applications are those derived from human immunodeficiency virus-1 (lentiviral vectors) and Moloney murine leukemia virus (gammaretroviral vectors). Integrome analyses revealed that gammaretroviral vectors preferentially integrate in the proximity of transcription start sites, CpG islands, and genes with implications in cancer, while lentiviral vectors tend to integrate in actively transcribed genes.2,3,4,5,6,7,8 In contrast, alpharetroviral vectors have a relatively neutral integration spectrum, without strong preferences with respect to the aforementioned genomic features.9,10,11 Thus, alpharetroviral vectors might have a favorable integration spectrum for clinical applications. However, previous studies used alpharetroviral vectors harboring viral coding sequences and intact long-terminal repeats (LTRs). While viral coding sequences are potentially immunogenic in host cells and increase the risk of vector mobilization, intact LTRs, containing transcriptional elements, are capable of activating cellular genes and contribute to insertional mutagenesis.6,12 Therefore, we have developed alpharetroviral self-inactivating (SIN) vectors, eliminating enhancer and promoter elements from the LTRs, and set up an advanced split-packaging system, additionally removing viral-coding sequences and retroviral splice sites from the vector.13
In the present study, we performed side-by-side comparisons of alpharetroviral, gammaretroviral, and lentiviral SIN vectors in a serial murine bone marrow (BM) transplantation model, which allowed us to evaluate the potential of alpharetroviral SIN vectors in relation to state-of-the-art clinically used vectors. We analyzed transgene expression levels, mean vector copy numbers (mVCNs), and integration site distributions in primary and secondary recipients (for a period of up to 31 and 17 weeks, respectively). The integrome studies were furthermore complemented by a correlation analysis of the transcriptome and intragenic insertions in engrafting cells. The combination of transgene expression and integration site analyses allowed us to address whether the integration pattern of alpharetroviral SIN vectors is compatible with long-term transgene expression in the progeny of serially transplanted HSCs. In addition, we performed sensitive genotoxicity studies using the in vitro immortalization (IVIM) assay to assess alpharetroviral SIN vector genotoxicity in comparison with established gammaretroviral and lentiviral SIN vectors.12,14 Our results suggest that alpharetroviral vectors are suitable for genetic modification of HSCs, supporting long-term transgene expression in an experimental model that is known to be subject to epigenetic silencing. We furthermore showed that the favorable alpharetroviral integration pattern also applied to the newly developed SIN vectors. In addition, we provide functional evidence that this favorable integration spectrum is reflected in reduced genotoxicity.
Results
Comparable sustained multilineage transgene expression in a serial murine BM transplantation model for alpharetroviral, gammaretroviral, and lentiviral SIN vectors
To evaluate the potential of alpharetroviral vectors for clinical applications, we assessed the abilities of alpharetroviral, gammaretroviral, and lentiviral SIN vectors to transduce HSCs in a serial murine BM transplantation model. For comparison of vector-mediated effects, we equipped all vectors with the same internal cassette. This consisted of the spleen focus forming virus (SFFV) promoter, known to cause insertional proto-oncogene upregulation and attract epigenetic silencers,15 enhanced green fluorescent protein (EGFP) as a sensitive reporter, and the modified woodchuck hepatitis virus post-transcriptional regulatory element (wPRE) to enhance vector titer and transgene expression.16 Gammaretroviral and lentiviral SIN vectors have been described.17,18,19 The previously published alpharetroviral SIN vector13 was slightly modified for this study. We extended the SIN deletion also removing the TATA box, thereby mimicking the gammaretroviral and lentiviral SIN vectors, which also lack TATA boxes in their LTRs (Figure 1). Viral particles for the three vector families were pseudotyped with the vesicular stomatitis virus envelope glycoprotein and concentrated via ultracentrifugation. Titers on murine SC-1 fibroblasts were similar, underlining the potency of our newly developed split-packaging system (alpharetroviral 1.23 × 109 transducing units /ml; gammaretroviral 1.27 × 109 transducing units/ml; lentiviral 6.12 × 108 transducing units/ml). A multiplicity of infection (MOI) of ten was applied to transduce murine lineage-negative cells, which had been prestimulated for 2 days in the HSC-expanding STIF cocktail.20,21 After transduction, the percentages of both viable and EGFP-expressing cells were similar in alpharetroviral, gammaretroviral, and lentiviral groups as measured by flow-cytometry (Figure 2a and Supplementary Figure S1). However, there was a difference in mean fluorescence intensities (MFIs), an indicator of transgene expression levels. The MFI was lowest in cells transduced with alpharetroviral SIN vectors and highest in cells transduced with gammaretroviral SIN vectors, potentially related to the different integration patterns and/ or different transcript stabilities.
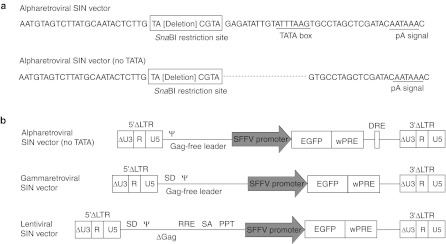
Figure 1.
Vectors used in this study (edited from ref. 13). (a) Nucleotide sequences of the unique 3′ (U3) region of alpharetroviral self-inactivating (SIN) vectors used in a previous study13 and in this study (noTATA). The site of sequence deletion is indicated by a dashed line. The nucleotide sequences of the introduced SnaBI restriction site, as well as those of the TATA box and the polyadenylation signal (pA signal), are shown. (b) Schema of lentiviral, gammaretroviral, and alpharetroviral SIN vectors used in this experiment. Indicated are the long-terminal repeat (LTR) (ΔU3, R, and U5), direct repeat element (DRE), packaging signal (ψ), splice donor, and acceptor site (SD and SA), spleen focus-forming virus (SFFV) promoter, enhanced green fluorescent protein (EGFP), and woodchuck hepatitis virus post-transcriptional regulatory element (wPRE), Rev-responsive element (RRE), polypurine tract (PPT), and remaining viral gag-coding sequences (Δgag).
Figure 2.
Enhanced green fluorescent protein (EGFP) expression in transduced cells. (a) EGFP expression in transduced murine lineage-negative cells. Indicated are percentages of EGFP+ cells and mean fluorescence intensities (MFIs). Exemplary flow-cytometry plots of untransduced cells are shown in comparison to cells transduced with the alpharetroviral self-inactivating (SIN) vector 1 day and 9 days after transduction. Also provided are time-series of flow-cytometric analyses of transduced cells by all three vectors from 1 day to 9 days after transduction. (b) EGFP+ cells in different lineages of peripheral blood. From left to right: EGFP+ cells in CD11b+ cells, in CD19+ cells, or in CD3ε+ cells. Depicted are mean values + SD; n = 8 for primary recipients and n = 5 for secondary recipients. (c) EGFP MFI in different lineages of peripheral blood. From left to right: EGFP MFI in CD11b+ cells, in CD19+ cells, or in CD3ε+ cells. Depicted are mean values + SD; n = 8 for primary recipients and n = 5 for secondary recipients. (d) EGFP+ cells and EGFP MFI in transduced human X-CGD PLB985 cells during long-term culture in vitro.
Transduced murine lineage-negative cells were then transplanted into lethally irradiated mice (n = 8 recipients for each group). After 31 weeks, primary transplanted mice were sacrificed and EGFP expression levels in BM cells were determined (Table 1). To reduce the number of experimental animals without compromising the pool of retroviral insertions and to focus on maintenance of gene expression in hematopoietic cells of mice showing good engraftment, we pooled the BM of the four mice with the highest percentage of EGFP-positive cells for secondary transplantation (n recipients = 5; EGFP-positive cells in selected mice: alpha: 18.5 ± 13.5%; gamma: 32.2 ± 13.6%; lenti: 28.4 ± 13.9%). Secondary transplanted mice were monitored for 17 weeks and then sacrificed.
Table 1. EGFP expression and mVCN in BM cells.
There was no indication of disease based on blood counts, spleen, and liver weights (Supplementary Figure S2). Peripheral blood was analyzed for transgene expression at various time-points (6 and 31 weeks after primary transplantation, 6 and 17 weeks after secondary transplantation) and antibody staining was performed to distinguish expression in myeloid (CD11b+), B (CD19+), and T cells (CD3ε+) (Figure 2b,c). In the three groups the mean percentages of EGFP-expressing cells in the myeloid lineage ranged from 43.5 to 76.7%, whereas the mean percentages of EGFP-expressing cells in B and T cells were lower, ranging from 11.5 to 39.3%. This trend was also reflected by the MFIs, which were highest in transgene-positive cells of the myeloid lineage. In flow-cytometric analyses of whole blood, we furthermore analyzed transgene expression in erythrocytes and in platelets (gated by forward and sideward scatter characteristics) (Supplementary Figure S3). Similar to our measurements in B and T cells, the mean percentages of EGFP-expressing cells in erythrocytes and platelets ranged from 14.6 to 55.1%. Irrespective of the analyzed lineage, the percentages of transgene-expressing cells tended to be lowest in the alpharetroviral group and highest in the lentiviral group, especially in primary recipients. Differences in MFI between the retroviral vectors, as observed in freshly transduced cells in vitro, apparently diminished in transplanted cells in vivo (Figure 2c).
The percentages of EGFP-positive cells in the BM roughly correlated with the determined mVCNs within the three different groups (Table 1). However, comparing the groups with one another, similar percentages of transgene-expressing cells were accompanied by lower mVCNs in the alpha- or gammaretroviral than in the lentiviral context. Despite variances in percentages of transgene-expressing cells, MFIs, and mVCNs, we observed comparable sustained multilineage transgene expression upon serial transplantation for all three vectors used in this study. This also demonstrated the alpharetroviral SIN vectors' ability to effectively transduce HSCs at low MOI, and to support long-term multilineage transgene expression in an experimental setting that is sensitive for epigenetic silencing of retroviral vectors.22
Furthermore, we monitored EGFP expression from alpharetroviral SIN vectors in bulk transduced human myelomonocytic cells X-CGD PLB985, which have been reported to silence retroviral vectors.23 EGFP expression in the alpharetroviral SIN vectors was either regulated by the SFFV promoter or the cellular EFS promoter (intron-less form of the human elongation factor-1α promoter), of which the latter one is also incorporated into state of the art clinical vectors. During an observation period of 20 weeks we observed relatively constant percentages of EGFP-positive cells (Figure 2d). The drop in MFI in the EFS context between day 100 and 110, reflects differences in the composition of the cell culture and not inactivation of the EFS promoter (data not shown), hence this experiment further shows the potential of alpharetroviral SIN vectors to support long-term transgene expression.
Identification of integration sites
To identify the integration sites of alpharetroviral SIN vectors in the progeny of transplanted murine HSC, we established a suitable ligation-mediated PCR (LM PCR) protocol with barcoded primers followed by high-throughput sequencing (as described in ref. 21). Genomic DNA samples were collected from transduced murine lineage-negative cells before transplantation and after further in vitro culture. In order to identify integration sites in transplanted mice, we collected genomic DNA from total BM at endpoints of both primary and secondary transplantations. It has been shown that the influence of the LM PCR-inherent restriction bias can be reduced by using more than one restriction enzyme for primary genomic DNA digest.24 Therefore, we used multiple enzymes to digest genomic DNA—three different ones for alpharetroviral samples (Tsp509I, MseI, MnlI) and two different ones for gammaretroviral (MseI, HaeIII) and lentiviral samples (Tsp509I, HaeIII) (for LM PCR gel pictures see Supplementary Figures S4 and S5). Usage of the same restriction enzymes for the three different groups was not possible due to different internal restriction sites, but we were able to use at least one overlapping restriction enzyme between two of the groups. The retrieved sequence reads were sorted according to barcode, clustered, trimmed, and aligned to the mouse genome.
Favorable integration site distribution in murine HSCs of alpharetroviral compared with gammaretroviral and lentiviral SIN vectors
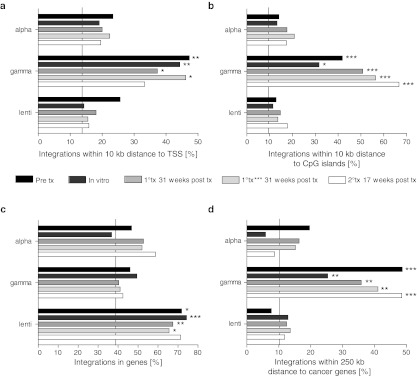
To characterize the integrome of the three different retroviral vectors at various time-points of the transplantation experiment, we performed LM PCRs and used QuickMap25 to analyze integrations in potential regions of the genome that would indicate a higher likelihood of deregulation of the surrounding genes, such as transcription start sites, CpG islands, intragenic regions, and genes with implications in cancer. To rule out that the usage of different restriction enzymes influenced integration characteristics, we compared integration characteristics obtained with different enzymes and found no major differences (data not shown). When we analyzed integrations within 10 kb distance to transcription start sites of protein-coding genes and integrations within 10 kb distance to CpG islands, we found gammaretroviral integrations to be over-represented compared with alpharetroviral and lentiviral integrations (Figure 3a,b). In contrast, intragenic insertions were over-represented in the lentiviral group compared with alpha- and gammaretroviral groups (Figure 3c). For analyses of integrations near genes with implications in cancer we identified integration sites within 250 kb distance to genes that are annotated in the retrovirus tagged cancer gene database (RTCGD) and the cancer-gene census database (CGC). While gammaretroviral insertions were clearly over-represented, both alpharetroviral and lentiviral insertions showed a reduced incidence of integrations near annotated cancer genes (Figure 3d).
Figure 3.
Integration site analyses. (a) Integrations near transcription start sites. Shown are the percentages of integration sites near transcriptions start sites of adjacent protein-coding genes within a distance of 10 kb. (b) Integrations near CpG islands. Shown are the percentages of integration sites near CpG islands within a distance of 10 kb. (c) Integrations in genes. (d) Integrations in proximity of genes with implications in cancer. Shown are the percentages of integrations in 250 kb proximity to retrovirus tagged cancer gene database (RTCGD) and cancer-gene census database (CGC) annotated cancer genes. Vertical lines indicate percentages of integrations for a random reference sample provided by QuickMap. n = numbers of nonredundant reads: alpha pre n = 107; alpha in vitro n = 68; alpha 1°tx n = 165; alpha 1°tx*** n = 125; alpha 2°tx n = 46; gamma pre n = 74; gamma in vitro n = 79; gamma 1°tx n = 67; gamma 1°tx*** n = 39; gamma 2°tx n = 33; lenti pre n = 39; lenti in vitro n = 70; lenti 1°tx n = 442; lenti 1°tx*** n = 206; lenti 2°tx n = 93. Statistical significance was tested with Fisher's exact test, corrected for multiple testing by the Benjamini–Hochberg method. Indicated are gammaretroviral or lentiviral data bars which are significantly different from alpharetroviral data bars. *P = 0.01–0.05, **P = 0.01–0.001, ***P < 0.001.
Even though the present in vivo study was designed to evaluate the transduction potential of the different vectors in long-term repopulating HSCs and associated integration characteristics, rather than genotoxic events (low MOI, no tumor-prone mouse model), we were interested in whether cells carrying integrations near genes with implications in cancer persisted at various time-points of the experiment. For visualization of such persisting integration sites we generated Venn diagrams and found most of the cancer-related overlapping integrations sites to be maintained between primary and secondary cohorts (Supplementary Figure S6). Among those, we identified gammaretroviral integrations near Lmo2—a retroviral integration site which has been previously associated with adverse events in clinical trials.26,27 We also identified another clinically relevant integration site in the Prdm16 locus15,28 (lentiviral primary cohort), but cells harboring this integration were undetectable after secondary transplantation. However, despite the persistence of some clones carrying integrations near annotated cancer genes in gammaretroviral, alpharetroviral, and lentiviral groups from primary to secondary transplantation, the overall frequency of integrations near such genes did not increase with secondary transplantation, thus marking the absence of clonal outgrowth in the BM.
Reduced frequency of alpharetroviral SIN vector integrations in highly expressed genes in engrafting cells compared with gammaretroviral and lentiviral SIN vectors
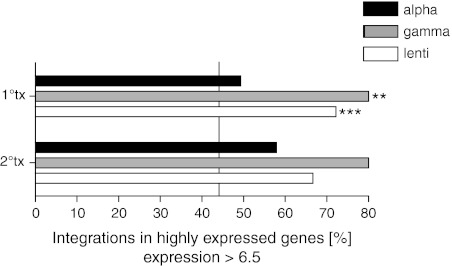
To further delineate integration characteristics, we differentiated integrations in highly transcribed genes from those in less active ones. For this purpose, we determined the transcriptome in long-term repopulating-HSCs (LT-HSCs) and compared it to in gene integrations in engrafting cells. LT-HSCs were obtained by sorting murine BM for the well-described cell-fraction with the following phenotype: lineage-negative, Sca1-high, c-Kit-high,29 CD34-low,30 CD135-low,31 and IL7Rα-negative.32 Even though we initially transduced lineage-negative cells, only the LT-HSC cell fraction contributes to the integrome after long-term engraftment and was thus used for correlation analysis of gene activity and integration targeting in engrafting cells. This analysis showed correlations of gene activity and intragenic integration targeting to be stronger for lentiviral and gammaretroviral vectors compared with alpharetroviral vectors (Figure 4), implying that genes highly expressed in immature hematopoietic cells are relatively disfavored targets for alpharetroviral integration.
Figure 4.
Integrations in highly expressed genes. Shown are integrations in highly expressed genes (Log2 expression value >6.5). Statistical significance was tested with Fisher's exact test. The vertical line indicates the percentage of integrations in highly expressed genes for random integration targeting. n = numbers of intragenic integrations/numbers of intragenic integrations on array: alpha 1°tx n = 78/73; alpha 2°tx n = 22/19; gamma 1°tx n = 26/25; gamma 2°tx n = 11/10; lenti 1°tx n = 295/266; lenti 2°tx n = 74/69. Indicated are gammaretroviral or lentiviral data bars which are significantly different from alpharetroviral data bars. **P = 0.01–0.001, ***P < 0.001.
Reduced genotoxicity of alpharetroviral SIN vectors in the IVIM assay compared with gammaretroviral and lentiviral SIN vectors
To assess the genotoxicity of alpharetroviral SIN vectors, we took advantage of the established IVIM assay.12,14 For a given vector the percentage of positive assays indicates the incidence of immortalizing events. The fitness of immortalized cells is described by correcting the replating frequency (number of positive wells) with the mVCN detected in samples obtained 4 days post-transduction.12,14
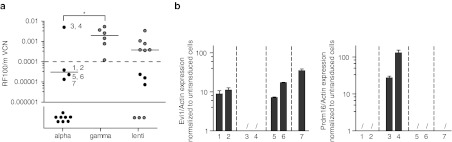
We compared immortalizing capacities of alpharetroviral, gammaretroviral, and lentiviral SIN vectors, all harboring the same internal cassette (SFFVpromoter.EGFP.wPRE), and found alpharetroviral vectors to show the lowest and gammaretoviral vectors to show the highest genotoxicity (Figure 5a). Directly comparing alpharetoviral with lentiviral SIN vectors we observed a 2.7-fold reduced incidence of immortalization and a trend toward lower fitness of immortalized cells in the alpharetroviral context. Further distinguishing robustly replating from weakly replating clones (dashed horizontal line at RF100/mVCN = 0.0001; Figure 5a), we found the occurrence of robustly replating clones to be reduced twofold using lentiviral instead of gammaretroviral SIN vectors, and to be further reduced sixfold using alpharetroviral instead of lentiviral SIN vectors. When we exchanged the internal SFFV promoter of the alpharetroviral SIN vector with the cellular EFS promoter, we were unable to obtain positive assays, despite higher mVCNs than in the SFFV promoter context (Table 2), as previously reported for gammaretroviral SIN vectors.33
Figure 5.
IVIM assay. (a) Replating frequencies normalized to mean vector copy numbers. We analyzed the replating frequencies (RF) of cells transduced with alpharetroviral, gammaretroviral, and lentiviral self-inactivating (SIN) vectors. Plotted are the RFs corrected for mean vector copy numbers (mVCNs) measured 4 days after transduction. Data points were either obtained in this study (black) or have been published previously (gray).14 Numbers next to alpharetroviral data-points are identifiers of wells with cells recovered after replating (Table 2). Horizontal bars indicate the median of all positive assays for a given vector. Statistical significance was tested with Fisher's exact test on the number of positive and negative assays. *P = 0.01–0.05. The horizontal dashed line at RF100/mVCN = 0.0001 separates robustly replating from weakly replating clones. (b) Expression levels of Evi1 or Prdm16 in recovered cells after replating. RNA was purified from expanded cells after replating, reverse transcribed, and subjected to quantitative PCR (qPCR) for detection of Evi1 or Prdm16 transcripts. Target gene expression levels were determined, normalized to actin expression levels, and are depicted in reference to untransduced cells harvested before replating. Data not in measurable range; dashed lines separate clones recovered from independent plates/assays. Depicted are technical triplicate mean values + SD.
Table 2. IVIM assay.
To further characterize the immortalizing events, we analyzed cells in positive alpharetroviral assays recovered from single wells after replating. The cells were expanded, genomic DNA was collected, and integration sites were determined by LM PCR and subsequent sequencing of PCR fragments purified from agarose gels. Again, different restriction enzymes were used for LM PCR in order to increase the recovery of integration sites24 (for LM PCR gel pictures see Supplementary Figure S7a). Wells from the alpharetroviral assay with the highest replating frequency showed an insertion close to the Prdm16 locus in reverse orientation (identifiers 3 and 4 in Figure 5a). We also found insertions close to the Evi1 locus in two out of seven wells analyzed (identifiers 5 and 7 in Figure 5a). Alpharetroviral insertions with respect to Evi1 or Prdm16 were in similar regions as gammaretroviral and lentiviral insertions; nevertheless they occurred at a reduced frequency and at different positions in the genome (Supplementary Figure S7b).
Next, we analyzed the transcriptional upregulation of Evi1 and Prdm16 in all samples by quantitative reverse transcription PCR targeting the 3′ untranslated region of Prdm16 and the exon 2/3 boundary of Evi1.34 Expression levels are depicted in reference to transcript levels of untransduced cells harvested before replating (Figure 5b). Prdm16 transcripts were upregulated in two wells (identifiers 3 and 4) which harbored an alpharetroviral integration in close proximity to this gene. In contrast, all other wells analyzed showed an upregulation of Evi1 transcripts, even though in some of them we were unable to retrieve an insertion close of the Evi1 locus. In none of the analyzed wells were Evi1 and Prdm16 transcripts upregulated simultaneously.
Altogether, the results of the IVIM assays revealed that alpharetroviral SIN vectors showed a reduced genotoxicity compared with their gammaretroviral and lentiviral counterparts and that alpharetroviral immortalizing events were linked to Evi1 or Prdm16 upregulation, which is the phenotype for which this assay is uniquely sensitive.12,14,35
Discussion
To our knowledge, this is the first report of a direct comparison of alpharetroviral, gammaretroviral, and lentiviral SIN vectors in transplanted HSCs, and in sensitive genotoxicity studies. To allow for comparison of vector-mediated effects, all vectors were equipped with the same internal cassette, mediating EGFP expression via the SFFV promoter. The SFFV internal promoter represents a worst case scenario for the risk of both, insertional gene activation and epigenetic silencing.15,23,33,36 We showed that initial transduction efficiencies in prestimulated murine lineage-negative cells were comparable between the three groups and that all vectors were able to transduce HSCs at low MOI as demonstrated by sustained transgene-expression in different lineages of peripheral blood in primary and secondary recipients. In general, mean percentages of EGFP-expressing cells and MFIs in the myeloid lineage were higher than those in B and T cells. This observation can partially be explained by lineage-dependent transcription factor binding motifs within the SFFV promoter.37 It is also possible that retroviral transduction of engrafting cells leads to a higher fraction of transgene-positive HSCs which are more committed to the myeloid lineage. Despite inter-lineage expression variability, we observed a trend of percentages of transgene expressing cells to be lowest in the alpharetroviral group and to be highest in the lentiviral group, especially in the first cohort. This could reflect the superior ability of lentiviral vectors to transduce nondividing cells, thus leading to higher transduction rates in HSCs. To increase the percentages of alpharetroviral and gammaretroviral transgene-positive HSCs in future transplantation studies, transduction of cell populations with higher enrichment of HSCs, the usage of improved prestimulation protocols, or viral envelope glycoproteins, might be of advantage.29,38,39
Measuring mVCNs and percentages of transgene-positive cells in the BM of primary and secondary transplanted mice, we found the percentages of transgene-positive cells to roughly correlate with mVCNs within the different groups. While this does not necessarily rule out variegation or extinction of transgene expression, it indicates that most silencing events can be attributed to transcriptional silencing occurring directly after integration. When comparing the three groups with one another, it became apparent that similar percentages of transgene-expressing cells were accompanied by higher mVCNs in the lentiviral context than in alpha- or gammaretroviral contexts. This suggests that alpharetroviral SIN vectors were not more prone to silencing than gammaretroviral and lentiviral SIN vectors. This could be attributed to the SIN vector architecture, potentially removing silencer elements from the vector and rendering transgene expression independent from LTR-mediated transcription (as reviewed in ref. 40). However, the data can also be interpreted in terms of vector-intrinsic properties and would be in line with previous reports of lentiviral vector transduction leading to multiple integrants per cell.41 On the one hand multiple copy integrations are certainly expedient for high transgene expression levels, but on the other hand they pose a risk in terms of insertional mutagenesis taking into consideration that more than one mutational event per cell is required for malignant cell transformation (as reviewed in ref. 42). Therefore, further investigations are needed to clearly distinguish vector-intrinsic properties from silencing mechanisms.
To characterize the retroviral vector integromes, we performed LM PCRs on genomic DNA collected from murine lineage-negative cells before transplantation and from total BM at endpoints of primary and secondary transplantations. We focused on genomic regions in which integration events are likely to influence gene expression and/ or cellular integrity. Alpharetroviral integration was characterized by a decreased percentage of integrations in the vicinity of transcription start sites, CpG islands, and genes with implications in cancer in contrast to gammaretroviral integrations. Intragenic insertions were found with a decreased percentage comparing alpharetroviral with lentiviral sample groups. This is in agreement with previous studies in vitro and in vivo.2,3,4,5,6,8,9,10,11 Hence, the favorable integration characteristics of the alpharetroviral vectors also applied to our SIN vectors, both in lineage-negative cells and in engrafted HSCs after serial BM transplantation. Importantly, despite the largely “extragenic” profile of alpharetroviral SIN vector integrations, long-term transgene expression occurred in an experimental setting that is known to be subject to epigenetic silencing.22
Comparing the LT-HSC transcriptome to intragenic integrations in engrafting cells, we observed stronger correlations in gammaretroviral and lentiviral contexts than in the alpharetroviral context. While the preferences for lentiviral and gammaretroviral integrations for actively transcribed genes have previously been reported,4,5,7,8,11 publications on alpharetroviral integration targeting in that respect are somewhat ambiguous. Studies in different model systems have reported high transcriptional activity and protein binding to exert inhibitory effects on alpharetroviral integraton,43,44 while studies in cell lines have shown alpharetroviral integration to be random or to be preferred in active genes.7,9,11 However, in contrast to these publications, cells analyzed in our study underwent selection processes associated with HSC survival and engraftment. It has been suggested that genes expressed for the maintenance of stemness, such as Evi1 and Hmga2, are at the same time potentially involved in leukemia development upon deregulation,45 and therefore alpharetroviral integration being comparably disfavored in these genes might be of advantage.
While the present in vivo study was not primarily designed to evaluate genotoxicity (low MOIs, no tumor-prone mouse model), but rather long-term gene marking of the different vectors in repopulating HSCs and associated integration characteristics, we performed sensitive in vitro studies to assess the vectors' genotoxic potential. In the IVIM assay, we compared alpharetroviral, gammaretroviral, and lentiviral SIN vectors and observed a reduced incidence of immortalization and a trend toward lower fitness of immortalized cells for alpharetroviral vectors. With all vectors harboring the same internal cassette, this difference can only be attributed to vector-mediated effects and is likely to be the result of differences in integration characteristics9,10,11 and can be further influenced by the occurrence of secondary mutations in the genome. When we exchanged the internal “high-risk” SFFV promoter of the alpharetroviral SIN vector with the cellular EFS promoter, we were unable to obtain positive assays, despite higher mVCNs than in the SFFV promoter context. This is in line with our previous finding that physiological promoters, such as the EFS promoter, reduce the genotoxicity of gammaretroviral SIN vectors below the detection level of the IVIM assay.33 Consistent with previous publications, the IVIM assay conditions introduced a bias for cells with insertions in close proximity of the Evi1 or Prdm16 locus and/ or the mutually exclusive upregulation of one of these genes.12,14,35 Nevertheless, as Evi1 integrations were associated with clinical adverse reactions,15,28 the assay represents a clinically relevant read-out. By revealing a decreased incidence of immortalization and a lower fitness of immortalized cells after alpharetroviral transduction compared with gammaretroviral and lentiviral transductions, we provide functional evidence that the favorable alpharetroviral integration spectrum is reflected in reduced genotoxicity.
Altogether, our results show that alpharetroviral SIN vectors are able to transduce repopulating HSCs at low MOI with a relatively neutral integration pattern, resulting in low genotoxicity in the IVIM assay. With genotoxicity posing one of the major challenges for human gene therapy using retroviral vectors, this suggests that the alpharetroviral SIN vector system is a promising alternative to commonly used lentiviral and gammaretroviral vector systems for HSC gene therapy and also for studies in basic stem cell biology in which insertional genotoxicity of gene delivery systems may produce an undesired experimental bias. However, more detailed studies on silencing and genotoxicity mechanisms in HSCs are needed to define which other modifications can be made to further enhance the vector system's safety. For example improved shielding of the retroviral cassette from the host genome by introducing stronger polyadenylation signals, insulators, and/ or ubiquitous chromatin opening elements might be advantageous, especially for target diseases requiring high levels of therapeutic transgene expression. Careful investigations in clinically relevant disease models are thus of importance to define the measures that need to be taken in order to approach the goal of human gene therapy with minimized adverse effects.
Materials and Methods
Cloning of vectors. The architecture of the alpharetroviral,13 gammaretroviral, and lentiviral vectors17,18,19 is described in detail in the results section and Figure 1b. We extended the SIN deletion of the alpharetroviral vector thereby removing the TATA box and mimicking the architecture of gammaretroviral and lentiviral SIN vectors. The TATA box was removed from the ΔU3 region of the alpharetroviral SIN vector pAlpha.SIN.SF.EGFP.wPRE13 using primers 5′_ASLV_noTATA_SnabI (5′-TGTACGTAGTGCCTAGCTCGATACAATAAAC-3′) and 3′_ASLV_SIN_XhoI (5′-CTCTCGAGAATGAAGCCTTCTGCTTCATGCA-3′) (SnabI and XhoI sites are underlined). The resulting fragment was subcloned, digested with SnabI and XhoI, as was the vector pAlpha.SIN.SF.EGFP.wPRE. Ligation of the PCR fragment to the 5,559 bp vector backbone resulted in pAlpha.SIN.SF.EGFP.wPRE (noTATA).
Cultivation of cell lines and production of viral supernatants. Human embryonic kidney 293T cells and murine SC1 fibroblasts were cultured in Dulbecco's modified Eagle's medium with stable glutamine (Biochrom, Berlin, Germany) supplemented with 10% fetal calf serum, 100 U/ml penicillin, 100 µg/ml streptomycin, and 1 mmol/l sodium pyruvate (all PAA, Pasching, Austria). The human X-CGD PLB985 cell line was cultivated in Roswell Park Memorial Institute 1640 medium (Invitrogen, Darmstadt, Germany) supplemented with 10% heat-inactivated fetal bovine serum (Pan Biotech, Aidenbach, Germany), 4 mmol/l L-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin (all PAA). Viral supernatants were produced by transient transfection of 293T cells as previously described for alpharetroviral, gammaretroviral, and lentiviral vectors, using 5 µg of vector plasmids and 1.5 µg of the vesicular stomatitis virus envelope glycoprotein envelope plasmid (pMD.G).13,16,17,19 Supernatants were collected 36 hours after transfection, filtered through Millex-GP 0.22 µm filters (Millipore, Schwalbach, Germany), concentrated via ultracentrifugation, and stored at −80 °C until further usage.
Enrichment and cultivation of murine lineage-negative BM cells. BM cells of C57Bl/6J mice (Janvier; Le Genest, St Isle, France) were flushed out of femurs and tibias and lineage-negative cells were isolated by magnetic sorting using lineage-specific antibodies (Lineage Cell Depletion Kit; Miltenyi Biotech, Bergisch Gladbach, Germany). Lineage-negative cells were cultured in STIF medium consisting of StemSpan serum-free expansion medium (Stemcell Technologies SARL, Koeln, Germany), supplemented with 200 U/ml penicillin, 200 µg/ml streptomycin, 1% glutamine, 10 ng/ml murine stem cell factor, 20 ng/ml murine thrombopoietin, 20 ng/ml murine insulin-like growth factor 2, and 10 ng/ml human fibroblast growth factor 1.20 All cytokines were purchased from Peprotech (Hamburg, Germany), except for murine insulin-like growth factor 2 (R&D Systems, Wiesbaden-Nordenstadt, Germany). Note that heparin at a concentration of 5 I.U./ml (Liquemin; Roche, Mannheim, Germany) was not added to the medium until after retroviral transduction.
Transduction of cell lines and murine lineage-negative BM cells. Transduction of murine SC1 fibroblasts was performed as described previously.13 The human X-CGD PLB985 cell line was transduced by spinfection (90 minutes, 900g, 32 °C) in medium supplemented with 8 µg/ml protamine sulfate (Sigma-Aldrich Chemie, Steinheim, Germany). For transduction of murine lineage-negative BM cells, cells were plated in pre-coated wells with Retronectin (Takara, Otsu, Japan) at a density of 1 × 106 cells/ml, and supernatants were added. The medium was supplemented with 4 µg/ml protamine sulfate (Sigma-Aldrich Chemie). After 24 hours, cells were dissociated using Gibco enzyme-free, phosphate-buffered saline -based cell dissociation buffer (Invitrogen) and either further cultured in vitro or subjected to transplantation. For further in vitro culture, the STIF medium was complemented by the addition of heparin (5 I.U./ml) (Liquemin; Roche). For transplantation, dissociated cells were washed and resuspended in Iscove's modified Dulbecco's medium (Biochrom).
Animal housing and irradiation procedure. Animal experiments were approved by the supervising animal research review board. C57Bl/6J mice were obtained from Janvier (Le Genest) and kept in specific pathogen-free conditions in the animal facility of Hannover Medical School. Male C57Bl/6J mice served as donors for BM cells, while female C57/Bl/6J mice served as both primary and secondary recipients. Recipient mice were conditioned 24 hours before transplantation by myeloablative irradiation (10 Gy), and were administered Ciprofloxacin (Bayer Vital, Leverkusen, Germany) at 100 mg/ml in drinking water for antimicrobial protection until three weeks after transplantation.
Flow cytometry. For the analysis of EGFP-expression only, cells were resuspended in phosphate-buffered saline containing 4% heat-inactivated fetal calf serum (PAA) and measured on a FACSCalibur flow cytometer (Becton Dickinson, Heidelberg, Germany). For an extended analysis of the different lineages of peripheral blood, erythrocytes were lysed using BD Pharm Lyse buffer (BD Pharmingen, San Diego, CA) for 10 minutes at room temperature. After washing with and resuspending in phosphate-buffered saline, unspecific Fc-binding was blocked by the addition of FcR purified α-mouse CD16/32 antibody. Cells were stained for surface markers CD11b, CD19, and CD3ε by the addition of conjugated antibodies APC-α-mouse CD11b, PerCP-Cy5.5 α-mouse CD19, PE-α-mouse CD3ε. All antibodies were purchased from eBiosciences, except for PerCP-Cy5.5 α-mouse CD19, which was purchased from BD Pharmingen. After 30 minutes incubation at 4 °C in the dark, cells were washed, resuspended in phosphate-buffered saline and measured on an LSR II flow cytometer (Becton Dickinson). A homogeneous cell population was gated as determined by scatter characteristics, and ≥15,000 events were monitored. Data was analyzed using CellQuest or FlowJo software (Tree Star, Ashland, OR).
Isolation of genomic DNA and determination of VCNs. For isolation of genomic DNA from cells, the QIAamp DNA Blood Kit (Qiagen, Hilden, Germany) was used according to manufacturer's instructions. VCNs of transduced cells were determined by quantitative PCR on an Applied Biosystems (Darmstadt, Germany) Step One Plus Real Time PCR System (Applied Biosystems) using the Quanti-Tect SYBR Green Kit (Qiagen) and a reference plasmid containing vector specific wPRE, and Flk sequence (kindly provided by M. Galla and T. Maetzig). CT values were determined using wPRE- and Flk-specific primers and VCNs were calculated using the efficiency-corrected ΔΔCT method.12,13
LM PCR and high-throughput sequencing. To identify integration sites, LM PCR was performed as described previously with minor modifications.46 100 ng of total genomic DNA was used along with primers alpha-LTRI: (Bio TEG) GCAATACTCTTGTACGTAGTG; alpha-LTRII: CGCCATTTGACCATTCACCACA; alpha-LTRIII: TTGG TGTGCACCTGGGTTGA; gamma-LTRI: (Bio TEG) CGCTAGCGA TATCGAATTCACAACC; gamma-LTRII: GTCCTCCGATTGACTGCG TCG; gamma-LTRIII: CCCAATAAAGCCTCTTGCTGT;34,46 lenti-LTRI: (Bio TEG) GAACCCACTGCTTAAGCCTCA; lenti-LTRII: AGCTTGCC TTGAGTGCTTCA; lenti-LTRIII: AGTAGTGTGTGCCCGTCTGT.47 Primer annealing temperatures for alpharetroviral samples were reduced to 58 °C. Different combinations of enzymes were used for primary genomic DNA digests, and respective internal control digests after first exponential PCR: alpha Tsp509I-NotI, MseI-NotI, MnlI-XcmI; gamma MseI-BglII, HaeIII-PasI; lenti Tsp509I-BssHII, HaeIII-BssHII. All enzymes were purchased from New England Biolabs (Frankfurt am Main, Germany), except for PasI, which was purchased from Fermentas (St Leon-Rot, Germany). For samples obtained from the IVIM assay purification and sequencing of dominant products was performed as described. In contrast, samples obtained from the mouse BM transplantation experiment were used for LM PCR with barcoded primers and subsequent high-throughput pyrosequencing (as described in ref. 21). For this purpose LTRIII and OCII primers were modified, and PCR products after the second exponential PCR were purified using the QIAquick PCR Purification Kit (Qiagen). Pyrosequencing was performed in the Department of Medicinal Microbiology at Hannover Medical School using GS FLX reagents (Roche). Raw sequence reads were sorted according to barcode, clustered, trimmed using IntegrationSeq, aligned to the NCBI m37 mouse genome assembly, and analyzed using QuickMap.25
LT-LSK sorting and gene expression array. BM was obtained from 10 C57Bl6/J mice (Janvier; Le Genest), conforming to the directions of the institutional board for animal experimentation. In short, femurs were crushed in a mortar after which the resulting suspension was filtered through a 70-µm filter. Erythrocytes were lysed using BD Pharm Lyse buffer (BD Pharmingen). The remaining mononuclear cells were stained with biotin labeled antibodies against IL7Rα (eBioscience, San Diego, CA) and lineage markers (Caltag, Hamburg, Germany), Sca-1 PerCP Cy5.5 (BD Pharmingen), cKit APC, CD34 FITC, CD135 PE and Fc blocking reagent (eBioscience). Biotin-labeled antibodies were counter stained with streptavidin PE-Cy7 (BD Pharmingen). The labeled cells were sorted using a FACS Aria (Becton Dickinson). LT-LSK cells (Lineage-negative, IL7Rα-negative, Sca-1-high, cKit-high, CD135-low, CD34-low) cells were isolated to >95% purity and cultured for 2 days in Stemspan serum-free expansion medium (Stem Cell Technologies, Vancouver, British Columbia, Canada) supplemented with 10 ng/ml murine stem cell factor (R&D, Minneapolis, MN), 20 ng/ml murine thrombopoietin (Peprotech), 10 ng/ml human fibroblast growth factor 1 (R&D), 100 ng/ml hIGFBP2 and 100 ng/ml Angptl5 (both Abnova, Heidelberg, Germany). Afterwards, RNA was isolated using RNEasy micro columns (Qiagen) according to the manufacturer's protocol. The resulting RNA was amplified using the Ovation RNA amplification system (v2; Nugen; AC Bemmel, the Netherlands) and fragmented and biotin labeled with the Encore labeling system (Nugen), after which the amplified RNA was hybridized to Affymetrix Mouse 430.2 3′ IVT arrays followed by washing, staining and scanning according to the manufacturer's protocols. Expression values were normalized and summarized using GCRMA.48 Genes with alpharetroviral insertions in the coding sequence of the gene were identified (Quickmap25) and expression of these genes was divided into low (Log2 expression value <6.5) or high (>6.5). The threshold for high expression was determined by analysis of distribution of expression values of all probesets on the array. When multiple probesets were annotated to the same gene, the probeset with the highest expression value was used. The percentage of integrations in highly expressed genes for random integration targeting was calculated as the percentage of genes on the arrays with an aggregated maximum of expression >6.5. The microarray data can be retrieved from Gene Expression Omnibus database using accession number GSE34731.
IVIM assay. The IVIM assay was performed as described previously.12,14 Four days after transduction VCN were determined by quantitative PCR and used for normalization of replating frequencies. Assays in which more than one positive well was observed after replating of untransduced cells were excluded from analysis. Cells recovered after replating in positive alpharetroviral assays were expanded for further analyses including determination of integration sites and Evi1 and Prdm16 transcript levels.
Isolation of RNA and determination of transcript levels. RNA was isolated using RNAzol (WAK Chemicals, Steinbach, Germany) according to the manufacturer's instructions. The RNA pellet was resuspended in RNAse-free water supplemented with 1 µl RiboLock RNase inhibitor (Fermentas) and was stored at −20 °C. For determination of Prdm16, Evi1, β-Actin transcript levels, RNA was reverse-transcribed using the QuantiTect reverse transcription kit (Qiagen). For transcript quantification cDNA was subjected to quantitative PCR using primers Prdm16 forward: CTCATCTGCCTCCACAGTGA; Prdm16 rev: GGACCACAATCCAGTGCTTT; Evi1 forward: GAGGCC GTAGAAATCGGAAGATCTTAGATGAG; Evi1 reverse: CTGGCATG CAACAAGGTTGTGCTGATC; for β-actin detection Quantitect primers were used (Qiagen). Transcript levels were calculated using the efficiency-corrected ΔΔCT method using a reference plasmid containing Prdm16, Evi1, and β-Actin sequences.49
SUPPLEMENTARY MATERIAL Figure S1. FACS gating strategies for murine lineage-negative and peripheral blood cells. Figure S2. Blood counts and organ weights. Figure S3. EGFP expression in erythrocytes and platelets in peripheral blood. Figure S4. LM PCR on alpharetroviral mouse samples. Figure S5. LM PCR on gammaretroviral and lentiviral mouse samples. Figure S6. Venn diagrams of annotated RTCGD and CGC cancer genes. Figure S7. Integration site analyses in IVIM samples.
Acknowledgments
We thank Julia Skokowa for help with microarray analyses, Olga Kustikova for advice on LM PCR, Adrian Schwarzer for consultation in statistics, Bernhard Schiedlmeier for help with sorting of primary cells, and Dirk Heckl for providing Prdm16 primers. We furthermore thank Sabine Knoess and Johanna Krause for technical assistance with the IVIM assay and Thomas Neumann for technical assistance with animal experiments, microarray analyses, and LM PCR. We are also very grateful to Rena Struss and Martin Hapke for help with animal work, the Department of Microbiology, especially Sabrina Woltemate, for help with pyrosequencing, and the Department of Radiotherapy for irradiation of mice (all Hannover Medical School). This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB738, SPP1230 and Cluster of Excellence REBIRTH (EXC 62/1)), the German Ministry for Research and Education (IFB-Tx (01EO0802)), the German Academic Exchange Service (DAAD (0315187)), the European Union (Integrated Project PERSIST), and the Madeleine Schickedanz-KinderKrebs-Stiftung. The authors declare that they have no conflict of interest, except that A.S., J.D.S., and C.B. are inventors on a patent application describing alpharetroviral SIN vectors.
Supplementary Material
FACS gating strategies for murine lineage-negative and peripheral blood cells.
Blood counts and organ weights.
EGFP expression in erythrocytes and platelets in peripheral blood.
LM PCR on alpharetroviral mouse samples.
LM PCR on gammaretroviral and lentiviral mouse samples.
Venn diagrams of annotated RTCGD and CGC cancer genes.
Integration site analyses in IVIM samples.
REFERENCES
- Kohn DB. Update on gene therapy for immunodeficiencies. Clin Immunol. 2010;135:247–254. doi: 10.1016/j.clim.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B., and, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Hematti P, Hong BK, Ferguson C, Adler R, Hanawa H, Sellers S.et al. (2004Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells PLoS Biol 2e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR., and, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B.et al. (2007Hot spots of retroviral integration in human CD34+ hematopoietic cells Blood 1101770–1778. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M.et al. (2009The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy J Clin Invest 119964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr SD, Leipzig J, Shinn P, Ecker JR., and, Bushman FD. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J Virol. 2005;79:12035–12044. doi: 10.1128/JVI.79.18.12035-12044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard BC, Dickerson D, Beebe K, Gooch C, Fletcher J, Okbinoglu T.et al. (2007Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells Mol Ther 151356–1365. [DOI] [PubMed] [Google Scholar]
- Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C.et al. (2004Genome-wide analyses of avian sarcoma virus integration sites J Virol 7811656–11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Renaud G, Gomes TJ, Golmes T, Ferris A, Hendrie PC.et al. (2008Reduced genotoxicity of avian sarcoma leukosis virus vectors in rhesus long-term repopulating cells compared to standard murine retrovirus vectors Mol Ther 161617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC.et al. (2004Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences PLoS Biol 2E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U, Bohne J, Schmidt M, von Kalle C, Knöss S, Schambach A.et al. (2006Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity Blood 1082545–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerth JD, Maetzig T, Galla M, Baum C., and, Schambach A. Self-inactivating alpharetroviral vectors with a split-packaging design. J Virol. 2010;84:6626–6635. doi: 10.1128/JVI.00182-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, Brugman MH.et al. (2009Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors Mol Ther 171919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A.et al. (2010Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease Nat Med 16198–204. [DOI] [PubMed] [Google Scholar]
- Schambach A, Bohne J, Baum C, Hermann FG, Egerer L, von Laer D.et al. (2006Woodchuck hepatitis virus post-transcriptional regulatory element deleted from X protein and promoter sequences enhances retroviral vector titer and expression Gene Ther 13641–645. [DOI] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D.et al. (1998A third-generation lentivirus vector with a conditional packaging system J Virol 728463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach A, Bohne J, Chandra S, Will E, Margison GP, Williams DA.et al. (2006Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells Mol Ther 13391–400. [DOI] [PubMed] [Google Scholar]
- Schambach A, Mueller D, Galla M, Verstegen MM, Wagemaker G, Loew R.et al. (2006Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors Gene Ther 131524–1533. [DOI] [PubMed] [Google Scholar]
- Zhang CC., and, Lodish HF. Murine hematopoietic stem cells change their surface phenotype during ex vivo expansion. Blood. 2005;105:4314–4320. doi: 10.1182/blood-2004-11-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetzig T, Brugman MH, Bartels S, Heinz N, Kustikova OS, Modlich U.et al. (2011Polyclonal fluctuation of lentiviral vector-transduced and expanded murine hematopoietic stem cells Blood 1173053–3064. [DOI] [PubMed] [Google Scholar]
- Challita PM., and, Kohn DB. Lack of expression from a retroviral vector after transduction of murine hematopoietic stem cells is associated with methylation in vivo. Proc Natl Acad Sci USA. 1994;91:2567–2571. doi: 10.1073/pnas.91.7.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentilin L, Qin G, Tafuro S, Dinauer MC, Baum C., and, Giacca M. Variegation of retroviral vector gene expression in myeloid cells. Gene Ther. 2000;7:153–166. doi: 10.1038/sj.gt.3301057. [DOI] [PubMed] [Google Scholar]
- Gabriel R, Eckenberg R, Paruzynski A, Bartholomae CC, Nowrouzi A, Arens A.et al. (2009Comprehensive genomic access to vector integration in clinical gene therapy Nat Med 151431–1436. [DOI] [PubMed] [Google Scholar]
- Appelt JU, Giordano FA, Ecker M, Roeder I, Grund N, Hotz-Wagenblatt A.et al. (2009QuickMap: a public tool for large-scale gene therapy vector insertion site mapping and analysis Gene Ther 16885–893. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E.et al. (2008Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1 J Clin Invest 1183132–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H.et al. (2008Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients J Clin Invest 1183143–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U.et al. (2006Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1 Nat Med 12401–409. [DOI] [PubMed] [Google Scholar]
- Osawa M, Nakamura K, Nishi N, Takahasi N, Tokuomoto Y, Inoue H.et al. (1996In vivo self-renewal of c-Kit+ Sca-1+ Lin(low/-) hemopoietic stem cells J Immunol 1563207–3214. [PubMed] [Google Scholar]
- Osawa M, Hanada K, Hamada H., and, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Adolfsson J, Borge OJ, Bryder D, Theilgaard-Mönch K, Astrand-Grundström I, Sitnicka E.et al. (2001Upregulation of Flt3 expression within the bone marrow Lin(-)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity Immunity 15659–669. [DOI] [PubMed] [Google Scholar]
- Kumar R, Fossati V, Israel M., and, Snoeck HW. Lin-Sca1+kit- bone marrow cells contain early lymphoid-committed precursors that are distinct from common lymphoid progenitors. J Immunol. 2008;181:7507–7513. doi: 10.4049/jimmunol.181.11.7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, Grassman E.et al. (2008Physiological promoters reduce the genotoxic risk of integrating gene vectors Mol Ther 16718–725. [DOI] [PubMed] [Google Scholar]
- Modlich U, Schambach A, Brugman MH, Wicke DC, Knoess S, Li Z.et al. (2008Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16 Leukemia 221519–1528. [DOI] [PubMed] [Google Scholar]
- Du Y, Jenkins NA., and, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Thornhill SI, Howe SJ, Ulaganathan M, Schambach A, Sinclair J.et al. (2007Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells Blood 1101448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlers A, Zipfel PF, Schwieger M, Ostertag W., and, Baum C. In vivo analysis of retroviral enhancer mutations in hematopoietic cells: SP1/EGR1 and ETS/GATA motifs contribute to long terminal repeat specificity. J Virol. 2002;76:303–312. doi: 10.1128/JVI.76.1.303-312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CC, Kaba M, Ge G, Xie K, Tong W, Hug C.et al. (2006Angiopoietin-like proteins stimulate ex vivo expansion of hematopoietic stem cells Nat Med 12240–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding AK, Maurice M, Morling FJ, Cosset FL., and, Russell SJ. Inverse targeting of retroviral vectors: selective gene transfer in a mixed population of hematopoietic and nonhematopoietic cells. Blood. 1998;91:1802–1809. [PubMed] [Google Scholar]
- Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- Woods NB, Muessig A, Schmidt M, Flygare J, Olsson K, Salmon P.et al. (2003Lentiviral vector transduction of NOD/SCID repopulating cells results in multiple vector integrations per transduced cell: risk of insertional mutagenesis Blood 1011284–1289. [DOI] [PubMed] [Google Scholar]
- Hanahan D., and, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Weidhaas JB, Angelichio EL, Fenner S., and, Coffin JM. Relationship between retroviral DNA integration and gene expression. J Virol. 2000;74:8382–8389. doi: 10.1128/jvi.74.18.8382-8389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield LF, Fraize CD., and, Coffin JM. Relationship between retroviral DNA-integration-site selection and host cell transcription. Proc Natl Acad Sci USA. 2005;102:1436–1441. doi: 10.1073/pnas.0409204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toren A, Bielorai B, Jacob-Hirsch J, Fisher T, Kreiser D, Moran O.et al. (2005CD133-positive hematopoietic stem cell “stemness” genes contain many genes mutated or abnormally expressed in leukemia Stem Cells 231142–1153. [DOI] [PubMed] [Google Scholar]
- Kustikova OS, Modlich U., and, Fehse B. Retroviral insertion site analysis in dominant haematopoietic clones. Methods Mol Biol. 2009;506:373–390. doi: 10.1007/978-1-59745-409-4_25. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Hoffmann G, Wissler M, Lemke N, Müssig A, Glimm H.et al. (2001Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples Hum Gene Ther 12743–749. [DOI] [PubMed] [Google Scholar]
- Wu Z, Fraley ME, Bilodeau MT, Kaufman ML, Tasber ES, Balitza AE.et al. (2004Design and synthesis of 3,7-diarylimidazopyridines as inhibitors of the VEGF-receptor KDR Bioorg Med Chem Lett 14909–912. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FACS gating strategies for murine lineage-negative and peripheral blood cells.
Blood counts and organ weights.
EGFP expression in erythrocytes and platelets in peripheral blood.
LM PCR on alpharetroviral mouse samples.
LM PCR on gammaretroviral and lentiviral mouse samples.
Venn diagrams of annotated RTCGD and CGC cancer genes.
Integration site analyses in IVIM samples.