Abstract
We have identified homologs of a human BMP receptor-associated molecule BRAM1 in Caenorhabditis elegans. One of them, BRA-1, has been found to bind DAF-1, the type I receptor in the DAF-7 transforming growth factor-β pathway through the conserved C-terminal region. As analyzed using a BRA-1∷GFP (green fluorescent protein) fusion gene product, the bra-1 gene is expressed in amphid neurons such as ASK, ASI, and ASG, where daf-1 is also expressed. A loss-of-function mutation in bra-1 exhibits robust suppression of the Daf-c phenotype caused by the DAF-7 pathway mutations. We propose that BRA-1 represents a novel class of receptor-associated molecules that negatively regulate transforming growth factor-β pathways.
Secreted signaling molecules of the transforming growth factor (TGF)-β superfamily induce diverse cellular responses including cell proliferation, cell differentiation, and apoptosis (1, 2). Recent studies have identified components essential for TGF-β signal transduction. These components include type I and type II serine/threonine (Ser/Thr) kinase receptors, SMADs, and transcriptional regulators that cooperate with SMADs (3).
Caenorhabditis elegans is one of the most thoroughly understood multicellular animals in terms of cellular development, anatomy, and genome content. Recently, its whole genome sequence was elucidated (4). This allows us to use genetic approaches for the understanding of conserved signaling pathways, including TGF-β signaling. In C. elegans, at least two TGF-β like signaling pathways exist: the dauer larva formation (daf) and the small (sma) pathways. The former pathway regulates formation of dauer larva in response to starvation or overcrowding conditions (5). Dauer constitutive (daf-c) or defective (daf-d) genes such as daf-1, daf-3, daf-4, daf-5, daf-7, daf-8, and daf-14 have been proposed to act in a common pathway in the regulation of dauer larva formation (6–11). The latter pathway appears to control body length as well as ray pattern formation in the male tail (12–15).
To understand the precise molecular mechanism of TGF-β signaling, we have been attempting to identify components that modulate TGF-β signaling. Using a mouse type I receptor (BMPRI/ALK3) (16) for a TGF-β ligand, BMP, in a yeast two-hybrid screen, we have recently isolated a human gene encoding a protein that binds to the BMP receptor. This protein, designated BMP receptor-associated molecule 1 (BRAM1) (17), was shown to be a cytoplasmic protein and to bind BMPRI-A specifically. However, its in vivo function was not clear.
In this article, we describe the gene bra-1, the homolog of BRAM1 in C. elegans. The bra-1 gene is expressed in amphid neurons where daf-1, the type I receptor (6) in the DAF-7 TGF-β pathway, is also expressed. BRA-1 has been found to bind DAF-1 through the conserved C-terminal region. Moreover, a loss-of-function mutation in bra-1 exhibits robust suppression of the Daf-c phenotype caused by the DAF-7 pathway mutations. We propose that BRA-1 represents a novel class of receptor-associated molecules that negatively regulate TGF-β pathways in C. elegans.
Materials and Methods
Strains and Genetics.
The techniques used for culturing C. elegans were essentially as described by Brenner (18).
The following strains were used in this work.: wild-type C. elegans variety Bristol strain (N2), bra-1(nk1)X,daf-1(m402)IV, daf-1(m213)IV, daf-1(m40)IV, daf-1(e1287)IV, daf-2 (e1370)III, daf-7(m62)III, daf-7(e1372)III, daf-11(m47)V, daf-14(m77)IV, mut-2(r459)I; dpy-19(n1347)III, dpy-9(e12)IV, unc-42(e270)V, unc-43(e408)IV, unc-45(e286)III.
Mammalian Cell Expression Plasmid Construction.
The cytoplasmic domains of DAF-1 were inserted into the mammalian expression vector pEBgs, which produces fusion proteins joined to glutathione S-transferase at the N terminus under the control of the EF-1 alpha gene promoter. SpeI/EcoRI fragments of daf-1 cDNA were ligated to in-frame with the SpeI/ClaI site of pEBgs. Full-length BRA-1-hemagglutinin (HA) was generated by XbaI/EcoRV; the digested fragment was ligated in-frame into the XbaI/EcoRV site of the HA-tag vector. The C-terminally deleted BRA-1 (amino acids 1–101L), which was the cDNA sequence corresponding to 1–101 of BRA-1 protein, was PCR amplified using two primers: 5′-CCGCTCTAGAACTAGTGGATCC-3′and 5′-AAAGATATCTGAGATTCTCCGCGTGCTTC-3′. The PCR product was digested with XbaI/EcoRV and cloned into the XbaI/EcoRV site of HA-tag vector. The N-terminally deleted (amino acids Glu-103–Gln-182) sequence, which was the cDNA sequence corresponding to the conserved C-terminal region, was PCR amplified using two primers: 5′-GAAGCACGCGGAGAATCTCATGG-3′ and 5′-AAAGATATCGTTGACTTTGAGTAGGCTCAG-3′. The PCR product was blunt ended and digested with EcoRV and cloned into the blunt-ended BamHI/EcoRV of the HA-tag vector. These plasmids were digested with XbaI/XhoI, and the inserts were cloned into the SalI/XbaI of pEF vector for mammalian expression, respectively.
Mammalian Cell Culture and Glutathione S-Transferase (GST) Pull-Down Assay.
COS7 cells were transiently transfected with the indicated constructs by the calcium phosphate method. Forty-eight hours after transfection, cells were lysed in lysis buffer (17), and 100-μl aliquots of lysates were incubated with 10 μl of glutathione-Sepharose beads at 4°C for 1 h. Precipitates were washed five times with 100 μl of lysis buffer and examined by Western blot analysis.
Generation of bra-1 Expression Constructs.
A translational bra-1∷GFP (green fluorescent protein) fusion construct containing a nuclear localization signal or not was made by cloning the 3.3-kbp SphI/SalI fragment derived from the cosmid F54B11 into the SphI/SalI site of pPD95.69. Extrachromosomal arrays were integrated into chromosomes using UV irradiation, followed by at least two backcrosses with N2.
Transformation of C. elegans.
Microinjection of DNAs into the gonadal syncytia of C. elegans hermaphrodites was carried out as described previously (19). Each transformation result was scored with multiple independent transgenic lines. All plasmids were injected at a concentration of 20 μg/ml. Injection was performed with coinjection of a rol-6(d) plasmid (pRF4) as a dominant marker for transformation (19) or myo-3∷gfp plasmid. A strain of N2 was used as the host strain unless noted otherwise.
Isolation of Tc1 Insertions in bra-1 and Deletion Derivatives.
Transposable elements were detected using nested PCR. The positions of the Tc1 insertions were confirmed by multiple PCR reactions with flanking primers, followed by direct sequencing of the PCR products from both insertion junctions. Tc1-specific primers Tc102 (5′-AGCCAGCTACAATGGCTTTC-3′), Tc103 (5′-GATGCAAACGGATACGCGAC-3′), Tc104 (5′-CCAAACAAATCCAGTGCAAC-3′), and Tc105 (5′-TGTCATTTCCTTGCAACCTC-3′) and gene-specific primers CBM-1 (5-GGCTCATGACCACATATCGA-3), CBM-2 (5-ACTCGCCTCATGGGCTCATT-3), CBM-3 (5-GATCTCTCCGAAATTGATCC-3), and CBM-4 (5-TCTCGAAGGAATTAACACAAGT-3) were used. The deletion mutant was isolated using a standard sib-selection procedure and was backcrossed ten times against N2 wild-type animals.
Examination of Dauer Formation Phenotypes.
The frequency of dauer formation was assessed under noninducing conditions essentially as described (10). Noninducing conditions were defined as uncrowded animals on well-seeded 6-cm NG agar plates. Between 4 and 12 adult hermaphrodites were placed on a plate at the test temperature. After allowing egg laying for a limited time (less than 12 h at 25°C), the parents were removed. As the progeny matured, the plates were checked frequently, and L4 non-dauers were counted.
Results
Isolation and Characterization of bra-1.
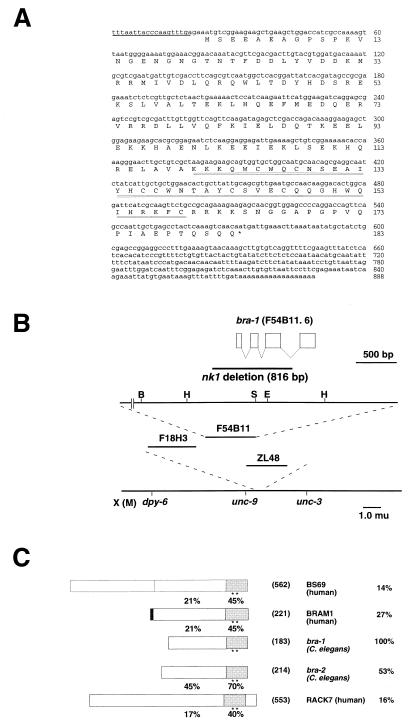
Using a C. elegans database (ACeDB), we found two C. elegans genes similar to human BRAM1 (hBRAM1) (17), which were designated bra-1 and bra-2. In this study, we investigated the function of bra-1. The bra-1 gene was mapped to F54B11.1 on chromosome X (Fig. 1B) where no gene defined by mutations appears to correspond to bra-1. Analysis of cDNA sequences indicates that bra-1 consists of four exons. The predicted BRA-1 protein consists of 183 amino acids and contains two Zn fingers as part of the MYND domain within the C-terminal region (Fig. 1A). In this C-terminal region, BRA-1 shows sequence similarity to BRA-2, an E1A binding protein BS69 (20), hBRAM1 (17), an alternatively spliced form of BS69, and a human protein kinase C-binding protein RACK7 (Fig. 1C). Higher molecular weight forms of BRA-1 or BRA-2 corresponding to mammalian BS69 were unlikely to be present since an RNA blot analysis using the mixed-stage population of C. elegans showed a single band of ≈0.9 kb, which is the expected size for bra-1 (888 bp) or bra-2 (909 bp) transcript (data not shown). It may be expected that bra-1 is involved in the regulation of one of either dauer formation (daf) or small (sma) pathways. The fact that BRA-1 and BRA-2 lack the N-terminal extension of BS69, which contains only the C-terminal portion, is physiologically important.
Figure 1.
Structural analysis of bra-1. (A) Nucleotide and predicted amino acid sequences of bra-1 cDNA. SL1 trans-spliced leader sequence is indicated with an underline. The conserved zinc-finger-like sequence is indicated with a double underline. (B) A physical and genetic map of the bra-1 region. The top line is the gene structure with exons shown in boxes separated by introns. The direction of transcription is from right to left. The deletion allele of the bra-1 gene was isolated by PCR screening of populations of Tc1 insertion strains for an imprecise Tc1 excision. The extent of the nk1 deletion, 816 bp, which correspond to cosmid F54B11 25420–26236, is shown. The second line is the partial restriction map of the genomic cosmid clone F54B11. B, BamHI; H, HindIII; S, SacI; E, EcoRI. The third line is the physical map showing the position of cosmid clone F54B11. The fourth line is the genetic map of a portion close to the center of the X chromosome. (C) Alignment of the predicted amino acid sequences of the structure of BS69 (human), BRAM1 (human), BRA-1 (C. elegans), BRA-2 (C. elegans), and RACK7 (human). The percent identities for the whole sequences with BRA-1 are indicated to the left. The percent identities of N-terminal and C-terminal regions are indicated next to respective regions. Conserved zinc-finger-like motifs are indicated with asterisks. Numbers of predicted amino acid residues are given in parentheses.
BRA-1∷GFP Fusion Gene Is Expressed in Amphid Neurons Such as ASK, ASI, and ASG, Where daf-1 Is Also Expressed.
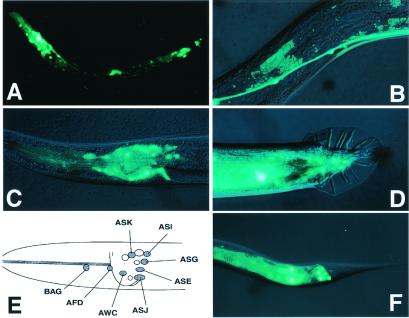
To examine the expression pattern of BRA-1, transgenic worms carrying a GFP reporter construct (BRA-1∷GFP) were generated and established the chromosomal integrated lines. BRA-1∷GFP was expressed strongly in amphid and phasmid neurons, and weakly in a few cells in the tail and in hypodermal cells. This expression pattern was constant throughout the larval and adult stages. Identified neurons include amphid neurons, AWC, AFD, ASI, ASG, ASK, and phasmid neurons PHA and PHB (Fig. 2). A similar expression pattern in these neurons was observed with DAF-1 (21), suggesting that BRA-1 and DAF-1 colocalize in the same set of cells. As shown previously in a laser cell ablation experiment, ADF, ASI, and ASG are responsible for inhibiting dauer formation, and ASJ is responsible for allowing animals to exit the dauer stage (22). The localized expression of BRA-1 in ASI, ASG, and ASJ supports the role of BRA-1 in the regulation of dauer larva formation.
Figure 2.
Expression of a BRA-1∷GFP Fusion. (A) BRA-1∷GFP expression of an L3 hermaphrodite. Strong expression was observed in the head region and the tail region. (B) Strong signals were seen in the ventral nerve cord. (C) Lateral view of the adult hermaphrodite showing GFP expression in the head. GFP fluorescence was observed in the cell bodies of BAG, AFD, ASK, ASI, ASG, ASE, ASJ, and AWC neurons. (D) Tail region of an adult male. GFP fluorescence was seen in unidentified male-specific neurons. (E) A diagram summarizing the BRA-1∷GFP-expressing neuron. (F) Tail region of adult hermaphrodite. GFP fluorescence was seen in the cell bodies of PHA and PHB phasmid neurons. Anterior is to the left, and dorsal is up.
BRA-1 Associates with the DAF-1 Type I Receptor Through the Conserved C-Terminal Region.
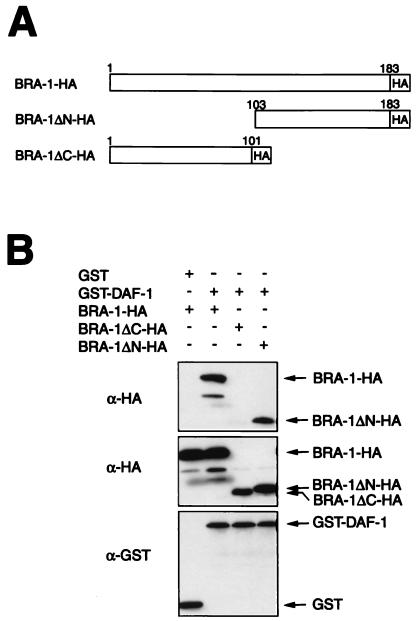
Since hBRAM1 was shown to bind BMP receptor-1A (17), and we also found that the expression pattern of bra-1 is similar to that of daf-1 type I receptor (Fig. 2), BRA-1 may bind a type I receptor DAF-1 as well, thereby modulating the activity of a TGF-β pathway. To test this possibility, we performed a GST pull-down assay using mammalian COS7 cells transiently transfected with cDNAs of GST-DAF-1 and (i) HA-tagged full-length, (ii) N-terminally deleted, or (iii) C-terminally deleted BRA-1 (Fig. 3A). As shown in Fig. 3B, the GST-DAF-1 fusion protein was efficiently immunoprecipitated with anti-HA antibody, demonstrating that BRA-1 physically interacts with the DAF-1 type I receptor. The N-terminal-deleted form of BRA-1, as in hBRAM1 (17), is sufficient to bind DAF-1, indicating the conserved role of the C-terminal region of hBRAM1 and BRA-1.
Figure 3.
BRA-1 associates with the DAF-1 type I receptor through the conserved C-terminal region in mammalian cells. (A) Schematic diagram of the structure of BRA-1 deletion mutations. (B) COS7 cells were transiently transfected with HA-tagged BRA-1, BRA-1ΔN, and BRA-1ΔC and GST alone or GST-DAF-1. Cell lysates were subjected to the GST pull-down procedure and then immunoblotted using an anti-HA antibody (Top). Expression of BRA-1-HA, BRA-1ΔN-HA, BRA-1ΔC-HA, GST, and GST-DAF-1 was measured by anti-HA or anti-GST immunoblotting of aliquots from cell lysates (Middle or Bottom).
bra-1 Negatively Regulates DAF-7 TGF-β Signaling.
The bra-1 (nk1) mutant was created with a method using Tc1 insertion and excision (23). nk1 is likely to be a null mutation as the majority of the coding region is deleted (Fig. 1B). The bra-1 (nk1) mutant exhibited a head-lifting phenotype; this is the typical phenotype observed in mutants, such as cat-2 (catecholamine abnormality) mutants, which are associated with the reduced dopamine level (24).
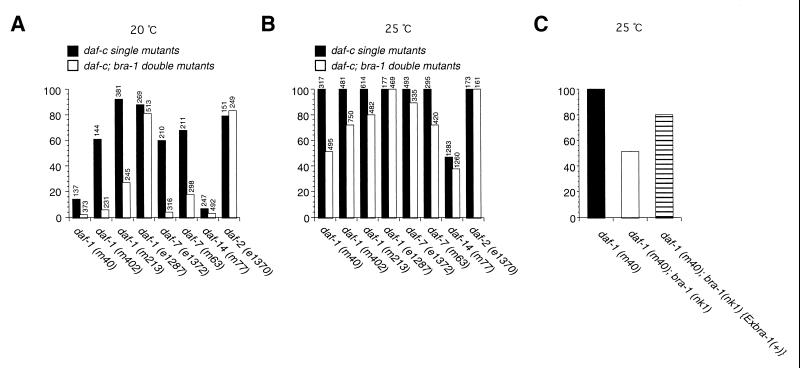
This phenotype, as well as the suppression phenotype described below, was rescued by a bra-1 transgene (Fig. 4C; data not shown), indicating that these phenotypes are caused by the bra-1 mutation. To investigate the involvement of bra-1 in regulating the DAF-7 TGF-β pathway, we generated a double mutant of bra-1 (nk1) with daf-1, a dauer constitutive (daf-c) mutant. With temperature-sensitive hypomorphic alleles of daf-1, m40, m402, and m213, a large fraction of the mutant worms enters the dauer stage in the presence of food at 20°C, and all worms undergo dauer formation at 25°C. However, the dauer larva formation was significantly reduced in double mutants containing bra-1 (nk1) and a mutation in daf-1 or daf-7. This suppression of dauer formation by bra-1 was not observed in the double mutant with daf-1 null allele e1287 (Fig. 4 A and B). Also, we found no genetic interaction of bra-1 with daf-2 that constitutes an independent daf pathway (Fig. 4 A and B). This result suggests that bra-1 functions as a negative regulator of the DAF-7 pathway, acting downstream of the DAF-7 ligand and the DAF-1 receptor. In contrast, a double mutant containing bra-1 (nk1) and a mutation in daf-14, which encodes a SMAD family of proteins, showed only a small change in frequency of dauer larva formation (Fig. 4 A and B). Together, these results suggest that BRA-1 acts downstream of the type I receptor but upstream of the SMADs in the DAF-7 pathway.
Figure 4.
Genetic interactions between bra-1 and daf-7 pathway genes. Percent dauer formation in DAF-7 pathway and daf-2 single mutants and double mutants with bra-1 (nk1) at 20°C (A) and 25°C (B). The total numbers of animals counted are indicated above the columns. Closed columns indicate the percent dauer formation of DAF-7 TGF-β pathway and daf-2 single mutants, and open columns indicate that of the double mutants with bra-1 (nk1). (C) Rescue experiment with a bra-1 genomic fragment and a daf-1 (m40), bra-1 (nk1) double mutant.
Discussion
BRAMs Are Conserved Proteins and Define a Novel Family of Proteins.
We identified two homologs of the human BRAM1 in C. elegans. These gene products share extremely conserved C-terminal region (Fig. 1C). The conserved region has two consensus sequences, which resemble a zinc-finger-related domain (MYND domain). The zinc-finger domain is known to be important for interaction of protein not only with DNA but also with protein. Since the C-terminal half of BRA-1 was shown to be necessary and sufficient for binding to DAF-1 (Fig. 3), this predicted zinc-finger motif may be conserved functional domain for association of BRAMs to type I receptors beyond species.
Upon completion of the C. elegans genome project, we know almost all of the structure of genes in C. elegans. In C. elegans, TGF-β type I receptor-related genes are found in only two, which are daf-1 (6) and sma-6 (15). BRAM1-related genes, which we found in C. elegans genome are BRA-1 and BRA-2. This may suggest that each type I receptor has one pathway-specific BRAM1-related molecule to associate with it and to be negatively regulated.
BRA-1 Is Involved in the DAF Pathway.
Recent studies on the TGF-β signaling pathway in C. elegans have revealed that there are at least two distinct pathways regulating neuronal activities related to dauer larva formation (6–11) and body length of the nematode (12–15). In our present study, we identified a negative regulator of the signaling from the type I receptor DAF-1 (6). DAF-7, the ligand in the DAF pathway, is expressed in the ASI amphid neuron and is thought to act in a neuroendocrine manner, regulating developmental and metabolic shifts in tissues that are remodeled during dauer formation (8). Here, we have demonstrated that bra-1, as well as daf-1, is expressed in amphid neurons including ASI neurons. Therefore, it is likely that dauer larva formation is regulated by interneuronal communications between ASI producing DAF-7 and other nearby amphid neurons expressing DAF-1 and BRA-1.
The bra-2, another homolog of BRAM1 in C. elegans, is expressed in pharyngeal muscle and intestine (K. M. unpublished result), and these expression patterns are similar to another type I receptor, sma-6 (15). These results may suggest possible involvement of bra-2 in the sma pathway, another TGF-β signaling in C. elegans.
Possible Mechanism of Negative Regulation of TGF-β Signaling by BRA-1.
In the TGF-β signaling system, association and phosphorylation of pathway-restricted Smads by the type I receptor is essential for activating the signaling pathway (3). However, little is known how Smad interaction with the receptor is controlled. Here, we describe the identification of novel type I receptor-associated protein BRA-1 in C. elegans. Genetic analysis of BRA-1 null mutant with previously known daf-7 TGF-β pathway mutants revealed that BRA-1 is epistatic to daf-7 and daf-1 and hypostatic to daf-14 (Fig. 4). Since daf-7, daf-1, and daf-14 are encoding ligand (8), type I receptor (7), and SMAD proteins (10), respectively, BRA-1 may be acting between type I receptor and the SMAD protein and may be negatively regulating this pathway in C. elegans. We also demonstrate that BRA-1 physically associates with DAF-1 type I receptor (Fig. 3). Absence of genetic interaction with a null mutant of daf-1 (e1287) also suggests that interaction of BRA-1 with DAF-1 is essential for the negative regulation. Taken these results together, we propose a hypothesis for a negative effect of BRA-1 on TGF-β signaling pathway and that BRA-1 blocks the physical interaction of type I receptor and SMAD proteins. To confirm this possibility, further analysis on protein–protein interactions is required.
Recently, a Smad2 binding protein, SARA (Smad anchor for receptor activation), was identified (25). SARA that is normally localized to inner plasma membrane binds Smad2 and recruits it into TGF-β receptor complex. Activation of TGF-β signaling upon ligand binding induces dissociation of Smad2 from SARA and the TGF-β receptor complex and leads to the formation of a Smad2/Smad4 complex and its nuclear translocation. Thus, SARA is a component of TGF-β signaling that functions to recruit Smad2 to the receptor by controlling the subcellular localization of the Smad. SARA per se interacts with the TGF-β receptor independently of Smad2 binding, although the association is enhanced in the presence of Smad2. Thus, a type I TGF-β receptor-binding protein SARA, defining a family of related proteins (25), is thought to be a positive regulator of the TGF-β signaling. In addition to SARA, we propose here that BRAMs may also represent a new class of TGF-β family signal mediator proteins. Although the precise mechanism of a negative effect of BRA-1 on dauer formation remains to be clarified, we hope the present study will provide a new aspect to the regulatory mechanism of TGF-β signaling system.
Acknowledgments
We thank the members of N. Ueno's lab, especially M. Mochii and S. Yoshida for helpful discussion, M. Ichikawa for helpful assistance, Y. Suzuki, and A. Wismann for critical reading of our manuscript. We thank A. Coulson for cosmid clones, Y. Kohara for yk clones, A. Fire for pPD95.69, I. Mori for the identification of the bra-1∷gfp expressing amphid neurons, and D. Riddle for daf-7 (m63), daf-1 (m402), daf-1 (e1287), and daf-1 (m213). Some nematode strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health, National Center for Research Resources. This research was supported by grants from the “Research for the Future” program of the Japan Society for the Promotion of Science (to N.U.).
Abbreviations
- TGF
transforming growth factor
- GST
glutathione S-transferase
- GFP
green fluorescent protein
- HA
hemagglutinin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kingsley D M. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 2.Hogan B L. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 3.Heldin C H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 4.The C. elegans Sequencing Consortium. Science. 1998;282:2012–2046. [Google Scholar]
- 5.Golden J W, Riddle D L. Proc Natl Acad Sci USA. 1984;81:819–823. doi: 10.1073/pnas.81.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georgi L L, Albert P S, Riddle D L. Cell. 1990;61:635–645. doi: 10.1016/0092-8674(90)90475-t. [DOI] [PubMed] [Google Scholar]
- 7.Estevez M, Attisano L, Wrana J L, Albert P S, Massague J, Riddle D L. Nature (London) 1993;365:644–649. doi: 10.1038/365644a0. [DOI] [PubMed] [Google Scholar]
- 8.Ren P, Lim C S, Johnsen R, Albert P S, Pilgrim D, Riddle D L. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- 9.Patterson G I, Koweek A, Wong A, Liu Y, Ruvkun G. Genes Dev. 1997;11:2679–2690. doi: 10.1101/gad.11.20.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue T, Thomas J H. Dev Biol. 2000;217:192–204. doi: 10.1006/dbio.1999.9545. [DOI] [PubMed] [Google Scholar]
- 11.Thomas J H, Birnby D A, Vowels J J. Genetics. 1993;134:1105–1117. doi: 10.1093/genetics/134.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savage C, Das P, Finelli A L, Townsend S R, Sun C Y, Baird S E, Padgett R W. Proc Natl Acad Sci USA. 1996;93:790–794. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morita K, Chow K L, Ueno N. Development (Cambridge, UK) 1999;126:1337–1347. doi: 10.1242/dev.126.6.1337. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki Y, Yandell M D, Roy P J, Krishna S, Savage-Dunn C, Ross R M, Padgett R W, Wood W B. Development (Cambridge, UK) 1999;126:241–250. doi: 10.1242/dev.126.2.241. [DOI] [PubMed] [Google Scholar]
- 15.Krishna S, Maduzia L L, Padgett R W. Development (Cambridge, UK) 1999;126:251–260. doi: 10.1242/dev.126.2.251. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki A, Thies R S, Yamaji N, Song J J, Wozney J M, Murakami K, Ueno N. Proc Natl Acad Sci USA. 1994;91:10255–10259. doi: 10.1073/pnas.91.22.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurozumi K, Nishita M, Yamaguchi K, Fujita T, Ueno N, Shibuya H. Genes Cells. 1998;3:257–264. doi: 10.1046/j.1365-2443.1998.00186.x. [DOI] [PubMed] [Google Scholar]
- 18.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hateboer G, Gennissen A, Ramos Y F, Kerkhoven R M, Sonntag-Buck V, Stunnenberg H G, Bernards R. EMBO J. 1995;14:3159–3169. doi: 10.1002/j.1460-2075.1995.tb07318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunther C V, Georgi L L, Riddle D L. Development (Cambridge, UK) 2000;127:3337–3347. doi: 10.1242/dev.127.15.3337. [DOI] [PubMed] [Google Scholar]
- 22.Bargmann C I, Horvitz H R. Science. 1991;251:1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- 23.Zwaal R R, Broeks A, van Meurs J, Groenen J T M, Plasterk R H A. Proc Natl Acad Sci USA. 1993;90:7431–7434. doi: 10.1073/pnas.90.16.7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loer C M, Kenyon C J. J Neurosci. 1993;13:5407–5417. doi: 10.1523/JNEUROSCI.13-12-05407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsukazaki T, Chiang T A, Davison A F, Attisano L, Wrana J L. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]