Abstract
NS5 methyltransferase (Mtase) has a crucial role in the replication of dengue virus. There are two active sites on NS5 Mtase i.e., SAM and RNA-cap binding sites. Inhibition of the NS5 Mtase activity is expected to prevent the propagation of dengue virus. This study was conducted to design cyclic peptide ligands as enzyme inhibitors of dengue virus NS5 Mtase through computational approach. Cyclopentapeptides were designed as ligand of SAM binding site as much as 1635 and 736 cyclopentpeptides were designed as ligand of RNA-cap binding site. Interaction between ligand and NS5 Mtase has been conducted on the Docking simulation. The result shows that cyclopentapeptide CTWYC was the best peptide candidate on SAM binding site, with estimated free binding energy -30.72 kca/mol. Cyclopentapeptide CYEFC was the best peptide on RNA-cap binding site with estimated free binding energy -22.89 kcal/mol. Both peptides did not have tendency toward toxicity properties. So it is expected that both CTWYC and CYEFC ligands could be used as a potential antiviral drug candidates, which can inhibit the SAM and RNA-cap binding sites of dengue virus NS5 Mtase.
Keywords: Dengue virus, cyclopentapeptide, NS5 methyltransferase, antiviral drug, inhibitor
Background
Dengue fever is a serious threat to global health issues. Geographic distribution of this disease has undergone tremendous expansion over the last 30 years. Approximately 100 countries are endemic for dengue fever and 40% of the world's population or about 2.5 billion people in the tropical and sub-tropics have an increased risk of catching the disease. More than 50 million of low-grade fever infections with 400,000 cases of dengue hemorrhagic fever are reported annually, which has caused many deaths of children in several countries in the Asian [1].
Dengue virus has four serotypes i.e., DENV-1, DENV-2, DENV-3, and DENV-4. The classification is based on the type of antibodies produced in the human body after infection. These four serotypes had the same morphology and genome but show different antigens so that a person can be infected with this virus more than once in the absence of complete cross-protection [2].
Nonstructural (NS) enzyme such as NS3 protease with NS2B cofactor, NS3 helicase/nucleoside triposfatase (NTPase)/ RNA 5 'triposfatase (RTPase), NS5 methyltransferase (Mtase), and NS5 RNA-dependent RNA polymerase (RdRp) were known to have an important role in the replication of dengue virus [3]. Currently, NS3 and NS5 of dengue virus enzyme are the most understood mechanism, making these enzyme as an ideal target for antiviral manufacture of dengue virus [4].
Several studies related to inhibition of the enzyme which is a potential target in the dengue virus have been carried out. Tambunan et al., [5] designed cyclic peptides CKRKC as potential inhibitors of NS2B-NS3 protease. Tambunan et al., [6] also designed of 49 cyclic peptides based on amino acid that could be recognized by the active site of NS2-NS3 protease and generating the best ligand CRKRC. Podvinec et al., [7] used the S-Adenosil-homocysteine (SAH), triphosphate ribavirin, and sinefungin as analogues inhibitors of the NS5 methyltransferase. The result showed that SAH has stability issues and indicate the nature of toxicity, ribavirin has a low activity, and sinefungin has low specificity and problems of nephrotoxicity. Ribavirin triphosphate (RTP) and RTPanalogues were designed as potential antiviral NS5 methyltransferase [8]. Lim et al., [9] discovered several potential compounds that are active against dengue virus NS5 methyltransferase through virtual screening using structurebased and ligand-based methods.
NS5 methyltransferase has two ties sides that are connected by a Y-shaped slit. The first binding site is the SAM (methyl donor) binding site and the second is the RNA-cap binding site that likely shallow and smaller in size [10]. This enzyme has two methylation processes, first step movement of a methyl group to the SAM binding site and then transfer the methyl group to the guanine bases of RNA. Although NS5 methyltransferase has two methylation processes, these are occurring at the same place [11]. RNA substrate is expected to change positions in order to receive the methyl group on the SAM binding site [12].
Currently, research on peptides for drug design and discovery are the most promising fields in the development of new drugs. More than 140 peptides were used as drugs and more than 400 peptides have entered on preclinical phase with average growth of more than 15% in a year [13]. This study was conducted to design cyclic peptide for two binding pocket of dengue virus NS5 methyltransferase, so it could be used as an inhibitor of dengue virus NS5 methyltransferase.
Methodology
Ligands design and preparation:
Determination of amino acid sequence was based on natural substrate NS5 methyltransferase that are regonized by the binding sites i.e., SAM binding site and RNA-cap binding site. Peptides were designed as cyclopentapeptides, which were joined by disulfide bonding at each terminal cystein and modeled into three-dimensional structure using ACDlabs. We designed cyclopentapeptides as ligand for SAM binding site and RNA-cap binding site. Preparation of cyclic peptides based on the polarity of amino acid residues at the active sites of the SAM and RNA-cap. Cyclopentapeptide optimization was carried out by choosing wash option, partial charge and energy minimization using MMFF94 forcefield, gas phase solvation and RMS gradient 0.001 kcal/ Å mol [14].
NS5 methyltransferase preparation:
Structure of NS5 methyltransferase with code 2P41 was obtained from PDB and was loaded into Molecular Operating Environment (MOE) 2008.10. The enzyme structure was repaired and optimized using the protonate3D option in MOE [15]. Hydrogen atoms were added by choosing partial charge option. Energy minimization was performed by employing MMFF94x force field, gas phase solvation and RMS gradient 0.05 kcal/ Å mol [14].
Molecular docking:
Molecular docking was performed by choosing Simulationdock option in MOE. Triangle matcher was generated as placement method. Triangle matcher method generates poses in a systematic manner and more accurate way than the alpha triangle method by aligning the ligand triplet of atoms with the triplet of alpha spheres in cavities of tight atomic packing [15]. A London dG scoring function was used to rank candidate poses. Forcefield was used in refinement and repetition was set to 100 with only one best pose to be retained.
Toxicity prediction:
Analysis of toxicity of the ligands were carried out on the best ligand docking results. Parameters that will be seen from the nature of the ligand are carcinogenicity and mutagenicity. The analysis was performed using software ToxTree v2.1.0 and Osiris Property Explorer. Toxicology analysis based on the rule Benigni / Bossa rulebase for mutagenicity and carcinogenicity developed by Romualdo Benigni and Cecilia Bossa from the Instituto Superiore in Sanita, Rome, Italy, and approved by the European Chemical Bureau, Institute for Health and Consumers Protection, European Commission- Joint Research Centre (JRC) in 2008..
Disscussion
Ligands Screening:
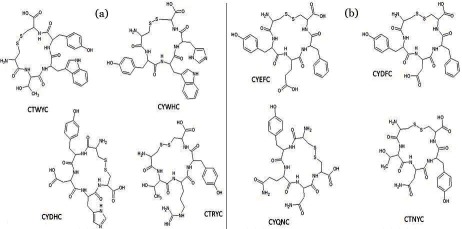
The inhibitors cyclopentapeptide designed was consisted of two cysteines at the end and three other amino acids are combined in the middle. Preparation of cyclic peptides by S-S bridge is intended to improve the stability of the ligand [16]. From combination of 20 amino acids based on polar and nonpolar, we obtained 1635 cyclopentapeptides as ligand for SAM binding site and 736 cyclopentapeptides for RNA-cap binding site. Screening of all ligands resulted in obtaining eight best ligands. The eight cyclopentapeptides that we designed are illustrated in (Figure 1). Four cyclopentapeptides as the best inhibitor on the SAM active side and four cyclopentapeptides on the RNA-cap pocket. Table 1 & 2 (see supplementary material) showed that CTWYC and CYEFC as the best inhibitor for SAM and RNA-cap site respectively.
Figure 1.
Illustrates (2D) eight cyclopentapeptides by S-S bridge, (a) Four cyclopentapeptides as the best inhibitor for the SAM active side and (b) Four cyclopentapeptides for the RNA-cap pocket.
SAM Binding Site:
Residues that are on the SAM binding site includes Ser56, Lys61, Cys82, Gly86, Trp87, Thr104, Lys105, Asp131, Val132, Phe133, Asp146, Ile147, Lys181, and Glu217. It had been reported that residues Lys61, Asp146, Lys181 and Glu217 are important on the SAM active site [17].
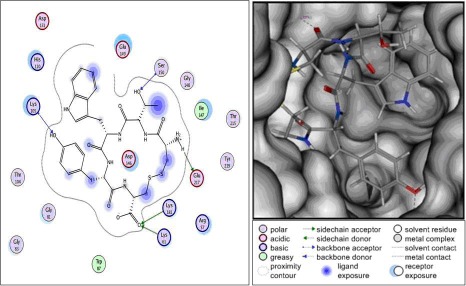
Result by screening of 1635 ligands was obtained CTWYC as the best inhibitor of the SAM site with binding free energy (ΔGbinding) of -30.72 kcal/mol. There are three polar interactions formed between CTWYC and SAM site (Figure 2), i.e., polar basic, polar acidic, and polar uncharged. Lys61, Lys105 and Lys181 are polar basic. The sidechain of Lys61 and Lys181 were interacted with CTWYC molecule by forming hydrogen bonds with the carboxyl site of cystein, and the backbond nitrogen of Lys105 was interacted by forming hydrogen bond with the 4–OH site of tyrosine. Glu217 is polar acidic, the backbond oxygen of Glu217 was interacted with CTWYC molecule by forming hydrogen bond with amino site of cystein. Ser150 is polar uncharged, the backbond nitrogen of Ser150 was interacted by forming hydrogen bond with the OH sidechain of threonine.
Figure 2.
Interactions formed between CTWYC and SAM binding site. There are three polar interactions by forming hydrogen bonds between CTWYC and SAM binding site.
Binding free energy (ΔGbinding) of cyclic peptide CTWYC while compared with SAM and SAH standards, has a value of ΔGbinding is much greater, it provides a sense that strength of CTWYC to inhibit the SAM active site was better stable. pKi value or affinity of CTWYC also larger from the other.
RNA-cap Site:
Residues has a role important at the RNA-cap site are Lys14, Leu17, Asn18, Leu20, Phe25, Lys29, Ser150, and Ser151 [17]. CYEFC cyclic peptide is the best inhibitor of RNA-cap site with binding free energy (ΔGbinding) of -22.89 kcal/mol. This result was obtained by screening of 736 cyclic peptides. There are three polar interactions formed between CYEFC and RNAcap site (Figure 3), i.e., polar basic, polar acidic, and polar uncharged. Lys29 is polar basic; the sidechain of Lys29 was interacted with CYEFC molecule by forming hydrogen bonds with the OH carboxyl of glutamate sidechain. Glu149 is polar acidic, the sidechain of Glu149 was interacted by forming hydrogen bonds with amina site of cystein. Asn18, Ser150, and Ser214 are polar uncharged. The backbond nitrogen of Asn18 was interacted with CYEFC molecule by forming hydrogen bonds with 4-OH of tyrosine. Ser150 was interacted with CYEFC by forming two hydrogen bonds, from the backbond and the sidechain of Ser150. The backbond of Ser150 was performed hydrogen bonds with amine site of cystein and the sidechain of Ser150 was performed hydrogen bonds with oxygen carboxyl of cystein. Ser214 was interacted by performing hydrogen bonds with carboxyl sidechain of glutamic acid.
Figure 3.
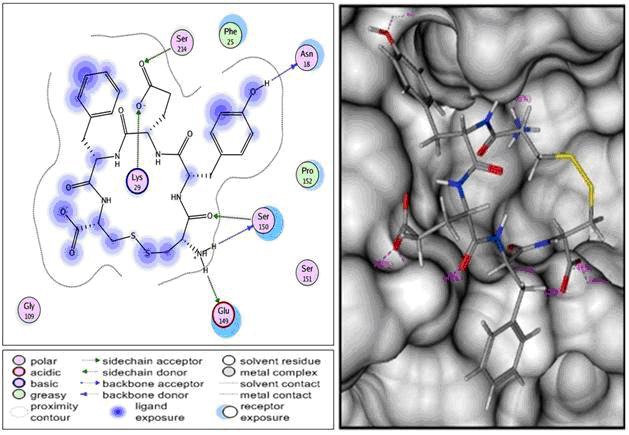
Interactions formed between CYEFC and RNA-cap site. There are three polar interactions by forming hydrogen bonds between CYEFC and RNA-cap site.
Binding free energy (ΔGbinding) of cyclic peptide CYEFC while compared with RTP standards, has a value of ΔGbinding is much greater, it provides a sense that strength of CTWYC to inhibit the RNA-cap site was better stable. pKi value or affinity of CYEFC also larger from the other but the value is below from the standard .
Toxicity Prediction:
Toxicological prediction using Toxtree, all the ligands for both of the targets SAM and RNA-cap sites did not have structural alerts (SAs) which are genotoxic and nongenotoxic properties. QSARs approach also showed that all ligands are not mutagenic or carcinogenic. The same results occured in the standard ligand RTP had no structural alerts (SAs) which are genotoxic and nongenotoxic. In other hand, standard ligand SAH was known to have structural alerts (SAs) which are nongenotoxic compounds and has potential as carcinogens. Predictions using the Osiris Property Explorer, SAM and SAH standards ligands have a higher tendency to mutagenic and effective reproductive properties, whereas the RTP standard ligand has no issues either on the mutagenic or tumorigenic properties, but has problems with the effective reproductive. All peptide ligands, for both targets did not have tendencies toward toxicity properties.
Conclusion
We have performed several cyclic peptides as potential inhibitor of dengue virus NS5 Mtase through virtual screening using docking-based methods. These peptides were predicted to bind by forming hydrogen bonds to SAM and RNA-cap sites with higher binding free energy (ΔGbinding) than standards. CTWYC cyclic peptide is the best inhibitor of the SAM site with binding free energy (ΔGbinding) of -30.72 kcal/mol. CYEFC cyclic peptide is the best inhibitor of RNAcap site with binding free energy (ΔGbinding) of -22.89 kcal/mol. CTWYC and CYEFC did not have a tendency towards toxicity properties. CTWYC and CYEFC expected that it could be used as potential antiviral drug candidate, which can inhibit the SAM and RNA-cap binding sites of dengue virus NS5 Mtase.
Supplementary material
Acknowledgments
This research is support by University of Indonesia (Riset Unggulan Universitas Indonesia tahap-1, 2012). The authors are grateful to Prof. T. Khayamian, Departement of Chemintry, Isfahan Technological University, Isfahan, Iran for his critical comments and excellent assistance in proof-reading the manuscript.
Footnotes
Citation:Idrus et al, Bioinformation 8(8): 348-352 (2012)
References
- 1.L Guglani, SK Kabra. Dengue Bull. 2005;29:58. [Google Scholar]
- 2.J Whitehorn, J Farrar. British Medical Bulletin. 2010;95:161. doi: 10.1093/bmb/ldq019. [DOI] [PubMed] [Google Scholar]
- 3.AM Kahn, et al. PLoS Negl Trop Dis. 2008;2:8 e272. doi: 10.1371/journal.pntd.0000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CG Noble, et al. Antiviral Res. 2010;85:3. [Google Scholar]
- 5.USF Tambunan, S Alamudi. Bioinformation. 2010;5:6. [Google Scholar]
- 6.USF Tambunan, et al. African Journal of Biotechnology. 2011;10:57. [Google Scholar]
- 7.Podvinec, et al. J Med Chem. 2010;53:4. doi: 10.1021/jm900776m. [DOI] [PubMed] [Google Scholar]
- 8.D Sivakumar, Sivaraman T. Med Chem. 2011;7:6. doi: 10.2174/157340611797928451. [DOI] [PubMed] [Google Scholar]
- 9.SV Lim, et al. BMC Bioinformatics. 2011;12:13. doi: 10.1186/1471-2105-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.SP Lim, et al. J Biol Chem. 2011;286:8. doi: 10.1074/jbc.M110.206227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MP Courageot, et al. J Virol. 2000;80:3. [Google Scholar]
- 12.SP Lim, et al. Antiviral Res. 2008;80:3. [Google Scholar]
- 13.A Huther, Dietrich U. AIDS Rev. 2007;9:4. [PubMed] [Google Scholar]
- 14.Manavalan, et al. BMC Struc Biol. 2010;10:1. [Google Scholar]
- 15.M Feher, William CI. J Chem Inf Model. 2009;49:7. doi: 10.1021/ci900060x. [DOI] [PubMed] [Google Scholar]
- 16.AJ Hell, et al. Pharm Res. 2009;29:9. [Google Scholar]
- 17.D Benarroch, et al. J Biol Chem. 2004;279:34. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.